Hyperspectral Superpixel-Wise Glioblastoma Tumor Detection in Histological Samples
Abstract
1. Introduction
2. Materials and Methods
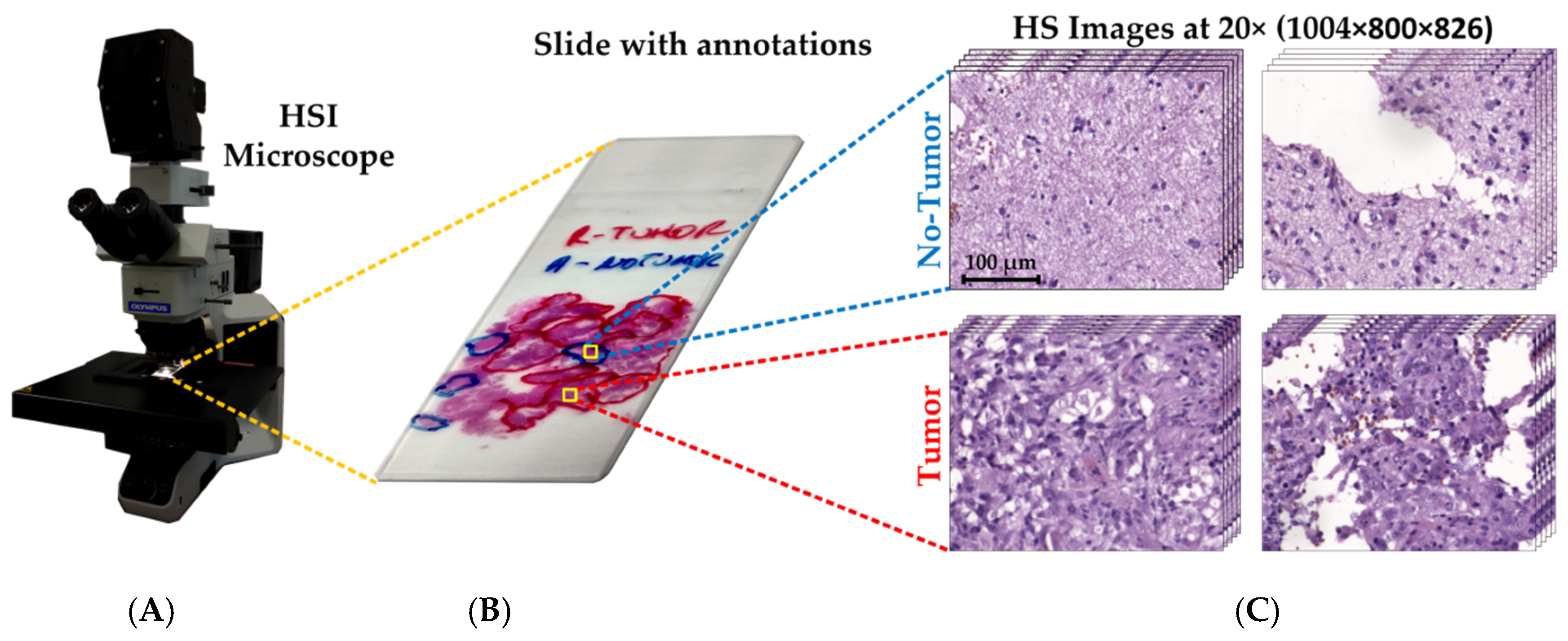
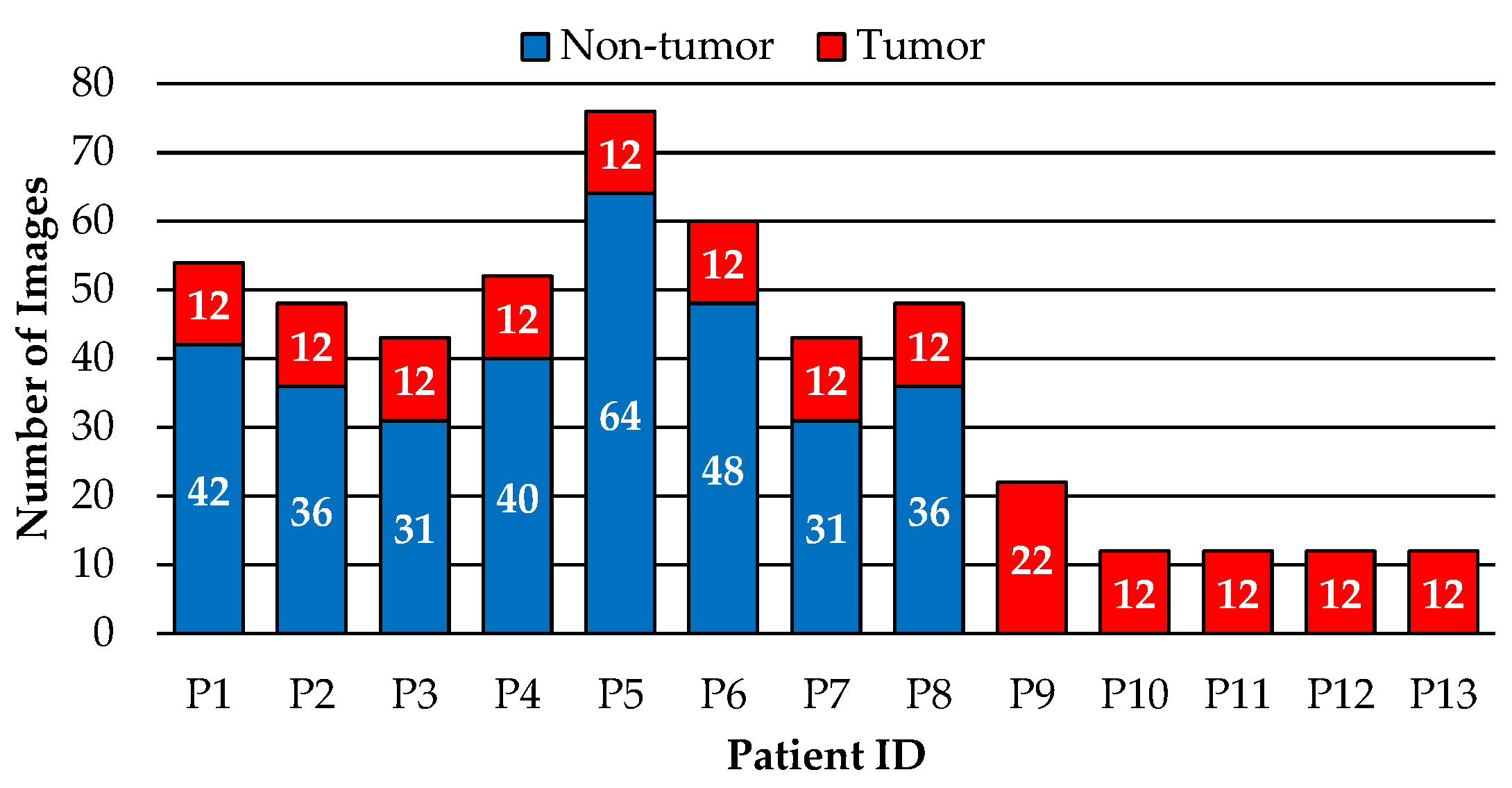
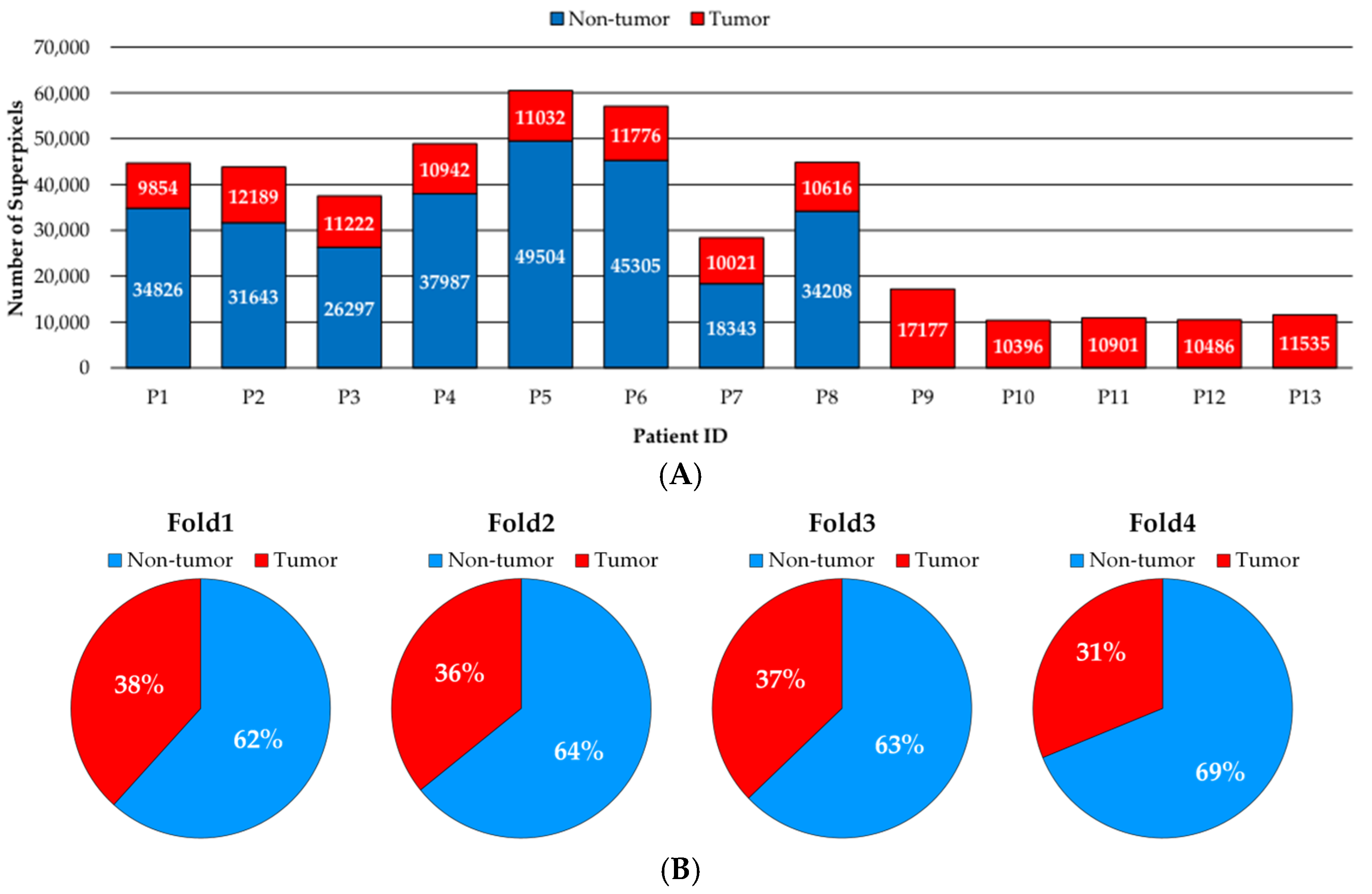
2.1. Dataset Description
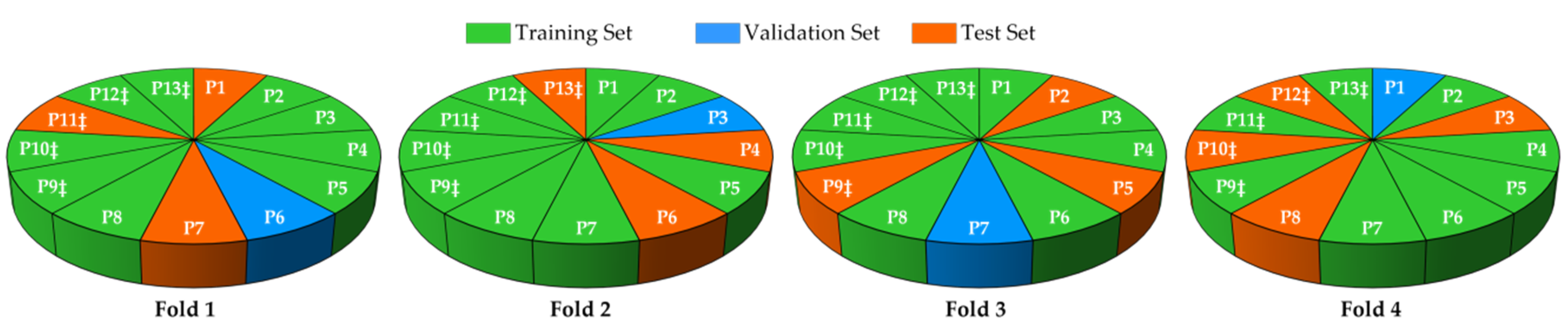
2.2. Data Partition
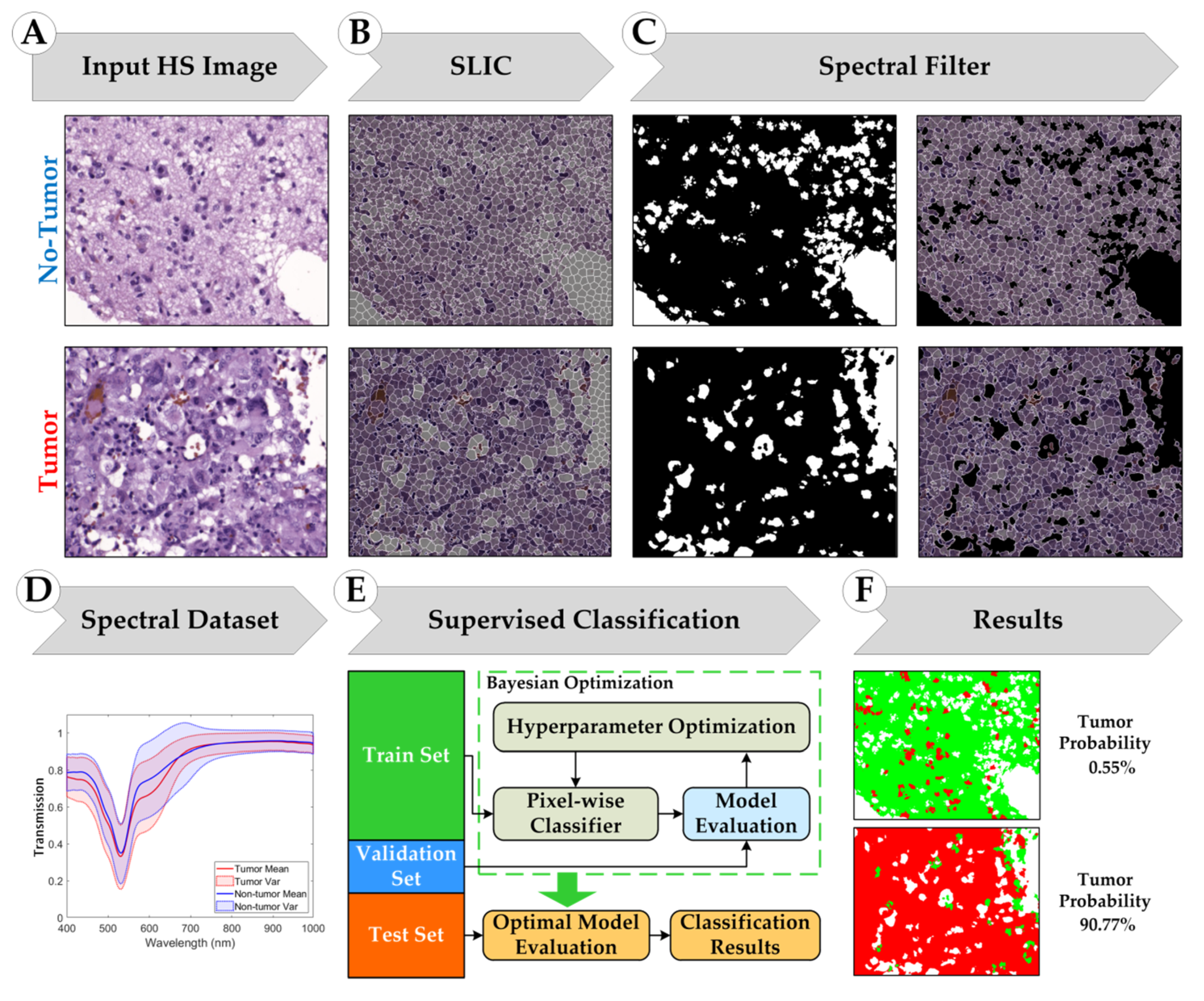
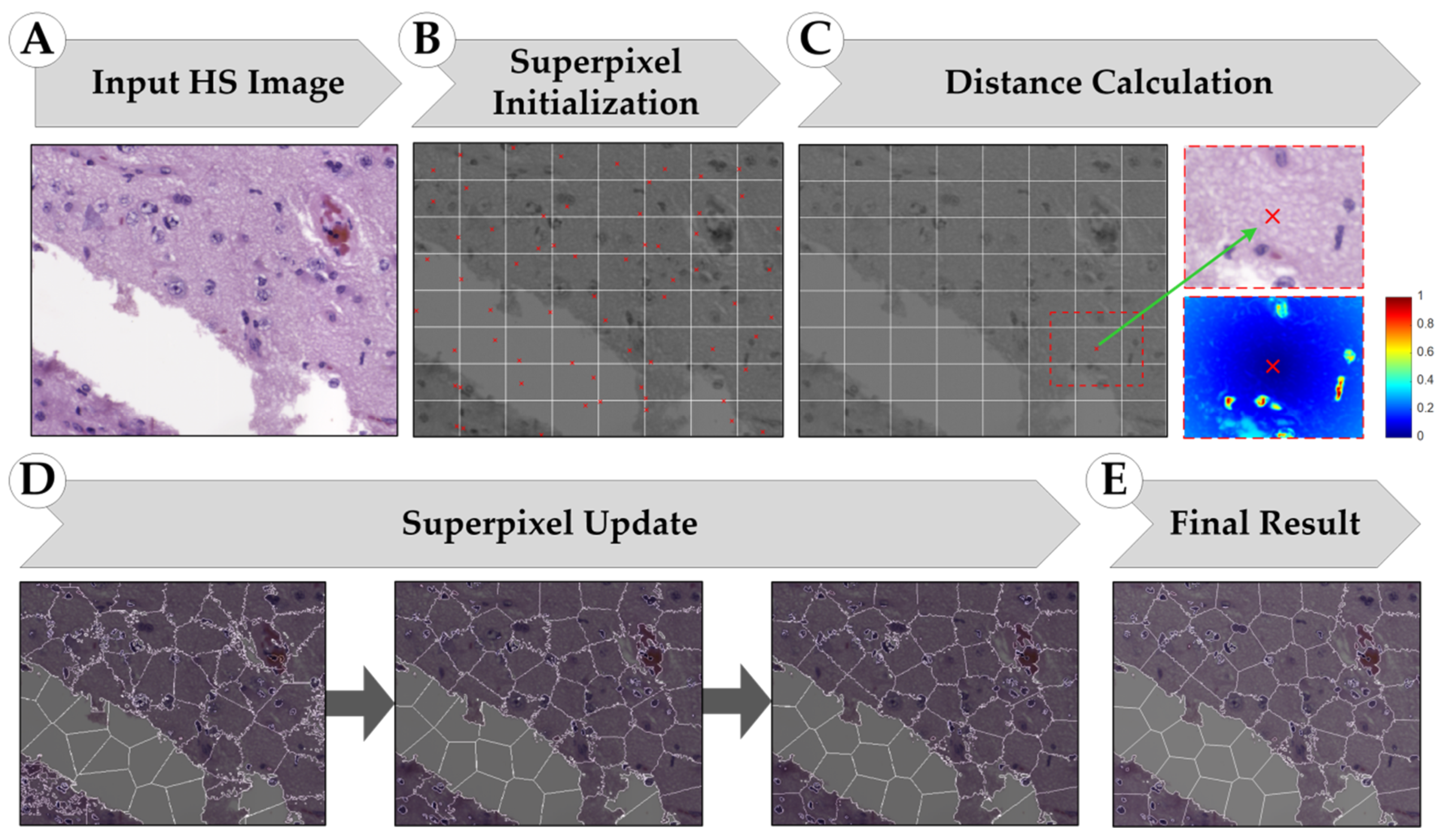
2.3. Superpixel-Based Processing Framework
2.3.1. Simple Linear Iterative Clustering (SLIC) Approach
2.3.2. Supervised Classification
2.4. Evaluation Metrics
3. Experimental Results
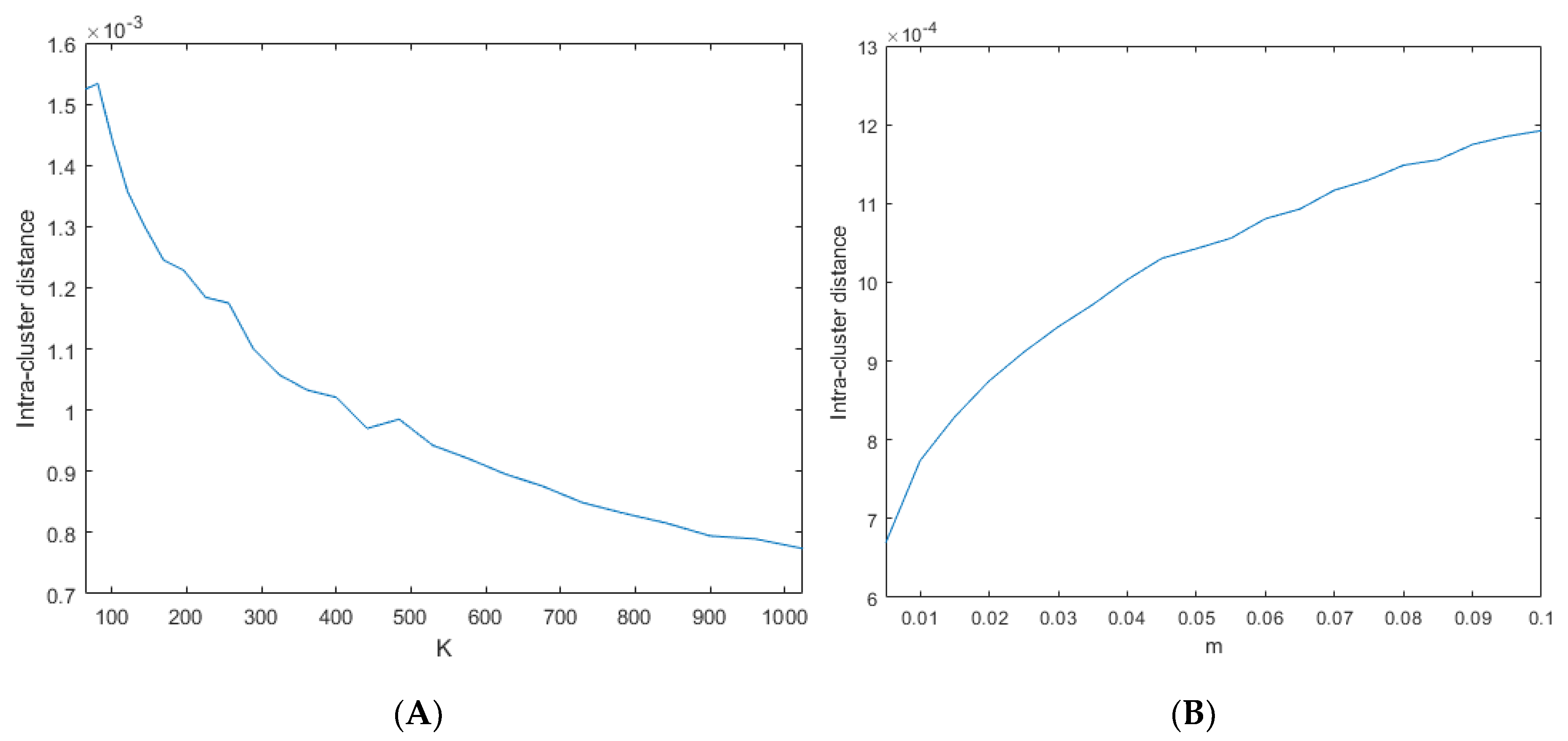
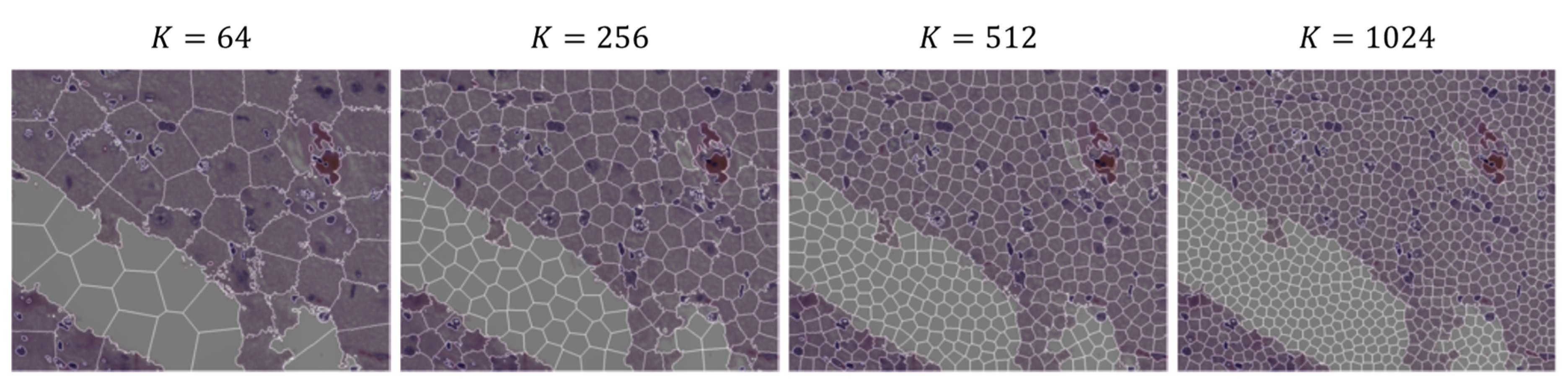
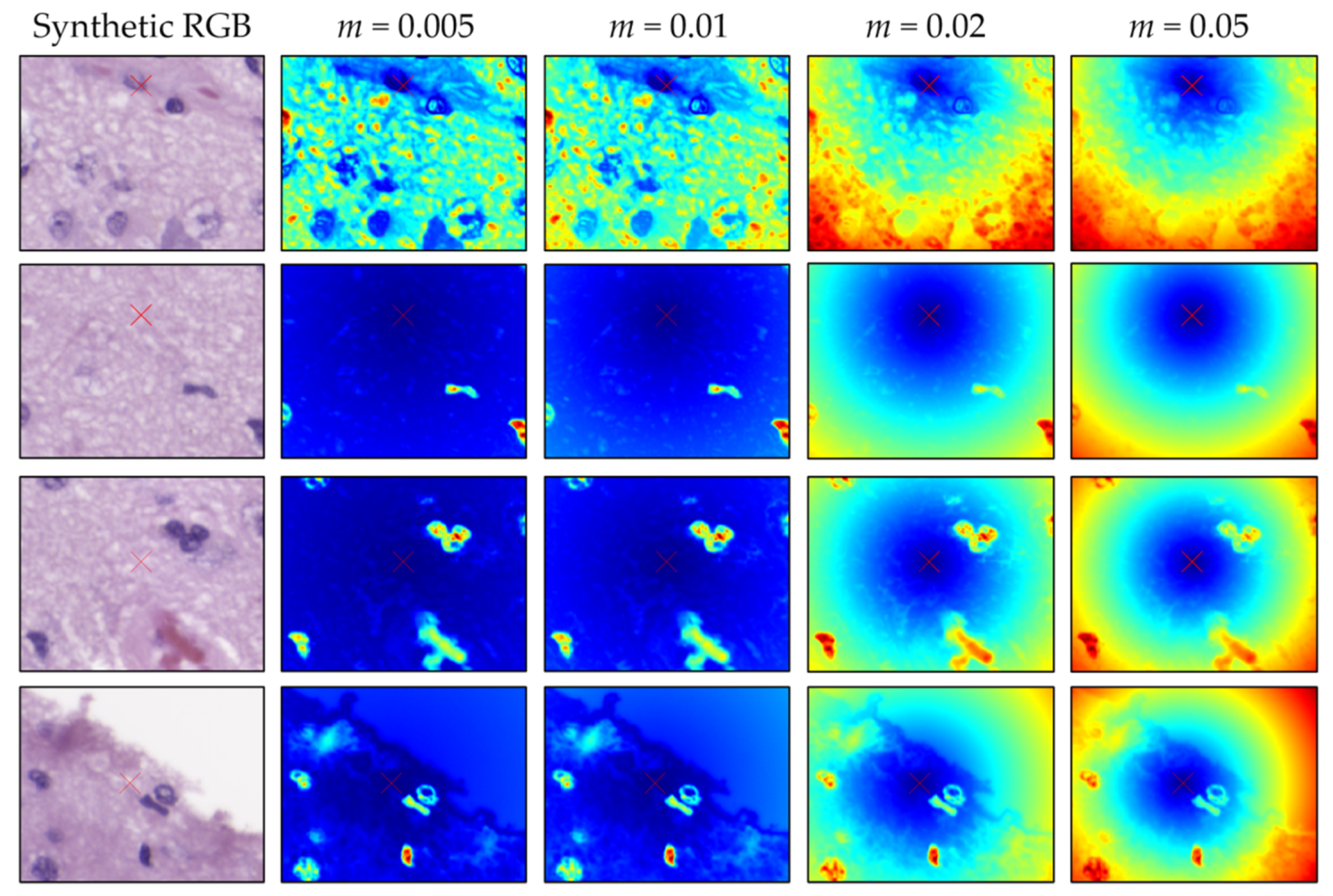
3.1. SLIC Hyperparameter Selection
3.2. Supervised Classification Results
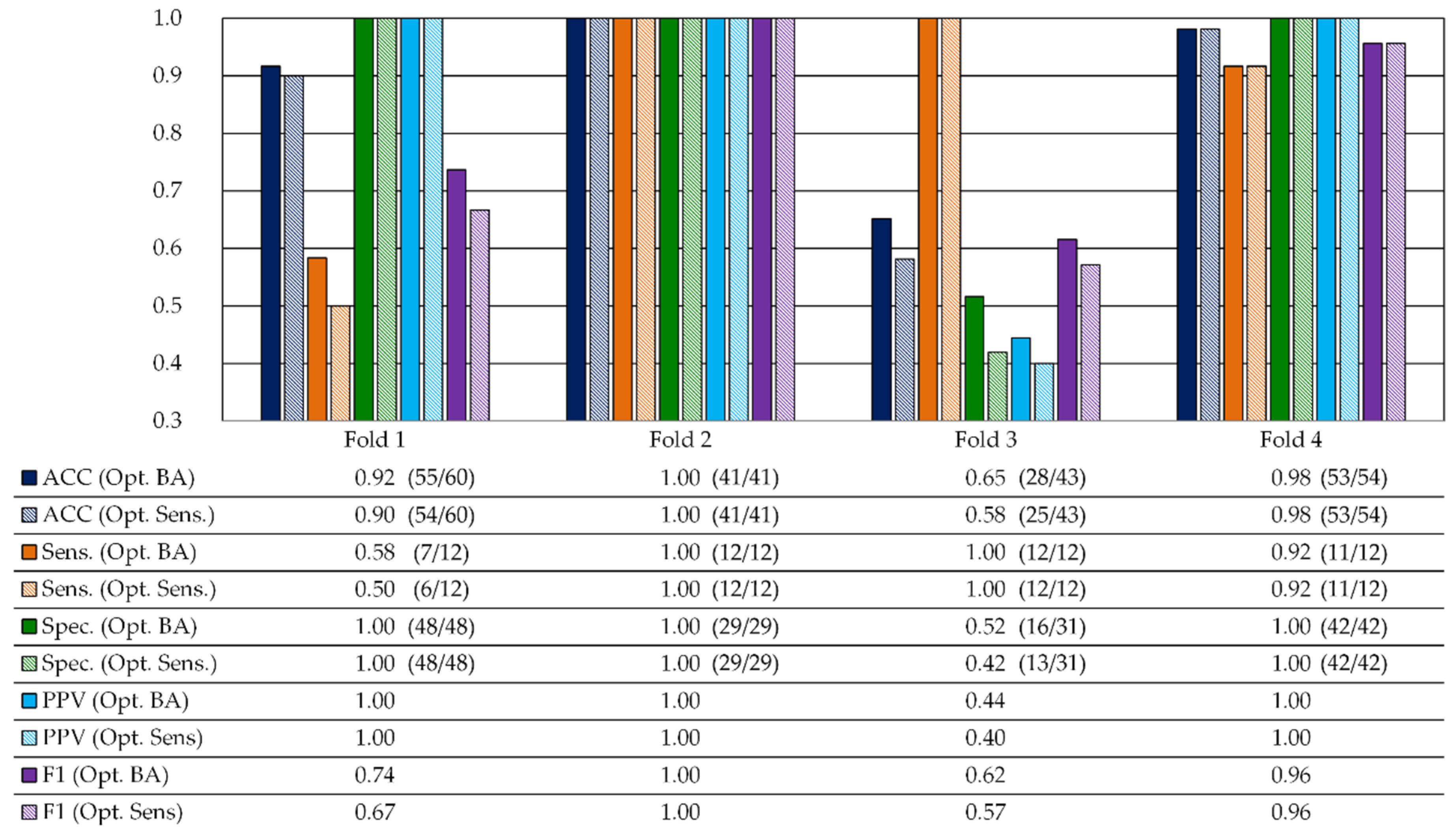
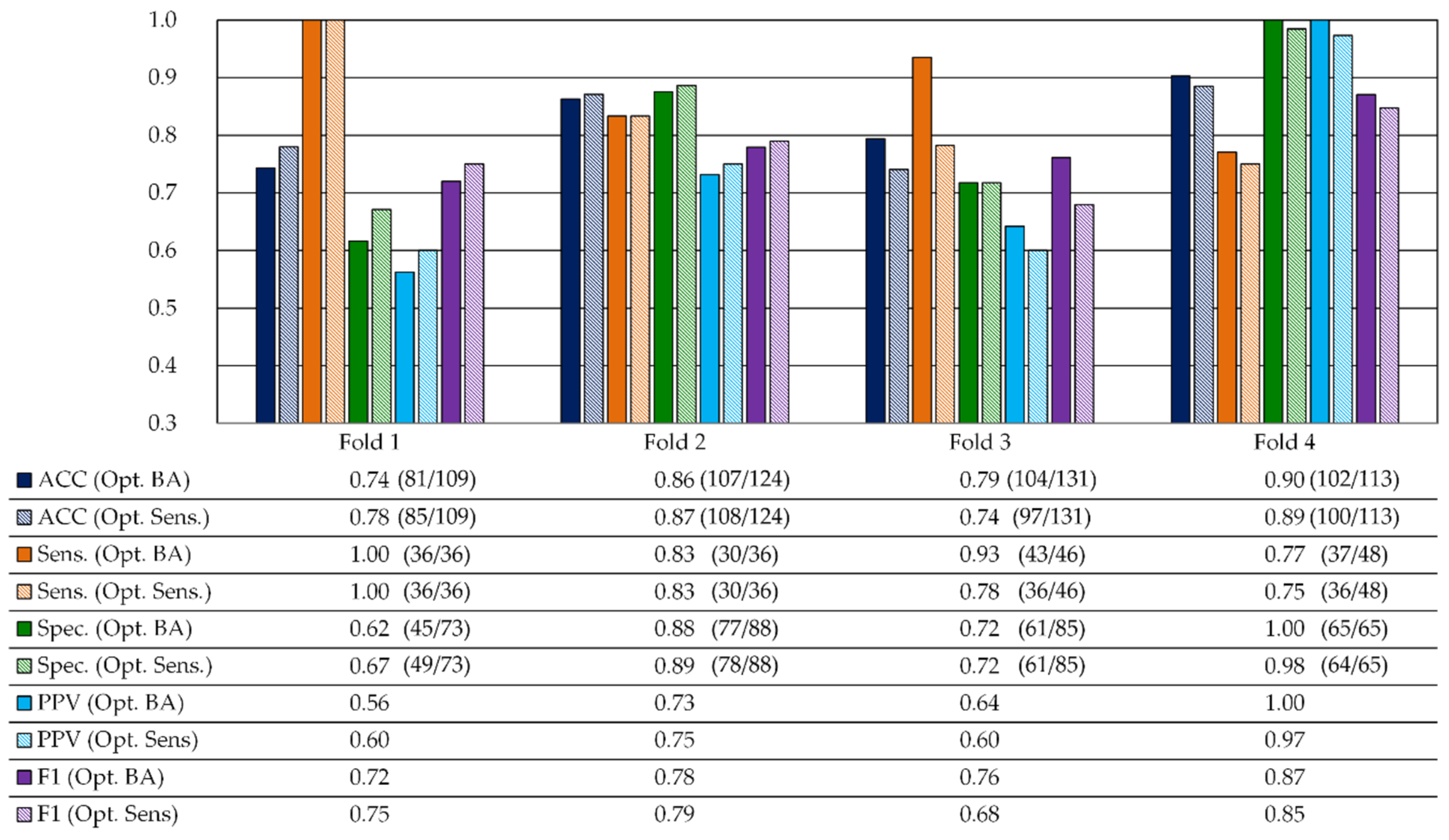
3.2.1. Validation Results
3.2.2. Quantitative Test Results
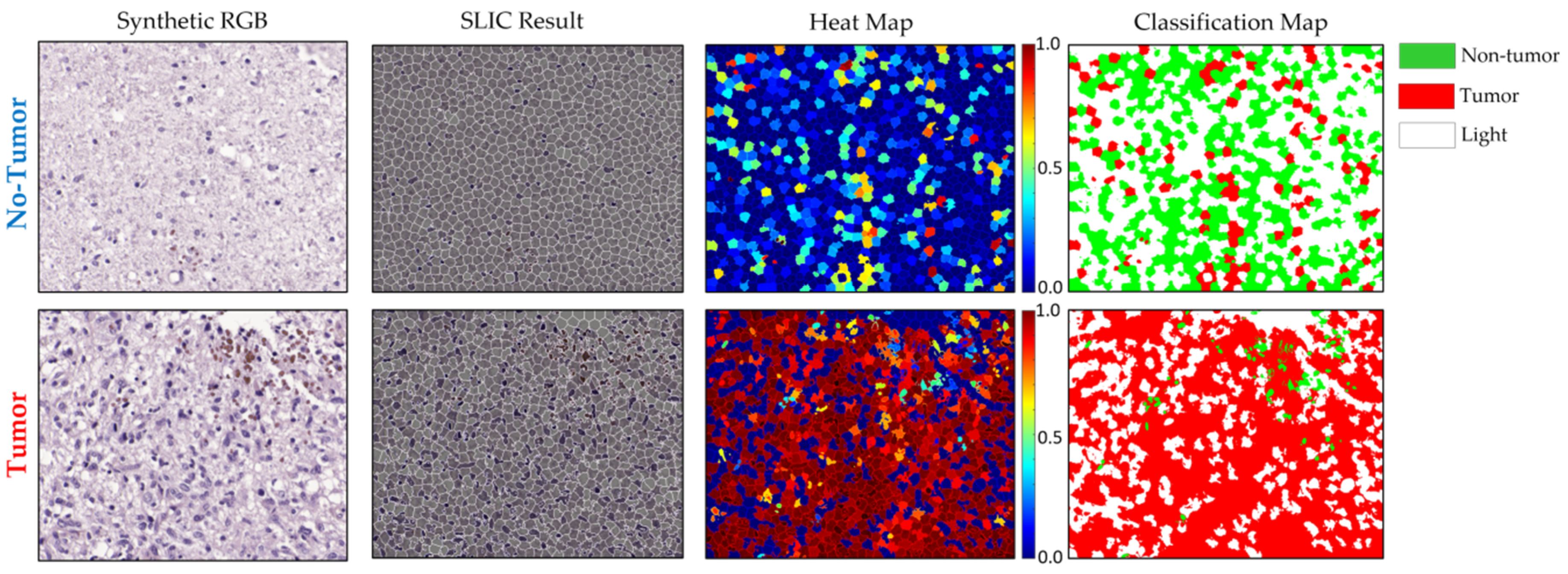
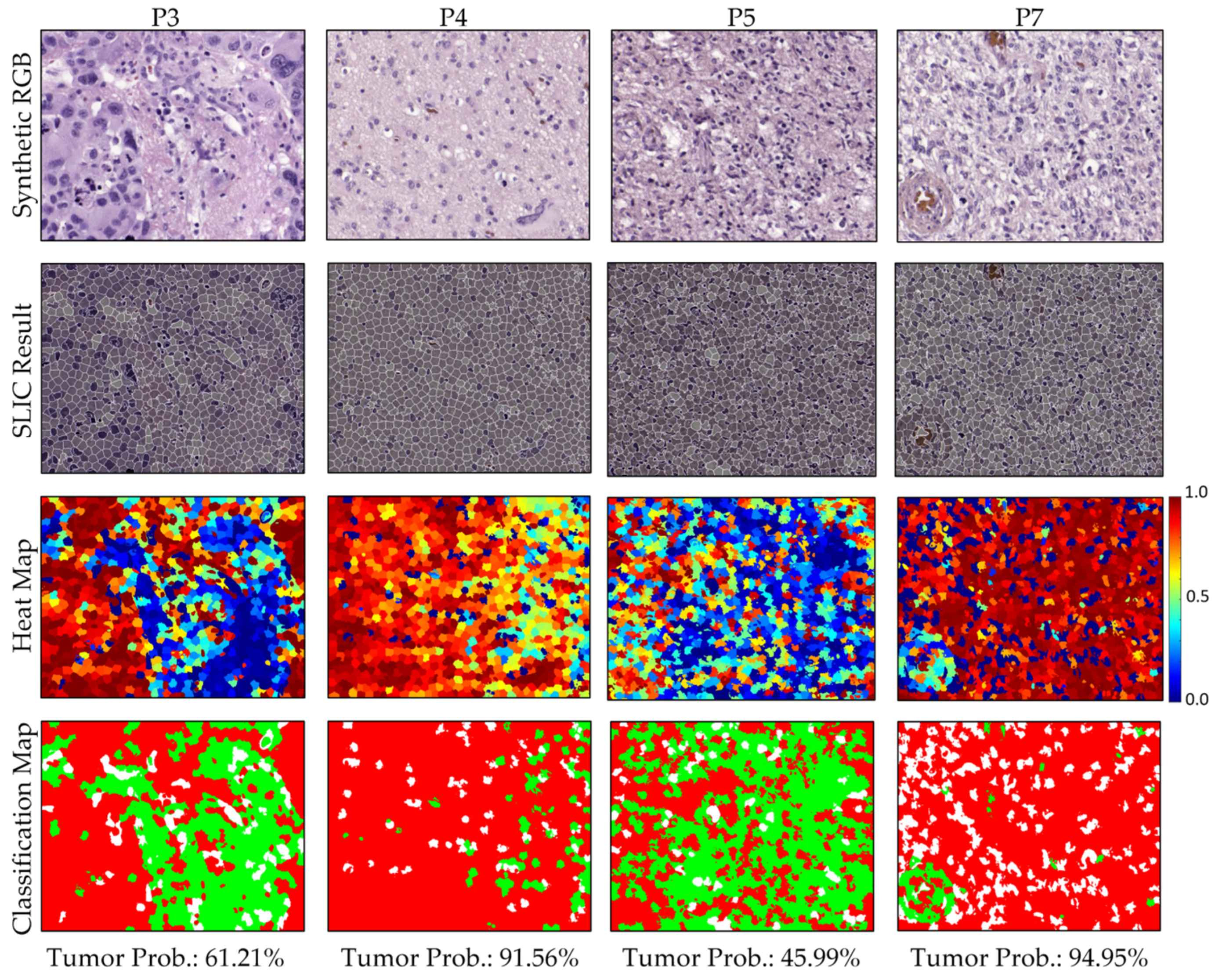
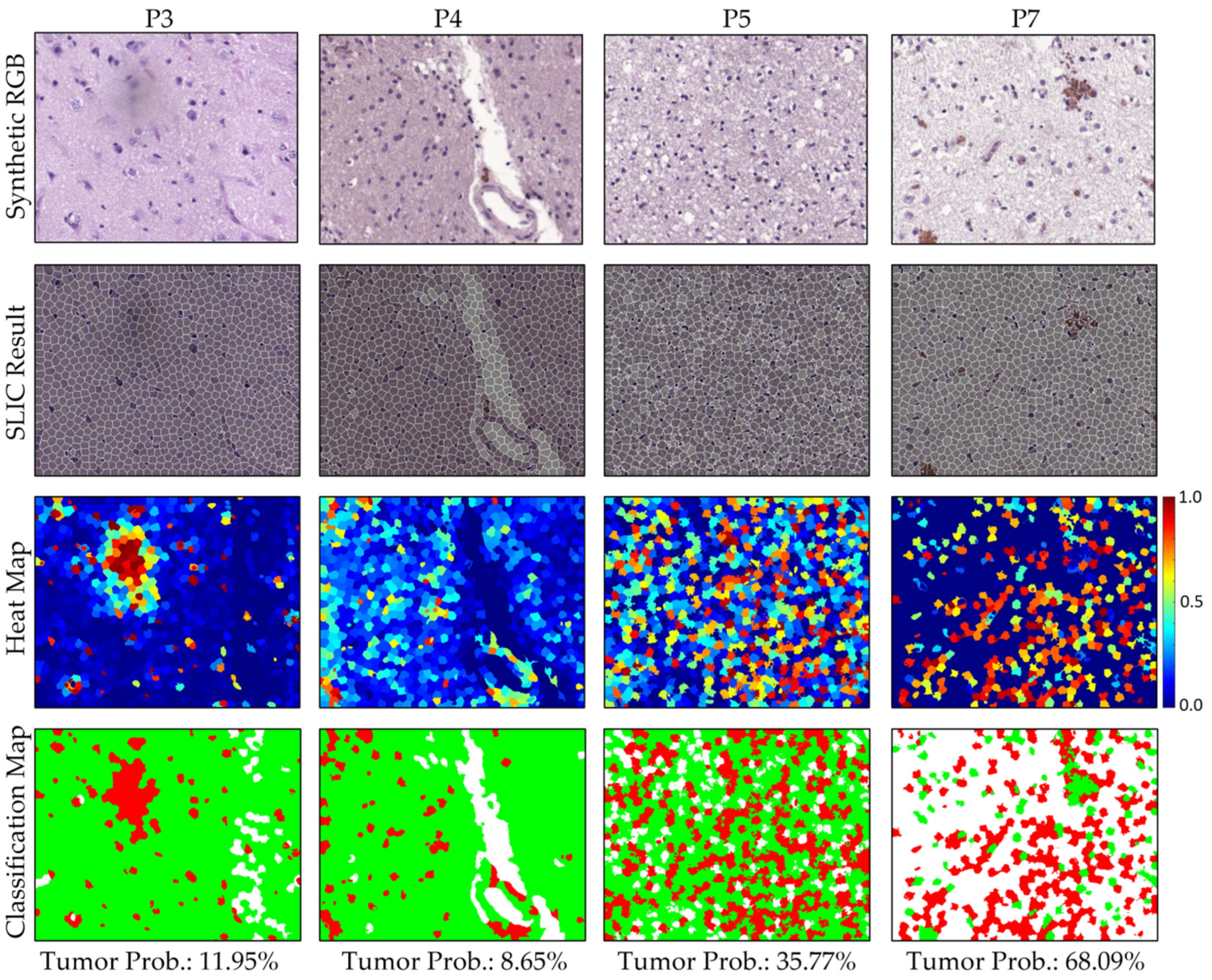
3.2.3. Qualitative Test Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Ethical Statements
References
- Van Es, S.L. Digital pathology: Semper ad meliora. Pathology 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Halicek, M.; Fabelo, H.; Ortega, S.; Callico, G.M.; Fei, B. In-Vivo and Ex-Vivo Tissue Analysis through Hyperspectral Imaging Techniques: Revealing the Invisible Features of Cancer. Cancers (Basel) 2019, 11, 756. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Halig, L.V.; Schuster, D.M.; Osunkoya, A.; Master, V.; Nieh, P.T.; Chen, G.Z.; Fei, B. Hyperspectral imaging and quantitative analysis for prostate cancer detection. J. Biomed. Opt. 2012, 17, 0760051. [Google Scholar] [CrossRef] [PubMed]
- Awan, R.; Al-Maadeed, S.; Al-Saady, R. Using spectral imaging for the analysis of abnormalities for colorectal cancer: When is it helpful? PLoS ONE 2018, 13, e0197431. [Google Scholar] [CrossRef]
- Septiana, L.; Suzuki, H.; Ishikawa, M.; Obi, T.; Kobayashi, N.; Ohyama, N.; Ichimura, T.; Sasaki, A.; Wihardjo, E.; Andiani, D. Elastic and collagen fibers discriminant analysis using H&E stained hyperspectral images. Opt. Rev. 2019, 26, 369–379. [Google Scholar]
- Khelifi, R.; Adel, M.; Bourennane, S. Multispectral texture characterization: Application to computer aided diagnosis on prostatic tissue images. EURASIP J. Adv. Signal Process. 2012, 2012, 118. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, J.; Zhou, M.; Li, Q.; Wang, Y.; Ang, Q.I.A.N.W.; Ang, J.I.W.; Hou, M.E.I.Z.; Ingli, Q.L.I.; Iting, Y.; et al. Spectral-spatial feature-based neural network method for acute lymphoblastic leukemia cell identification via microscopic hyperspectral imaging technology. Biomed. Opt. Express 2017, 8, 3017–3028. [Google Scholar] [CrossRef] [PubMed]
- Haj-Hassan, H.; Chaddad, A.; Harkouss, Y.; Desrosiers, C.; Toews, M.; Tanougast, C. Classifications of multispectral colorectal cancer tissues using convolution neural network. J. Pathol. Inform. 2017, 8, 1. [Google Scholar]
- Malon, C.; Cosatto, E. Classification of mitotic figures with convolutional neural networks and seeded blob features. J. Pathol. Inform. 2013, 4, 9. [Google Scholar] [CrossRef]
- Ortega, S.; Halicek, M.; Fabelo, H.; Camacho, R.; Plaza, M.D.L.L.; Godtliebsen, F.; Callicó, G.M.; Fei, B. Hyperspectral Imaging for the Detection of Glioblastoma Tumor Cells in H&E Slides Using Convolutional Neural Networks. Sensors 2020, 20, 1911. [Google Scholar]
- Yu, H.; Yang, Z.; Tan, L.; Wang, Y.; Sun, W.; Sun, M.; Tang, Y. Methods and datasets on semantic segmentation: A review. Neurocomputing 2018, 304, 82–103. [Google Scholar] [CrossRef]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, K.; Dong, Y.; Wu, K.; Hu, X. Semisupervised Classification Based on SLIC Segmentation for Hyperspectral Image. In IEEE Geoscience and Remote Sensing Letters; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Zu, B.; Xia, K.; Li, T.; He, Z.; Li, Y.; Hou, J.; Du, W. SLIC Superpixel-Based l2,1-Norm Robust Principal Component Analysis for Hyperspectral Image Classification. Sensors 2019, 19, 479. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhao, L.; Ren, L. Different Versions of Entropy Rate Superpixel Segmentation For Hyperspectral Image. In Proceedings of the 2019 IEEE 4th International Conference on Signal and Image Processing (ICSIP), Wuxi, China, 19–21 July 2019; pp. 1050–1054. [Google Scholar]
- Xie, F.; Lei, C.; Jin, C.; An, N. A Novel Spectral–Spatial Classification Method for Hyperspectral Image at Superpixel Level. Appl. Sci. 2020, 10, 463. [Google Scholar] [CrossRef]
- Alkhatib, M.Q.; Velez-Reyes, M. Improved Spatial-Spectral Superpixel Hyperspectral Unmixing. Remote Sens. 2019, 11, 2374. [Google Scholar] [CrossRef]
- Chung, H.; Lu, G.; Tian, Z.; Wang, D.; Chen, Z.G.; Fei, B. Superpixel-Based Spectral Classification for the Detection of Head and Neck Cancer with Hyperspectral Imaging. In Proceedings of the SPIE Medical Imaging, San Diego, CA, USA, 27 February–03 March 2016; Gimi, B., Krol, A., Eds.; SPIE: Bellingham, WA, USA, 2016; Volume 9788, p. 978813. [Google Scholar]
- Bejnordi, B.E.; Litjens, G.; Hermsen, M.; Karssemeijer, N.; van der Laak, J.A.W.M. A Multi-Scale Superpixel Classification Approach to the Detection of Regions of Interest in Whole Slide Histopathology Images. In Proceedings of the SPIE Medical Imaging, Orlando, Florida, USA, 21–26 February 2015; Gurcan, M.N., Madabhushi, A., Eds.; SPIE: Bellingham, WA, USA, 2015; Volume 9420, p. 94200H. [Google Scholar]
- Turkki, R.; Linder, N.; Kovanen, P.; Pellinen, T.; Lundin, J. Antibody-supervised deep learning for quantification of tumor-infiltrating immune cells in hematoxylin and eosin stained breast cancer samples. J. Pathol. Inform. 2016, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Zormpas-Petridis, K.; Failmezger, H.; Raza, S.E.A.; Roxanis, I.; Jamin, Y.; Yuan, Y. Superpixel-Based Conditional Random Fields (SuperCRF): Incorporating Global and Local Context for Enhanced Deep Learning in Melanoma Histopathology. Front. Oncol. 2019, 9, 1045. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella, D.; Webster, B.; Hiroko, K.C.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W.; et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary; Springer: Berlin/Heidelberg, Germany, 2016; Volume 131, pp. 803–820. [Google Scholar]
- Carter, E.C.; Schanda, J.D.; Hirschler, R.; Jost, S.; Luo, M.R.; Melgosa, M.; Ohno, Y.; Pointer, M.R.; Rich, D.C.; Vienot, F.; et al. CIE 015:2018 Colorimetry, 4th ed.; Technical Report; International Commission on Illumination: Vienna, Austria, 2018. [Google Scholar]
- Van der Meer, F. The effectiveness of spectral similarity measures for the analysis of hyperspectral imagery. Int. J. Appl. Earth Obs. Geoinf. 2006, 8, 3–17. [Google Scholar] [CrossRef]
- Ghamisi, P.; Plaza, J.; Chen, Y.; Li, J.; Plaza, A.J. Advanced Spectral Classifiers for Hyperspectral Images: A review. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–32. [Google Scholar] [CrossRef]
- Vapnik, V. Support vector machine. Mach. Learn. 1995, 20, 273–297. [Google Scholar]
- Mercier, G.; Lennon, M. Support Vector Machines for Hyperspectral Image Classification with Spectral-based kernels. In Proceedings of the International Geoscience and Remote Sensing Symposium (IGARSS), Toulouse, France, 21–25 July 2003; Volume 1, pp. 288–290. [Google Scholar]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Chollet, F. Keras. Available online: https://keras.io (accessed on 28 October 2019).
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian Optimization of Machine Learning Algorithms. arXiv 2012, arXiv:1206.2944. [Google Scholar]
- Saulig, N.; Lerga, J.; Milanovic, Z.; Ioana, C. Extraction of Useful Information Content From Noisy Signals Based on Structural Affinity of Clustered TFDs’ Coefficients. IEEE Trans. Signal Process. 2019, 67, 3154–3167. [Google Scholar] [CrossRef]
- Hržić, F.; Štajduhar, I.; Tschauner, S.; Sorantin, E.; Lerga, J. Local-Entropy Based Approach for X-Ray Image Segmentation and Fracture Detection. Entropy 2019, 21, 338. [Google Scholar] [CrossRef]
- Mandić, I.; Peić, H.; Lerga, J.; Štajduhar, I. Denoising of X-ray Images Using the Adaptive Algorithm Based on the LPA-RICI Algorithm. J. Imaging 2018, 4, 34. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Spectral-Spatial Feature Extraction for Hyperspectral Image Classification: A Dimension Reduction and Deep Learning Approach. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4544–4554. [Google Scholar] [CrossRef]
- Patil, C.; Baidari, I. Estimating the Optimal Number of Clusters k in a Dataset Using Data Depth. Data Sci. Eng. 2019, 4, 132–140. [Google Scholar] [CrossRef]
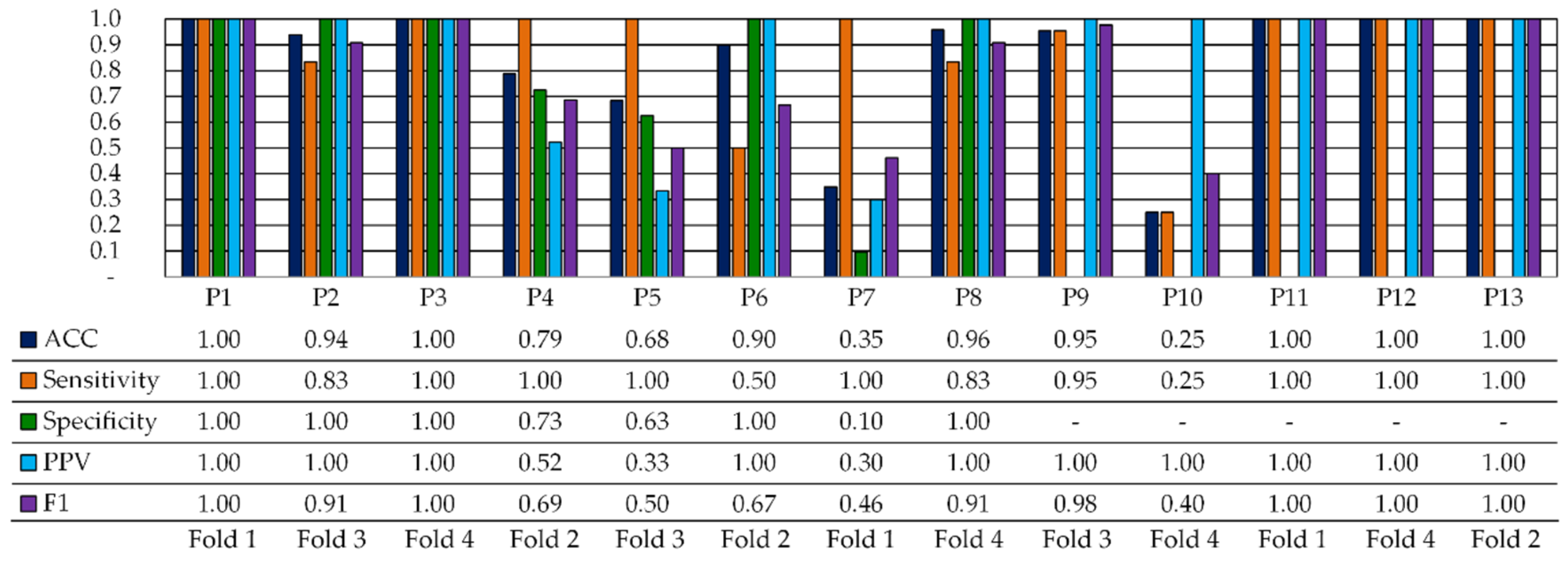
| No Opt. | Opt. Metric (BA) | Opt. Metric (Sensitivity) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Partition | ACC | Sens. | Spec. | PPV | F1 | ACC | Sens. | Spec. | PPV | F1 | ACC | Sens. | Spec. | PPV | F1 |
| Fold 1 | 0.65 | 0.12 | 0.78 | 0.13 | 0.13 | 0.88 | 0.54 | 0.97 | 0.84 | 0.66 | 0.87 | 0.47 | 0.97 | 0.81 | 0.60 |
| Fold 2 | 0.81 | 0.57 | 0.91 | 0.73 | 0.64 | 0.95 | 0.89 | 0.98 | 0.96 | 0.92 | 0.95 | 0.88 | 0.99 | 0.96 | 0.92 |
| Fold 3 | 0.39 | 0.92 | 0.10 | 0.36 | 0.51 | 0.61 | 0.99 | 0.41 | 0.48 | 0.64 | 0.58 | 0.99 | 0.35 | 0.45 | 0.62 |
| Fold 4 | 0.80 | 0.48 | 0.89 | 0.56 | 0.52 | 0.89 | 0.88 | 0.89 | 0.69 | 0.78 | 0.88 | 0.89 | 0.88 | 0.68 | 0.77 |
| Avg. | 0.66 | 0.52 | 0.67 | 0.44 | 0.45 | 0.83 | 0.82 | 0.81 | 0.74 | 0.75 | 0.82 | 0.81 | 0.80 | 0.73 | 0.73 |
| Std. | 0.20 | 0.33 | 0.39 | 0.26 | 0.22 | 0.15 | 0.19 | 0.27 | 0.21 | 0.13 | 0.17 | 0.23 | 0.30 | 0.22 | 0.15 |
| Proposed Superpixel-Based Approach (Results at Image Level) | Proposed Superpixel-Based Approach (Results at Superpixel Level) | Previous CNN-Based Approach [10] (Results at Patch Level) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Partition | ACC | Sensitivity | Specificity | ACC | Sensitivity | Specificity | ACC | Sensitivity | Specificity |
| P1 | 1.00 | 1.00 | 1.00 | 0.91 | 0.91 | 0.90 | 0.93 | 0.91 | 0.96 |
| P2 | 0.94 | 0.83 | 1.00 | 0.79 | 0.50 | 0.97 | 0.89 | 0.99 | 0.83 |
| P3 | 1.00 | 1.00 | 1.00 | 0.90 | 0.88 | 0.92 | 0.85 | 0.91 | 0.8 |
| P4 | 0.79 | 1.00 | 0.73 | 0.78 | 0.87 | 0.75 | 0.57 | 0.57 | 0.58 |
| P5 | 0.68 | 1.00 | 0.63 | 0.54 | 0.67 | 0.51 | 0.69 | 0.81 | 0.64 |
| P6 * | 0.90 | 0.50 | 1.00 | 0.85 | 0.35 | 0.98 | - | - | - |
| P7 | 0.35 | 1.00 | 0.10 | 0.53 | 0.95 | 0.30 | 0.66 | 0.71 | 0.63 |
| P8 | 0.96 | 0.83 | 1.00 | 0.89 | 0.75 | 0.93 | 0.96 | 0.96 | 0.96 |
| P9 | 0.95 | 0.95 | - | 0.92 | 0.92 | - | 0.99 | 0.99 | - |
| P10 | 0.25 | 0.25 | - | 0.41 | 0.41 | - | 0.89 | 0.89 | - |
| P11 | 1.00 | 1.00 | - | 0.98 | 0.98 | - | 0.92 | 0.92 | - |
| P12 | 1.00 | 1.00 | - | 0.83 | 0.83 | - | 0.92 | 0.92 | - |
| P13 | 1.00 | 1.00 | - | 0.97 | 0.97 | - | 0.99 | 0.99 | - |
| Avg. | 0.83 | 0.91 | 0.78 | 0.79 | 0.80 | 0.76 | 0.86 | 0.88 | 0.77 |
| Std. | 0.27 | 0.22 | 0.34 | 0.19 | 0.19 | 0.26 | 0.14 | 0.13 | 0.16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega, S.; Fabelo, H.; Halicek, M.; Camacho, R.; Plaza, M.d.l.L.; Callicó, G.M.; Fei, B. Hyperspectral Superpixel-Wise Glioblastoma Tumor Detection in Histological Samples. Appl. Sci. 2020, 10, 4448. https://doi.org/10.3390/app10134448
Ortega S, Fabelo H, Halicek M, Camacho R, Plaza MdlL, Callicó GM, Fei B. Hyperspectral Superpixel-Wise Glioblastoma Tumor Detection in Histological Samples. Applied Sciences. 2020; 10(13):4448. https://doi.org/10.3390/app10134448
Chicago/Turabian StyleOrtega, Samuel, Himar Fabelo, Martin Halicek, Rafael Camacho, María de la Luz Plaza, Gustavo M. Callicó, and Baowei Fei. 2020. "Hyperspectral Superpixel-Wise Glioblastoma Tumor Detection in Histological Samples" Applied Sciences 10, no. 13: 4448. https://doi.org/10.3390/app10134448
APA StyleOrtega, S., Fabelo, H., Halicek, M., Camacho, R., Plaza, M. d. l. L., Callicó, G. M., & Fei, B. (2020). Hyperspectral Superpixel-Wise Glioblastoma Tumor Detection in Histological Samples. Applied Sciences, 10(13), 4448. https://doi.org/10.3390/app10134448









