Time-Series Classification Based on Fusion Features of Sequence and Visualization
Abstract
1. Introduction
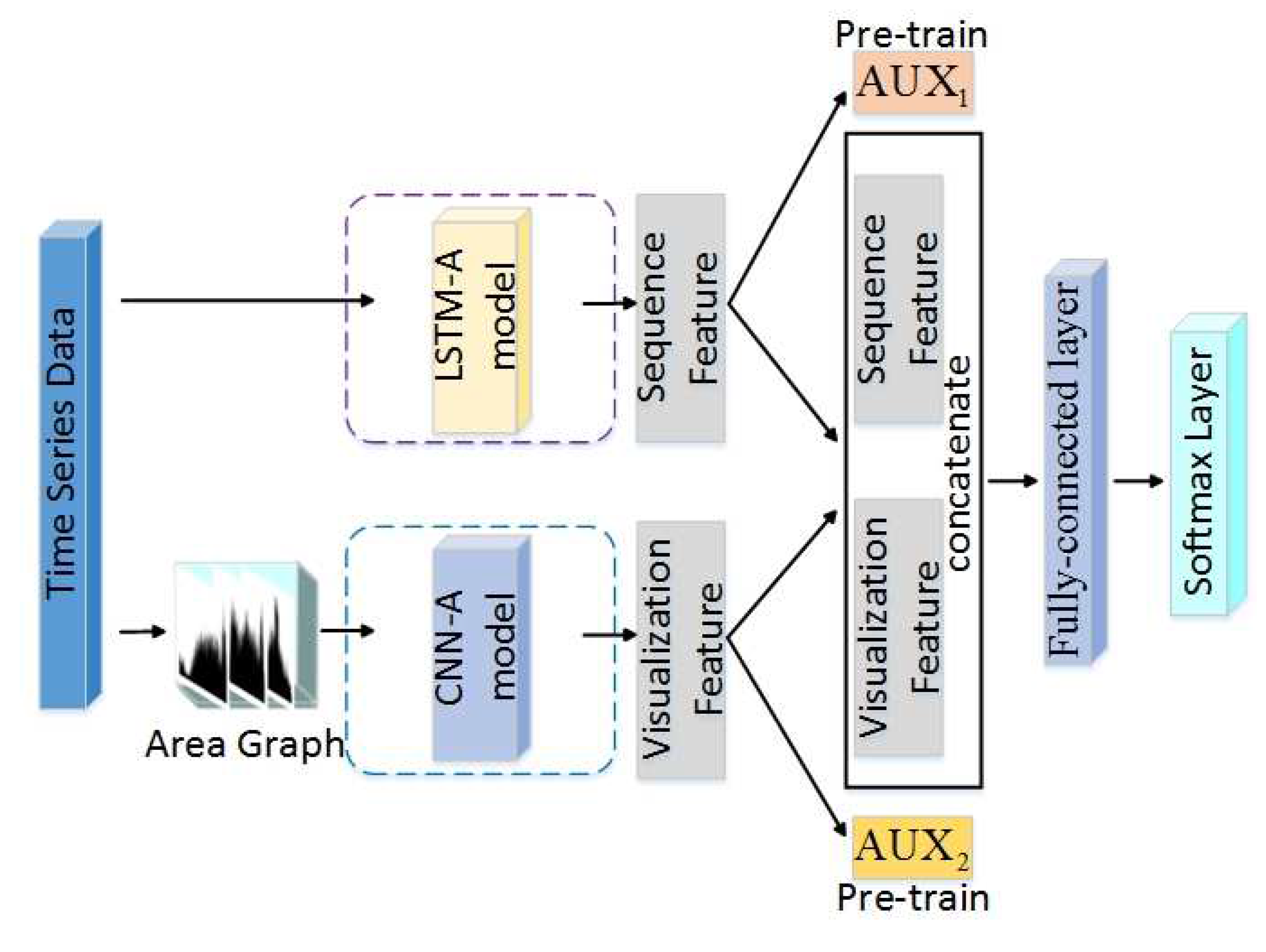
- The framework imitates the mechanism of the human brain, in that the human brain’s cognition is a combination of the human body’s multiple senses. Thus, we use a DNN to learn features from different aspects to enhance the feature space. Then, based on the fusion of features, we carry out the TSC task and gain promising results.
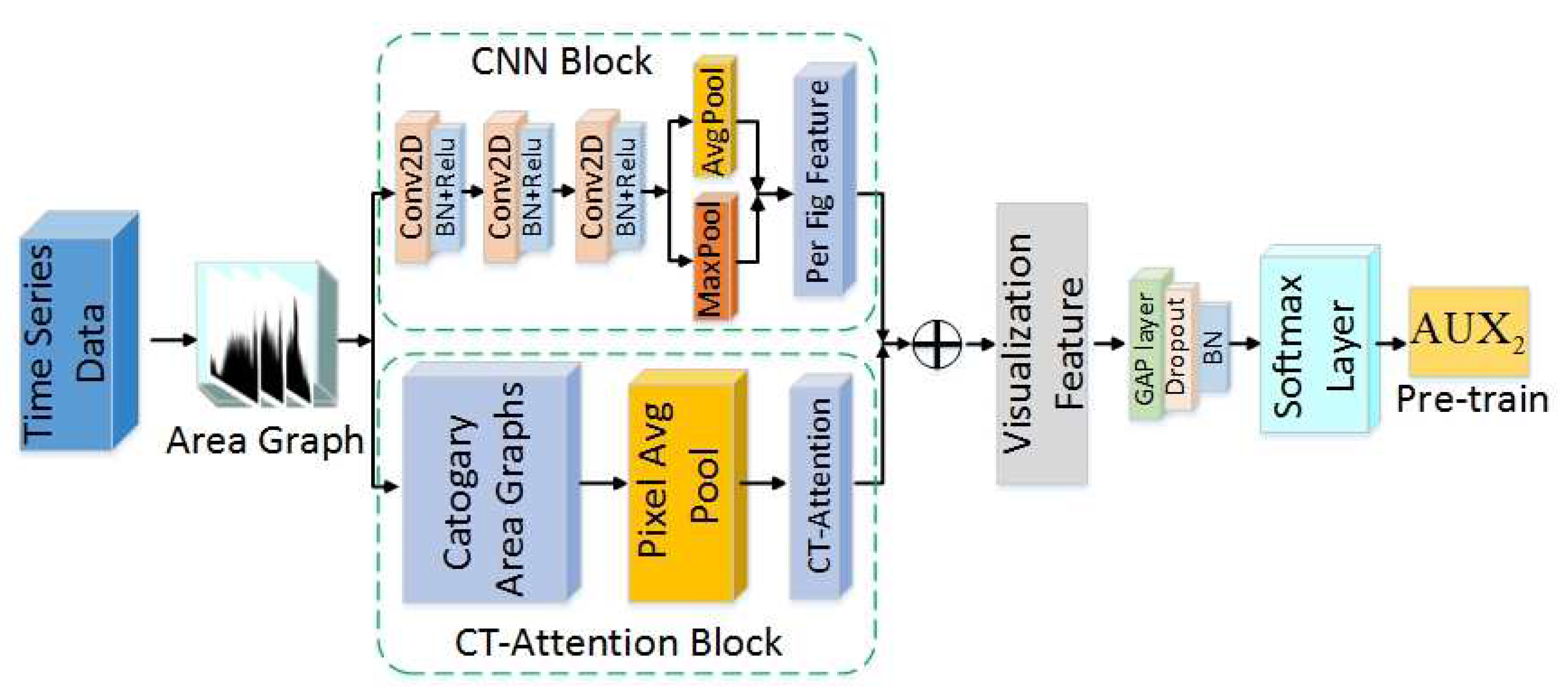
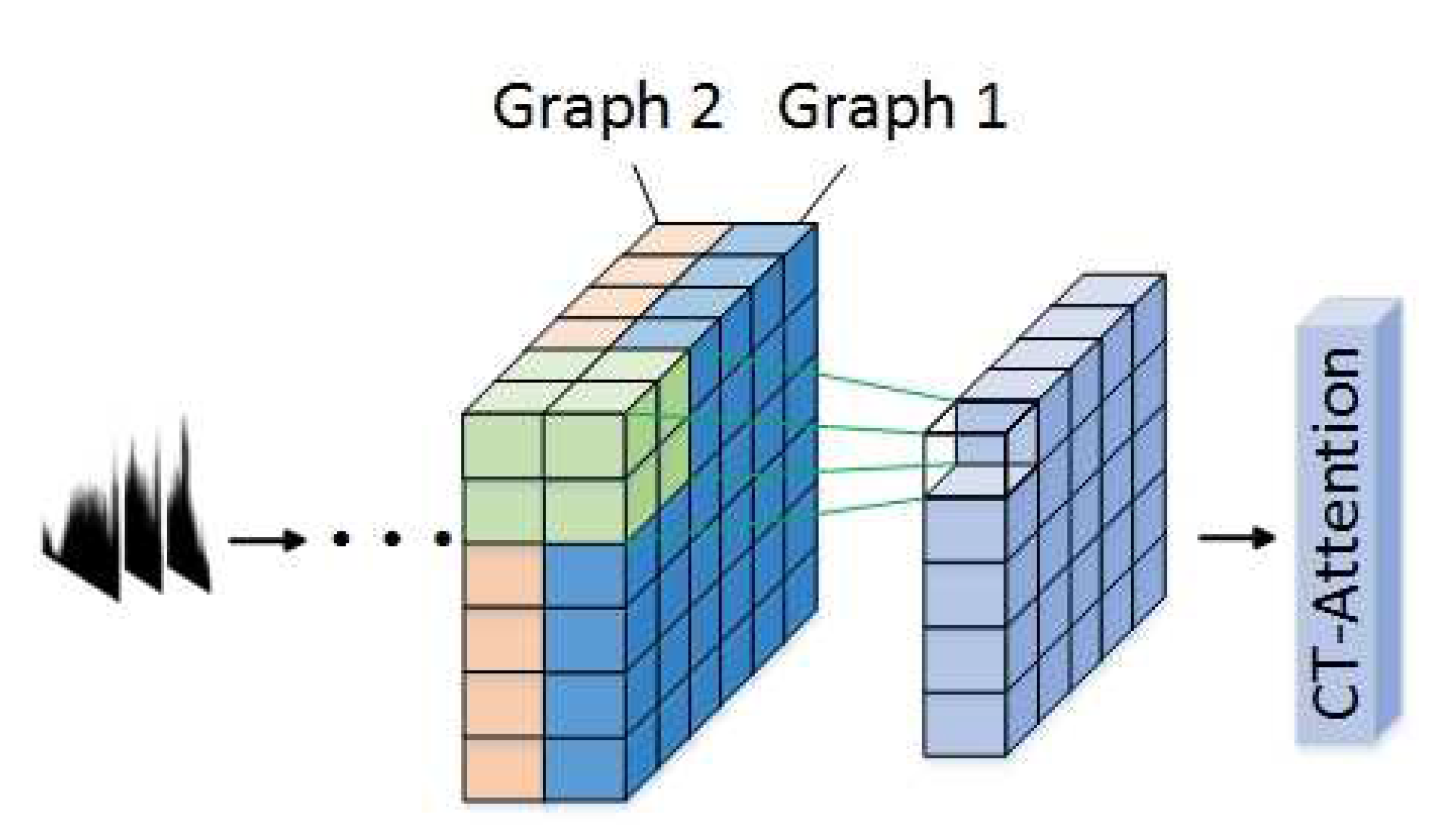
- The framework uses well-trained LSTM-A and CNN-A to extract sequence features and visualization features, as well as combining an attention mechanism to extract the key features that contribute to classification. In particular, an innovational category trend attention (CT-Attention) is gained from data belonging to the same category in an innovative way.
- The framework transforms TS data into Area Graphs. Compared with the existing visualization methods (such as RP), this conversion method is simpler and avoids the loss of some information in complex conversions as well as additional computational cost.
2. Related Work
2.1. Related Work on TSC
2.2. Related Work on Feature Extraction through DNN from Different Aspects
3. Model Design
3.1. Data Notation
3.2. LSTM-A
3.3. CNN-A
4. Experiment
4.1. Experiment Setting and Model Configuration
- CPU: Intel(R) Xeon(R) Gold 6140 CPU @ 2.30GHz (2 CPUs, 18 cores per CPU).
- GPU: NVIDIA Corporation GK210GL [Tesla K80] (8 GPUs).
- RAM: 256GB and 128GB video RAM for GPUs.
- OS: Ubuntu 18.04.3 LTS.
4.2. Integrated Model Evaluation
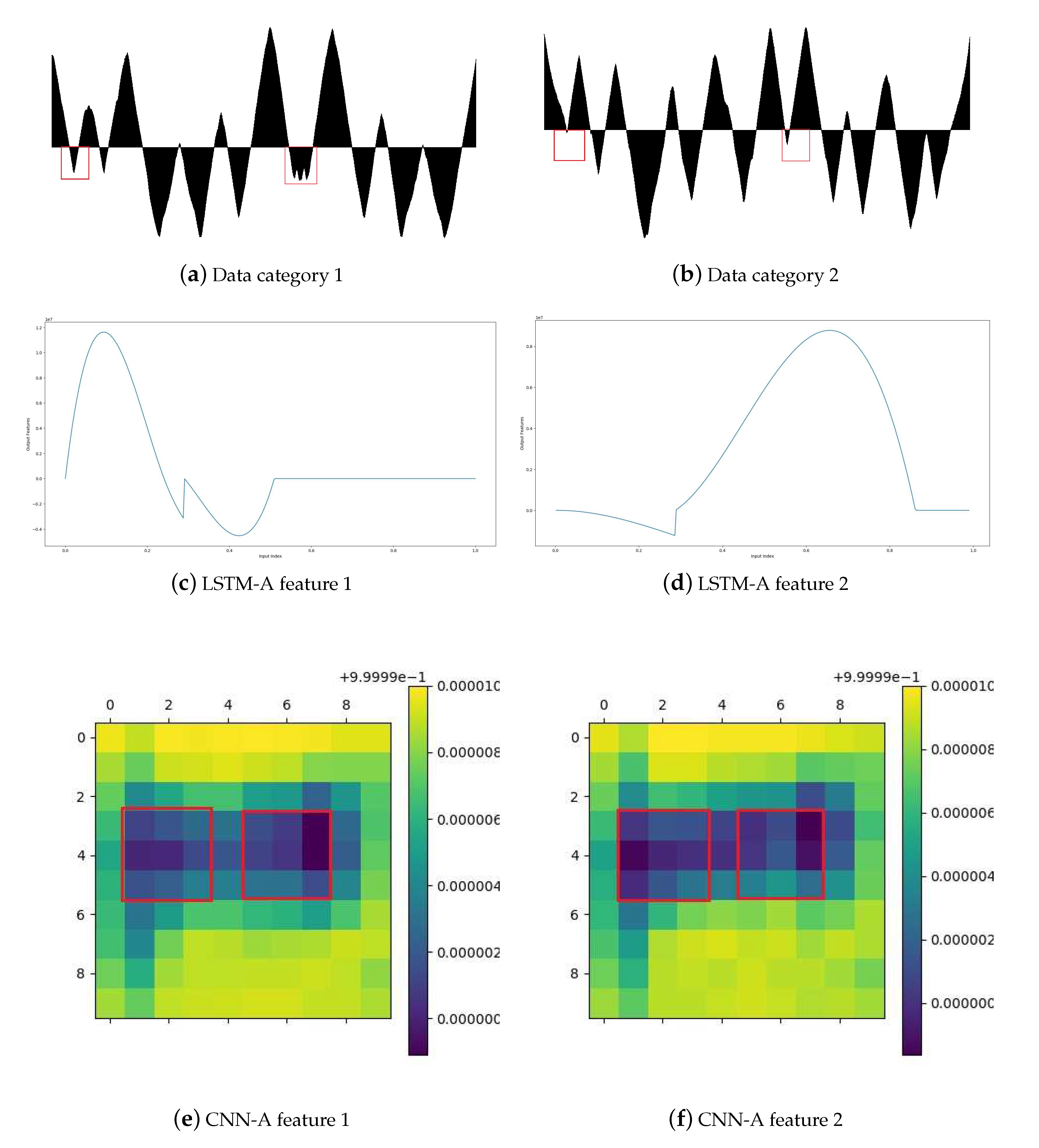
4.3. Sub-Model Evaluation
5. Conclusions and Future Work
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Esling, P.; Agon, C. Time-series data mining. ACM Comput. Surv. (CSUR) 2012, 45, 12. [Google Scholar] [CrossRef]
- Berndt, D.J.; Clifford, J. Using Dynamic Time Warping to Find Patterns in Time Series; KDD Workshop: Seattle, WA, USA, 1994; Volume 10, pp. 359–370. [Google Scholar]
- Ye, L.; Keogh, E. Time series shapelets: a new primitive for data mining. In Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Paris, France, 28 June–1 July 2009; pp. 947–956. [Google Scholar]
- Lines, J.; Taylor, S.; Bagnall, A. Hive-cote: The hierarchical vote collective of transformation-based ensembles for time series classification. In Proceedings of the 2016 IEEE 16th International Conference on Data Mining (ICDM), Barcelona, Spain, 12–15 December 2016; pp. 1041–1046. [Google Scholar]
- Graves, A. Supervised sequence labelling. In Supervised Sequence Labelling with Recurrent Neural Networks; Springer: New York, NY, USA, 2012; pp. 5–13. [Google Scholar]
- Wu, W.; Meng, Y.; Han, Q.; Li, M.; Li, X.; Mei, J.; Nie, P.; Sun, X.; Li, J. Glyce: Glyph-vectors for Chinese Character Representations. arXiv 2019, arXiv:1901.10125. [Google Scholar]
- Berat Sezer, O.; Murat Ozbayoglu, A. Financial Trading Model with Stock Bar Chart Image Time Series with Deep Convolutional Neural Networks. arXiv 2019, arXiv:1903.04610. [Google Scholar]
- Wang, Z.; Oates, T. Encoding time series as images for visual inspection and classification using tiled convolutional neural networks. In Proceedings of the Workshops at the Twenty-Ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015. [Google Scholar]
- Wang, Z.; Oates, T. Imaging time-series to improve classification and imputation. In Proceedings of the Twenty-Fourth International Joint Conference on Artificial Intelligence, Buenos Aires, Argentina, 25–31 July 2015. [Google Scholar]
- Tripathy, R.; Acharya, U.R. Use of features from RR-time series and EEG signals for automated classification of sleep stages in deep neural network framework. Biocybern. Biomed. Eng. 2018, 38, 890–902. [Google Scholar] [CrossRef]
- Hatami, N.; Gavet, Y.; Debayle, J. Classification of time-series images using deep convolutional neural networks. In Tenth International Conference on Machine Vision (ICMV 2017); International Society for Optics and Photonics: Bellingham, WA, USA, 2018; Volume 10696, p. 106960Y. [Google Scholar]
- Wang, Z.; Oates, T. Spatially encoding temporal correlations to classify temporal data using convolutional neural networks. arXiv 2015, arXiv:1509.07481. [Google Scholar]
- Schonfeld, E.; Ebrahimi, S.; Sinha, S.; Darrell, T.; Akata, Z. Generalized zero-and few-shot learning via aligned variational autoencoders. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 8247–8255. [Google Scholar]
- Zhang, H.; Zhang, J.; Koniusz, P. Few-shot Learning via Saliency-guided Hallucination of Samples. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 2770–2779. [Google Scholar]
- Dau, H.A.; Keogh, E.; Kamgar, K.; Yeh, C.C.M.; Zhu, Y.; Gharghabi, S.; Ratanamahatana, C.A.; Yanping, B.H.; Begum, N.; Bagnall, A.; et al. The UCR Time Series Classification Archive. Available online: https://www.cs.ucr.edu/~eamonn/time_series_data_2018/ (accessed on 31 October 2018).
- Ding, H.; Trajcevski, G.; Scheuermann, P.; Wang, X.; Keogh, E. Querying and mining of time series data: experimental comparison of representations and distance measures. Proc. VLDB Endow. 2008, 1, 1542–1552. [Google Scholar] [CrossRef]
- Lucas, B.; Shifaz, A.; Pelletier, C.; O’Neill, L.; Zaidi, N.; Goethals, B.; Petitjean, F.; Webb, G.I. Proximity forest: An effective and scalable distance-based classifier for time series. Data Min. Knowl. Discov. 2019, 33, 607–635. [Google Scholar] [CrossRef]
- Hills, J.; Lines, J.; Baranauskas, E.; Mapp, J.; Bagnall, A. Classification of time series by shapelet transformation. Data Min. Knowl. Discov. 2014, 28, 851–881. [Google Scholar] [CrossRef]
- Grabocka, J.; Schilling, N.; Wistuba, M.; Schmidt-Thieme, L. Learning time-series shapelets. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 392–401. [Google Scholar]
- Schäfer, P. The BOSS is concerned with time series classification in the presence of noise. Data Min. Knowl. Discov. 2015, 29, 1505–1530. [Google Scholar] [CrossRef]
- Deng, H.; Runger, G.; Tuv, E.; Vladimir, M. A time series forest for classification and feature extraction. Inf. Sci. 2013, 239, 142–153. [Google Scholar] [CrossRef]
- Baydogan, M.G.; Runger, G.; Tuv, E. A bag-of-features framework to classify time series. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2796–2802. [Google Scholar] [CrossRef]
- Schäfer, P.; Leser, U. Fast and accurate time series classification with weasel. In Proceedings of the 2017 ACM on Conference on Information and Knowledge Management, Singapore, 6–10 November 2017; pp. 637–646. [Google Scholar]
- Bostrom, A.; Bagnall, A. Binary shapelet transform for multiclass time series classification. In Transactions on Large-Scale Data-and Knowledge-Centered Systems XXXII; Springer: New York, NY, USA, 2017; pp. 24–46. [Google Scholar]
- Flynn, M.; Large, J.; Bagnall, T. The Contract Random Interval Spectral Ensemble (c-RISE): The Effect of Contracting a Classifier on Accuracy. In International Conference on Hybrid Artificial Intelligence Systems; Springer: New York, NY, USA, 2019; pp. 381–392. [Google Scholar]
- Wang, Z.; Song, W.; Liu, L.; Zhang, F.; Xue, J.; Ye, Y.; Fan, M.; Xu, M. Representation learning with deconvolution for multivariate time series classification and visualization. arXiv 2016, arXiv:1610.07258. [Google Scholar]
- Mittelman, R. Time-series modeling with undecimated fully convolutional neural networks. arXiv 2015, arXiv:1508.00317. [Google Scholar]
- Wang, S.; Hua, G.; Hao, G.; Xie, C. A cycle deep belief network model for multivariate time series classification. Math. Probl. Eng. 2017, 2017. [Google Scholar] [CrossRef]
- Mehdiyev, N.; Lahann, J.; Emrich, A.; Enke, D.; Fettke, P.; Loos, P. Time series classification using deep learning for process planning: a case from the process industry. Procedia Comput. Sci. 2017, 114, 242–249. [Google Scholar] [CrossRef]
- Aswolinskiy, W.; Reinhart, R.F.; Steil, J. Time series classification in reservoir-and model-space. Neural Process. Lett. 2018, 48, 789–809. [Google Scholar] [CrossRef]
- Bianchia, F.M.; Scardapaneb, S.; Løksea, S.; Jenssena, R. Reservoir computing approaches for representation and classification of multivariate time series. arXiv 2018, arXiv:1803.07870. [Google Scholar]
- Lines, J.; Bagnall, A. Time series classification with ensembles of elastic distance measures. Data Min. Knowl. Discov. 2015, 29, 565–592. [Google Scholar] [CrossRef]
- Bagnall, A.; Lines, J.; Hills, J.; Bostrom, A. Time-series classification with COTE: the collective of transformation-based ensembles. IEEE Trans. Knowl. Data Eng. 2015, 27, 2522–2535. [Google Scholar] [CrossRef]
- Shifaz, A.; Pelletier, C.; Petitjean, F.; Webb, G.I. Ts-chief: A scalable and accurate forest algorithm for time series classification. Data Min. Knowl. Discov. 2020, 1–34. [Google Scholar] [CrossRef]
- Dempster, A.; Petitjean, F.; Webb, G.I. ROCKET: Exceptionally fast and accurate time series classification using random convolutional kernels. arXiv 2019, arXiv:1910.13051. [Google Scholar]
- Lubba, C.H.; Sethi, S.S.; Knaute, P.; Schultz, S.R.; Fulcher, B.D.; Jones, N.S. catch22: CAnonical Time-series CHaracteristics. Data Min. Knowl. Discov. 2019, 33, 1821–1852. [Google Scholar] [CrossRef]
- Cui, Z.; Chen, W.; Chen, Y. Multi-scale convolutional neural networks for time series classification. arXiv 2016, arXiv:1603.06995. [Google Scholar]
- Wang, Z.; Yan, W.; Oates, T. Time series classification from scratch with deep neural networks: A strong baseline. In Proceedings of the 2017 international joint conference on neural networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 1578–1585. [Google Scholar]
- Du, Q.; Gu, W.; Zhang, L.; Huang, S.L. Attention-based LSTM-CNNs For Time-series Classification. In Proceedings of the 16th ACM Conference on Embedded Networked Sensor Systems, Shenzhen, China, 4–7 November 2018; pp. 410–411. [Google Scholar]
- Karim, F.; Majumdar, S.; Darabi, H.; Harford, S. Multivariate lstm-fcns for time series classification. Neural Netw. 2019, 116, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Karim, F.; Majumdar, S.; Darabi, H.; Chen, S. LSTM fully convolutional networks for time series classification. IEEE Access 2017, 6, 1662–1669. [Google Scholar] [CrossRef]
- Fawaz, H.I.; Lucas, B.; Forestier, G.; Pelletier, C.; Schmidt, D.F.; Weber, J.; Webb, G.I.; Idoumghar, L.; Muller, P.A.; Petitjean, F. InceptionTime: Finding AlexNet for Time Series Classification. arXiv 2019, arXiv:1909.04939. [Google Scholar]
- Kim, P. Matlab deep learning. Mach. Learn. Neural Netw. Artif. Intell. 2017, 130. [Google Scholar] [CrossRef]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Schuster, M.; Paliwal, K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997, 45, 2673–2681. [Google Scholar] [CrossRef]
- Greff, K.; Srivastava, R.K.; Koutník, J.; Steunebrink, B.R.; Schmidhuber, J. LSTM: A search space odyssey. IEEE Trans. Neural Netw. Learn. Syst. 2016, 28, 2222–2232. [Google Scholar] [CrossRef]
- Jozefowicz, R.; Zaremba, W.; Sutskever, I. An empirical exploration of recurrent network architectures. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 2342–2350. [Google Scholar]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural machine translation by jointly learning to align and translate. arXiv 2014, arXiv:1409.0473. [Google Scholar]
- Documentation, K. Available online: https://keras.io/api/ (accessed on 14 November 2017).
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Liu, C.L.; Hsaio, W.H.; Tu, Y.C. Time series classification with multivariate convolutional neural network. IEEE Trans. Ind. Electron. 2018, 66, 4788–4797. [Google Scholar] [CrossRef]


| Name | Configuration (the Parameter Value Was Selected from , According to the Optimal Accuracy on the Validation Set in Different Experiments). |
|---|---|
| TSC-FF | |
| Fully-connected Layer | {64, 128, 256} |
| LSTM-A | |
| LSTM layer1 | {64, 128, 256} |
| LSTM layer2 | {64, 128, 256} |
| LSTM layer3 | {64, 128, 256} |
| Dropout | {0.2, 0.5, 0.8} |
| Batch Normalization | momentum = 0.99, epsilon = 0.001 |
| Fully-connected Layer | {64, 128, 256} |
| CNN-A | |
| Conv2D1 | filter number {32, 64, 128}, kernel size {5 × 5, 8 × 8, 10 × 10}, strides {1, 2, 4} |
| Conv2D2 | filter number {32, 64, 128}, kernel size {5 × 5, 8 × 8, 10 × 10}, strides {1, 2, 4} |
| Conv2D3 | filter number {32, 64, 128}, kernel size {5 × 5, 8 × 8, 10 × 10}, strides {1, 2, 4} |
| Dropout | {0.2, 0.5, 0.8} |
| Batch Normalization | momentum = 0.99, epsilon = 0.001 |
| Avg. Pool | pool size {5 × 5, 8 × 8} |
| Max Pool | pool size {5 × 5, 8 × 8} |
| Fully-connected Layer | {64, 128, 256} |
| DTW | PF | BOSS | ST | STC | WS | TSF | RISE | HCT | CHI | RK | C2 | RN | FCN | IcT | TSC-FF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACSF1 | 0.5600 | 0.6383 | 0.7683 | 0.7826 | 0.8383 | 0.8180 | 0.6350 | 0.7600 | 0.8500 | 0.8070 | 0.8070 | 0.7777 | 0.8240 | 0.8800 | 0.8267 | 0.8552 |
| Adiac | 0.5857 | 0.7222 | 0.7490 | 0.7826 | 0.7932 | 0.7988 | 0.7119 | 0.7580 | 0.7962 | 0.7797 | 0.7720 | 0.6847 | 0.8154 | 0.8570 | 0.8223 | 0.7875 |
| ArroHea | 0.7771 | 0.8836 | 0.8688 | 0.7371 | 0.8067 | 0.8484 | 0.7968 | 0.8282 | 0.8760 | 0.8811 | 0.8590 | 0.7503 | 0.8587 | 0.8800 | 0.8804 | 0.8025 |
| Beef | 0.6667 | 0.5944 | 0.6122 | 0.9000 | 0.7356 | 0.7400 | 0.6889 | 0.7422 | 0.7356 | 0.6322 | 0.7600 | 0.4733 | 0.6767 | 0.7500 | 0.6822 | 0.8444 |
| BeetleFl | 0.7500 | 0.8600 | 0.9433 | 0.9000 | 0.9333 | 0.8867 | 0.8333 | 0.8717 | 0.9633 | 0.9583 | 0.8850 | 0.8400 | 0.8533 | 0.9500 | 0.8933 | 0.8889 |
| BirChick | 0.8500 | 0.9033 | 0.9833 | 0.8000 | 0.8700 | 0.8650 | 0.8150 | 0.8683 | 0.9400 | 0.9633 | 0.8817 | 0.8933 | 0.9450 | 0.9500 | 0.9517 | 0.9222 |
| BME | 0.7933 | 0.9991 | 0.8658 | 0.8267 | 0.9298 | 0.9478 | 0.9624 | 0.7860 | 0.9822 | 0.9964 | 0.9973 | 0.9049 | 0.9991 | 0.7533 | 0.9964 | 0.8921 |
| Car | 0.7333 | 0.8056 | 0.8483 | 0.9167 | 0.8583 | 0.8344 | 0.7661 | 0.7533 | 0.8689 | 0.8789 | 0.9117 | 0.7461 | 0.9083 | 0.9170 | 0.9011 | 0.9500 |
| CBF | 0.5922 | 0.9936 | 0.9989 | 0.9744 | 0.9853 | 0.9798 | 0.9719 | 0.9490 | 0.9983 | 0.9984 | 0.9959 | 0.9537 | 0.9882 | 1.0000 | 0.9961 | 1.0000 |
| Chinatow | 0.8406 | 0.9480 | 0.8771 | 0.9145 | 0.9630 | 0.9573 | 0.9530 | 0.8885 | 0.9628 | 0.9618 | 0.9669 | 0.9345 | 0.9701 | 0.9826 | 0.9649 | 0.9861 |
| ChlCon | 0.6831 | 0.6311 | 0.6582 | 0.6997 | 0.7352 | 0.7549 | 0.7231 | 0.7648 | 0.7339 | 0.6608 | 0.7961 | 0.5980 | 0.8410 | 0.8430 | 0.8636 | 0.7246 |
| CCECGTor | 0.7109 | 0.9377 | 0.9148 | 0.9543 | 0.9778 | 0.9850 | 0.9582 | 0.9474 | 0.9937 | 0.9534 | 0.8641 | 0.8030 | 0.7679 | 0.8130 | 0.8328 | 0.9511 |
| Coffee | 0.9286 | 0.9917 | 0.9857 | 0.9643 | 0.9893 | 0.9893 | 0.9869 | 0.9845 | 0.9929 | 0.9905 | 1.0000 | 0.9798 | 0.9964 | 1.0000 | 0.9988 | 1.0000 |
| Compu | 0.6680 | 0.7143 | 0.8005 | 0.7360 | 0.7991 | 0.7785 | 0.6488 | 0.7789 | 0.8111 | 0.7539 | 0.8429 | 0.7803 | 0.8604 | 0.8480 | 0.8656 | 0.7200 |
| CricketX | 0.6308 | 0.8004 | 0.7624 | 0.7718 | 0.7921 | 0.7757 | 0.6927 | 0.7062 | 0.8162 | 0.8304 | 0.8390 | 0.6089 | 0.8081 | 0.8150 | 0.8532 | 0.8077 |
| CricketY | 0.5590 | 0.7998 | 0.7504 | 0.7795 | 0.7779 | 0.7796 | 0.6859 | 0.7091 | 0.8098 | 0.8170 | 0.8450 | 0.5905 | 0.8102 | 0.7920 | 0.8600 | 0.8000 |
| CricketZ | 0.5564 | 0.8028 | 0.7693 | 0.7872 | 0.8072 | 0.7899 | 0.7059 | 0.7216 | 0.8339 | 0.8382 | 0.8532 | 0.6282 | 0.8132 | 0.8130 | 0.8611 | 0.8282 |
| Crop | 0.6488 | 0.7531 | 0.6857 | 0.7089 | 0.7367 | 0.7238 | 0.7456 | 0.7300 | 0.7682 | 0.7621 | 0.7517 | 0.6531 | 0.7638 | 0.7537 | 0.7932 | 0.7622 |
| DiaSizRed | 0.9346 | 0.9568 | 0.9452 | 0.9248 | 0.8593 | 0.9081 | 0.9417 | 0.9321 | 0.9143 | 0.9459 | 0.9580 | 0.9247 | 0.3062 | 0.9300 | 0.9507 | 0.9471 |
| DiPhOuAG | 0.7319 | 0.8022 | 0.8206 | 0.7754 | 0.7962 | 0.7928 | 0.8094 | 0.8216 | 0.8240 | 0.8281 | 0.8115 | 0.7830 | 0.7760 | 0.8350 | 0.7657 | 0.7661 |
| DiPhOuCor | 0.6763 | 0.8234 | 0.8117 | 0.7698 | 0.8273 | 0.8192 | 0.8058 | 0.8112 | 0.8236 | 0.8193 | 0.8244 | 0.8121 | 0.8092 | 0.8120 | 0.8156 | 0.7120 |
| DiPhTW | 0.5899 | 0.6921 | 0.6715 | 0.6619 | 0.6899 | 0.6787 | 0.6911 | 0.6945 | 0.6962 | 0.6918 | 0.7012 | 0.6811 | 0.6671 | 0.7900 | 0.6659 | 0.6875 |
| Earthqu | 0.6763 | 0.7496 | 0.7460 | 0.7410 | 0.7420 | 0.7475 | 0.7475 | 0.7482 | 0.7475 | 0.7482 | 0.7484 | 0.7388 | 0.7170 | 0.8010 | 0.7321 | 0.8160 |
| ECG200 | 0.8300 | 0.8730 | 0.8783 | 0.8300 | 0.8390 | 0.8590 | 0.8600 | 0.8510 | 0.8587 | 0.8550 | 0.8990 | 0.7887 | 0.8837 | 0.9000 | 0.8957 | 0.8916 |
| ECG5000 | 0.9196 | 0.9395 | 0.9401 | 0.9438 | 0.9418 | 0.9459 | 0.9434 | 0.9367 | 0.9456 | 0.9485 | 0.9474 | 0.9362 | 0.9370 | 0.9410 | 0.9421 | 0.9481 |
| ECGFiveD | 0.6864 | 0.8828 | 0.9923 | 0.9837 | 0.9779 | 0.9935 | 0.9520 | 0.9729 | 0.9938 | 0.9944 | 0.9960 | 0.8159 | 0.9510 | 0.9850 | 0.9959 | 0.9991 |
| ElecDev | 0.5709 | 0.8424 | 0.7974 | 0.7470 | 0.8818 | 0.8717 | 0.7931 | 0.8240 | 0.8797 | 0.8650 | 0.8932 | 0.8721 | 0.8884 | 0.7230 | 0.8902 | 0.7413 |
| EOGHoSi | 0.5470 | 0.8244 | 0.7067 | 0.7761 | 0.7599 | 0.7473 | 0.7060 | 0.6249 | 0.7957 | 0.8537 | 0.8138 | 0.6730 | 0.8647 | 0.7298 | 0.8836 | 0.8358 |
| EOGVeSi | 0.5801 | 0.7776 | 0.6603 | 0.7515 | 0.7132 | 0.6975 | 0.6726 | 0.6035 | 0.7630 | 0.8095 | 0.7811 | 0.6149 | 0.7517 | 0.7254 | 0.8145 | 0.7722 |
| EthLevel | 0.2680 | 0.3241 | 0.5087 | 0.6256 | 0.8557 | 0.7051 | 0.6693 | 0.6611 | 0.8490 | 0.6056 | 0.6253 | 0.3974 | 0.8525 | 0.6420 | 0.8755 | 0.8442 |
| FacAll | 0.8728 | 0.9771 | 0.9701 | 0.7787 | 0.9537 | 0.9731 | 0.9500 | 0.9649 | 0.9797 | 0.9827 | 0.9880 | 0.8108 | 0.9819 | 0.9290 | 0.9833 | 0.8705 |
| FaFour | 0.6250 | 0.9455 | 0.9955 | 0.8523 | 0.6564 | 0.9811 | 0.9068 | 0.8769 | 0.9731 | 0.9996 | 0.9314 | 0.6795 | 0.9250 | 0.9320 | 0.9386 | 0.9091 |
| FacUCR | 0.8434 | 0.9561 | 0.9513 | 0.9059 | 0.9101 | 0.9562 | 0.9040 | 0.8920 | 0.9613 | 0.9729 | 0.9713 | 0.7087 | 0.9643 | 0.9480 | 0.9769 | 0.9593 |
| FiftyW | 0.6923 | 0.8259 | 0.7059 | 0.7055 | 0.7366 | 0.7770 | 0.7217 | 0.6666 | 0.7716 | 0.8427 | 0.8251 | 0.5978 | 0.7240 | 0.6790 | 0.8268 | 0.8041 |
| Fish | 0.8971 | 0.9339 | 0.9697 | 0.9886 | 0.9499 | 0.9509 | 0.8305 | 0.8590 | 0.9794 | 0.9815 | 0.9741 | 0.7726 | 0.9699 | 0.9710 | 0.9726 | 0.9886 |
| FordA | 0.7174 | 0.8500 | 0.9214 | 0.9712 | 0.9342 | 0.9687 | 0.8158 | 0.9400 | 0.9441 | 0.9474 | 0.9424 | 0.9092 | 0.9315 | 0.9060 | 0.9591 | 1.0000 |
| FordB | 0.6654 | 0.8394 | 0.9074 | 0.8074 | 0.9198 | 0.9371 | 0.7907 | 0.9173 | 0.9280 | 0.9195 | 0.9230 | 0.8701 | 0.9128 | 0.8830 | 0.9409 | 1.0000 |
| FrRegTra | 0.9046 | 0.9424 | 0.9881 | 0.9951 | 0.9993 | 0.9906 | 0.9971 | 0.9523 | 0.9991 | 0.9985 | 0.9944 | 0.9982 | 0.9967 | 0.9965 | 0.9958 | 0.9989 |
| FrSmaTra | 0.7147 | 0.8234 | 0.9616 | 0.9689 | 0.9886 | 0.9006 | 0.9614 | 0.8787 | 0.9837 | 0.9955 | 0.9872 | 0.9598 | 0.9495 | 0.6902 | 0.9489 | 0.9668 |
| GunPoint | 0.9933 | 0.9913 | 0.9964 | 1.0000 | 0.9864 | 0.9931 | 0.9553 | 0.9809 | 0.9982 | 1.0000 | 0.9920 | 0.9431 | 0.9909 | 1.0000 | 0.9951 | 1.0000 |
| GPAgSpa | 0.9505 | 0.9969 | 0.9949 | 0.9805 | 0.9660 | 0.9813 | 0.9777 | 0.9863 | 0.9966 | 0.9996 | 0.9935 | 0.9439 | 0.9944 | 0.9424 | 0.9839 | 1.0000 |
| GPMaVeFe | 0.9468 | 0.9994 | 0.9996 | 0.9915 | 0.9865 | 0.9939 | 0.9960 | 0.9911 | 0.9999 | 0.9994 | 0.9999 | 0.9935 | 0.9901 | 0.9494 | 0.9983 | 0.9957 |
| GPOlVeYo | 0.8560 | 1.0000 | 0.9992 | 0.9945 | 0.9783 | 0.9860 | 1.0000 | 0.9998 | 1.0000 | 1.0000 | 0.9896 | 0.9642 | 1.0000 | 0.8952 | 1.0000 | 0.9264 |
| Ham | 0.4762 | 0.7835 | 0.8375 | 0.6857 | 0.8108 | 0.8213 | 0.7994 | 0.8197 | 0.8400 | 0.8051 | 0.8552 | 0.6940 | 0.8073 | 0.7620 | 0.8502 | 0.7128 |
| HandOut | 0.7378 | N/A | 0.9148 | 0.9324 | 0.9214 | 0.9454 | 0.9066 | 0.8810 | 0.9166 | N/A | N/A | 0.8684 | 0.9030 | 0.7760 | N/A | 0.9069 |
| Haptics | 0.2857 | 0.4583 | 0.4674 | 0.5227 | 0.5417 | 0.4492 | 0.4656 | 0.4788 | 0.5394 | 0.5233 | 0.5342 | 0.4587 | 0.4965 | 0.5510 | 0.5366 | 0.5213 |
| Herring | 0.5000 | 0.5745 | 0.5958 | 0.6719 | 0.6328 | 0.6021 | 0.6042 | 0.5984 | 0.6120 | 0.5974 | 0.6250 | 0.5557 | 0.5969 | 0.7030 | 0.6250 | 0.6491 |
| HouTwe | 0.8571 | 0.9370 | 0.9560 | 0.9204 | 0.9751 | 0.8106 | 0.8378 | 0.9297 | 0.9787 | 0.9703 | 0.9627 | 0.9462 | 0.9555 | 0.8908 | 0.9535 | 0.9613 |
| InlSkate | 0.4527 | 0.5610 | 0.5056 | 0.3727 | 0.4393 | 0.6746 | 0.3728 | 0.3904 | 0.5152 | 0.5719 | 0.4379 | 0.4724 | 0.4101 | 0.4110 | 0.5344 | 0.4616 |
| IERegTra | 1.0000 | 1.0000 | 0.9968 | 0.9957 | 0.9934 | 0.9718 | 1.0000 | 0.9929 | 1.0000 | 1.0000 | 0.9870 | 0.9288 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| IESmaTra | 1.0000 | 1.0000 | 0.9849 | 0.8736 | 0.8877 | 0.9067 | 0.9959 | 0.9722 | 1.0000 | 1.0000 | 0.9343 | 0.8372 | 0.3963 | 0.3574 | 1.0000 | 0.5150 |
| InWinSou | 0.2434 | 0.6069 | 0.5118 | 0.6268 | 0.6286 | 0.6195 | 0.6035 | 0.6364 | 0.6403 | 0.6322 | 0.6567 | 0.5592 | 0.4914 | 0.4020 | 0.6273 | 0.6269 |
| IPoDem | 0.9135 | 0.9560 | 0.8709 | 0.9475 | 0.9538 | 0.9468 | 0.9595 | 0.9445 | 0.9582 | 0.9624 | 0.9616 | 0.8775 | 0.9571 | 0.9700 | 0.9603 | 0.9633 |
| LarKitApp | 0.7680 | 0.8101 | 0.8358 | 0.8587 | 0.9299 | 0.7927 | 0.6363 | 0.7358 | 0.9201 | 0.8602 | 0.9300 | 0.8863 | 0.9539 | 0.8960 | 0.9524 | 0.8723 |
| Lightn2 | 0.6721 | 0.8486 | 0.8191 | 0.7377 | 0.6585 | 0.6273 | 0.7645 | 0.6820 | 0.7732 | 0.7689 | 0.7765 | 0.7448 | 0.8005 | 0.8030 | 0.8164 | 0.8155 |
| Lightn7 | 0.5753 | 0.7922 | 0.6712 | 0.7260 | 0.7434 | 0.7128 | 0.7205 | 0.6977 | 0.7575 | 0.7936 | 0.7977 | 0.6461 | 0.8100 | 0.8630 | 0.8215 | 0.8115 |
| Mallat | 0.8972 | 0.9720 | 0.9498 | 0.9642 | 0.9080 | 0.9664 | 0.9357 | 0.9541 | 0.9672 | 0.9767 | 0.9572 | 0.9061 | 0.9690 | 0.9800 | 0.9625 | 0.9542 |
| Meat | 0.6667 | 0.9872 | 0.9806 | 0.8500 | 0.9678 | 0.9767 | 0.9839 | 0.9867 | 0.9861 | 0.9844 | 0.9889 | 0.9428 | 0.9939 | 0.9670 | 0.9839 | 0.9333 |
| MedIma | 0.6632 | 0.7714 | 0.7158 | 0.6697 | 0.7098 | 0.7088 | 0.7462 | 0.6667 | 0.7404 | 0.7991 | 0.8051 | 0.7568 | 0.7922 | 0.7920 | 0.7963 | 0.7578 |
| MiPhOC | 0.6907 | 0.6589 | 0.6563 | 0.7938 | 0.6675 | 0.6604 | 0.6595 | 0.6998 | 0.6978 | 0.6944 | 0.7108 | 0.6881 | 0.5965 | 0.7680 | 0.5946 | 0.7663 |
| MiPhAG | 0.5584 | 0.8241 | 0.8095 | 0.6429 | 0.8315 | 0.8283 | 0.7995 | 0.8055 | 0.8129 | 0.8058 | 0.8345 | 0.7727 | 0.8236 | 0.7950 | 0.8340 | 0.7000 |
| MiPhTW | 0.5000 | 0.5491 | 0.5323 | 0.5195 | 0.5786 | 0.5543 | 0.5686 | 0.5851 | 0.5838 | 0.5732 | 0.5898 | 0.5574 | 0.5312 | 0.6120 | 0.5266 | 0.5745 |
| MSRegTra | 0.7518 | 0.9638 | 0.9179 | 0.9381 | 0.9602 | 0.9633 | 0.9230 | 0.9377 | 0.9659 | 0.9714 | 0.9657 | 0.9215 | 0.9701 | 0.8668 | 0.9663 | 0.9690 |
| MSSmaTra | 0.8043 | 0.9281 | 0.8717 | 0.9061 | 0.9368 | 0.9245 | 0.8402 | 0.8983 | 0.9447 | 0.9473 | 0.9307 | 0.8752 | 0.9162 | 0.9105 | 0.9134 | 0.9386 |
| MotStra | 0.7157 | 0.9150 | 0.8442 | 0.8970 | 0.9089 | 0.9048 | 0.8555 | 0.8780 | 0.9365 | 0.9301 | 0.9055 | 0.8485 | 0.9031 | 0.9500 | 0.8806 | 0.8943 |
| NonECG1 | 0.6972 | 0.8789 | 0.8416 | 0.9496 | 0.9359 | 0.9296 | 0.8848 | 0.9007 | 0.9278 | 0.9167 | 0.9022 | 0.8481 | 0.9442 | 0.9610 | 0.8744 | 0.8818 |
| NonECG2 | 0.8265 | 0.8585 | 0.9027 | 0.9511 | 0.9501 | 0.9362 | 0.9156 | 0.9184 | 0.9507 | 0.9736 | 0.9387 | 0.8772 | 0.9501 | 0.9550 | 0.9526 | 0.9424 |
| OliveOil | 0.8667 | 0.8291 | 0.8756 | 0.9000 | 0.8789 | 0.9133 | 0.8933 | 0.8933 | 0.8833 | 0.8253 | 0.8447 | 0.7456 | 0.8622 | 0.8330 | 0.8615 | 0.8667 |
| OSULeaf | 0.8802 | 0.3204 | 0.9691 | 0.9669 | 0.9563 | 0.8518 | 0.6433 | 0.6543 | 0.9748 | 0.3698 | 0.2751 | 0.7242 | 0.9747 | 0.9880 | 0.3346 | 0.9699 |
| PhOutCor | 0.7401 | 0.2755 | 0.8174 | 0.7634 | 0.8337 | 0.8217 | 0.8057 | 0.8125 | 0.8265 | 0.9601 | 0.1955 | 0.7919 | 0.8477 | 0.8260 | 0.9221 | 0.8096 |
| Phoneme | 0.2563 | 0.7157 | 0.2546 | 0.3207 | 0.3565 | 0.2595 | 0.1937 | 0.3466 | 0.3712 | 0.9689 | 0.9085 | 0.3002 | 0.3457 | 0.3450 | 0.9332 | 0.3058 |
| PigAir | 0.2260 | N/A | 0.9418 | 0.9656 | 0.9774 | 0.5378 | 0.2801 | 0.1825 | 0.9577 | N/A | N/A | 0.2769 | 0.4370 | 0.2067 | N/A | 0.5370 |
| PigArt | 0.6538 | N/A | 0.9676 | 0.9584 | 0.9457 | 0.9378 | 0.3072 | 0.7946 | 0.9668 | N/A | N/A | 0.8236 | 0.9537 | 0.8798 | N/A | 0.9719 |
| PigCVP | 0.4231 | 0.5005 | 0.9595 | 0.9291 | 0.8997 | 0.8979 | 0.1782 | 0.6825 | 0.9530 | 0.9614 | 0.8817 | 0.4761 | 0.4540 | 0.2500 | 0.9053 | 0.5253 |
| Plane | 1.0000 | 1.0000 | 0.9981 | 1.0000 | 0.9990 | 0.9949 | 0.9959 | 0.9965 | 1.0000 | 1.0000 | 1.0000 | 0.9883 | 1.0000 | 1.0000 | 0.9968 | 1.0000 |
| PowCons | 0.7611 | 0.9874 | 0.8900 | 0.8906 | 0.9406 | 0.9194 | 0.9931 | 0.9580 | 0.9924 | 0.9794 | 0.9561 | 0.8863 | 0.8861 | 0.8389 | 0.9861 | 0.9601 |
| PPOAG | 0.8179 | 0.8402 | 0.8285 | 0.8832 | 0.8455 | 0.8449 | 0.8450 | 0.8572 | 0.8559 | 0.8463 | 0.8524 | 0.8584 | 0.8174 | 0.8490 | 0.8221 | 0.8538 |
| PPOCor | 0.8195 | 0.8659 | 0.8655 | 0.8439 | 0.8953 | 0.8763 | 0.8489 | 0.8737 | 0.8852 | 0.8755 | 0.8993 | 0.8337 | 0.9056 | 0.9000 | 0.9063 | 0.8761 |
| ProPhTW | 0.7512 | 0.7907 | 0.7686 | 0.8049 | 0.8076 | 0.8015 | 0.8016 | 0.8132 | 0.8161 | 0.8111 | 0.8036 | 0.7863 | 0.7894 | 0.8100 | 0.7807 | 0.8043 |
| RefDev | 0.4080 | 0.6724 | 0.7828 | 0.5813 | 0.7772 | 0.7397 | 0.6116 | 0.6518 | 0.7909 | 0.7268 | 0.7300 | 0.7088 | 0.7868 | 0.5330 | 0.7594 | 0.5697 |
| Rock | 0.6200 | 0.7747 | 0.8027 | 0.8525 | 0.8673 | 0.8547 | 0.7600 | 0.7820 | 0.8553 | 0.8320 | 0.8047 | 0.7053 | 0.4160 | 0.3800 | 0.6273 | 0.6133 |
| ScreTyp | 0.4267 | 0.5720 | 0.5848 | 0.5200 | 0.7179 | 0.5959 | 0.4718 | 0.6059 | 0.7242 | 0.5942 | 0.6090 | 0.6007 | 0.7553 | 0.6670 | 0.7056 | 0.5392 |
| SeHaGen | 0.7450 | 0.9631 | 0.8877 | 0.8464 | 0.9323 | 0.7814 | 0.9474 | 0.8700 | 0.9692 | 0.9389 | 0.9231 | 0.8706 | 0.8089 | 0.7433 | 0.8846 | 0.9521 |
| SeHaMov | 0.6600 | 0.8909 | 0.6649 | 0.7094 | 0.7883 | 0.4603 | 0.8641 | 0.7240 | 0.8890 | 0.8850 | 0.6526 | 0.6825 | 0.4210 | 0.4200 | 0.5510 | 0.5916 |
| SeHaSub | 0.7022 | 0.9381 | 0.8351 | 0.8369 | 0.9133 | 0.7935 | 0.9160 | 0.8241 | 0.9506 | 0.9331 | 0.9119 | 0.8487 | 0.5547 | 0.5711 | 0.7639 | 0.6121 |
| ShapSim | 0.5389 | 0.7893 | 1.0000 | 0.9556 | 0.9996 | 0.9974 | 0.5137 | 0.7676 | 1.0000 | 1.0000 | 0.9981 | 0.9937 | 0.7272 | 0.8670 | 0.9235 | 0.8062 |
| ShapAll | 0.8350 | 0.8901 | 0.9085 | 0.8417 | 0.8653 | 0.9160 | 0.8043 | 0.8501 | 0.9313 | 0.9411 | 0.9232 | 0.8293 | 0.9331 | 0.8980 | 0.9382 | 0.8148 |
| SmKApp | 0.6533 | 0.7381 | 0.7471 | 0.7920 | 0.8128 | 0.8052 | 0.7882 | 0.8060 | 0.8283 | 0.8380 | 0.8183 | 0.8170 | 0.7966 | 0.8030 | 0.7706 | 0.8146 |
| SmoSub | 0.7867 | 0.9980 | 0.4073 | 0.7800 | 0.9358 | 0.8553 | 0.9873 | 0.8484 | 0.9862 | 0.9973 | 0.9749 | 0.8553 | 0.9929 | 0.8467 | 0.9849 | 0.9833 |
| SonSur1 | 0.7304 | 0.9201 | 0.8977 | 0.8436 | 0.8009 | 0.9093 | 0.8637 | 0.8670 | 0.8263 | 0.8897 | 0.9581 | 0.8834 | 0.9604 | 0.9680 | 0.9542 | 0.7630 |
| SonSur2 | 0.8583 | 0.8990 | 0.8794 | 0.9339 | 0.9370 | 0.9353 | 0.8743 | 0.9125 | 0.9367 | 0.9011 | 0.9350 | 0.9023 | 0.9689 | 0.9620 | 0.9513 | 0.9016 |
| StaCurv | 0.9622 | 0.9796 | 0.9766 | 0.9785 | 0.9786 | 0.9804 | 0.9704 | 0.9744 | 0.9796 | 0.9811 | 0.9810 | 0.9702 | 0.9720 | 0.9670 | 0.9781 | 0.9799 |
| Strberry | 0.9622 | 0.9605 | 0.9705 | 0.9622 | 0.9717 | 0.9786 | 0.9675 | 0.9730 | 0.9751 | 0.9740 | 0.9787 | 0.9229 | 0.9749 | 0.9690 | 0.9755 | 0.9709 |
| SweLeaf | 0.8848 | 0.9531 | 0.9202 | 0.9280 | 0.9340 | 0.9576 | 0.8979 | 0.9229 | 0.9494 | 0.9619 | 0.9632 | 0.8805 | 0.9586 | 0.9660 | 0.9698 | 0.8779 |
| Symbols | 0.9709 | 0.9670 | 0.9632 | 0.8824 | 0.9014 | 0.9532 | 0.8779 | 0.9127 | 0.9685 | 0.9708 | 0.9686 | 0.9480 | 0.9467 | 0.9620 | 0.9695 | 0.8000 |
| SynCont | 0.5667 | 0.9983 | 0.9666 | 0.9833 | 0.9919 | 0.9870 | 0.9916 | 0.6779 | 0.9942 | 0.9990 | 0.9978 | 0.9670 | 0.9944 | 0.9900 | 0.9958 | 1.0000 |
| ToeSeg1 | 0.7851 | 0.8355 | 0.9249 | 0.9649 | 0.9534 | 0.9430 | 0.6671 | 0.8804 | 0.9596 | 0.9598 | 0.9329 | 0.8127 | 0.9542 | 0.9690 | 0.9532 | 0.9793 |
| ToeSeg2 | 0.6846 | 0.8859 | 0.9615 | 0.9077 | 0.9451 | 0.9285 | 0.8026 | 0.9118 | 0.9682 | 0.9626 | 0.9326 | 0.8351 | 0.9531 | 0.9150 | 0.9636 | 0.9318 |
| Trace | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.9920 | 0.9830 | 1.0000 | 1.0000 | 1.0000 | 0.9997 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| TwLeECG | 0.9947 | 0.9818 | 0.9847 | 0.9974 | 0.9991 | 0.9975 | 0.8706 | 0.9107 | 0.9963 | 0.9901 | 0.9985 | 0.8539 | 0.9994 | 1.0000 | 0.9948 | 0.9288 |
| TwoPatt | 0.9975 | 1.0000 | 0.9917 | 0.9550 | 0.9903 | 0.9814 | 0.9938 | 0.4390 | 0.9995 | 1.0000 | 1.0000 | 0.8486 | 0.9998 | 0.8970 | 1.0000 | 1.0000 |
| UMD | 0.8472 | 0.9544 | 0.9664 | 0.9514 | 0.9368 | 0.9322 | 0.8333 | 0.5412 | 0.9674 | 0.9833 | 0.9829 | 0.8694 | 0.9525 | 0.9458 | 0.9799 | 0.8885 |
| UWavAll | 0.6770 | 0.9731 | 0.9449 | 0.8029 | 0.9580 | 0.9578 | 0.9624 | 0.9174 | 0.9666 | 0.9707 | 0.9773 | 0.8264 | 0.8690 | 0.8260 | 0.9512 | 0.9705 |
| UWavX | 0.5840 | 0.8314 | 0.7526 | 0.7303 | 0.8202 | 0.8176 | 0.8000 | 0.6339 | 0.8335 | 0.8468 | 0.8571 | 0.7685 | 0.7904 | 0.7540 | 0.8336 | 0.8460 |
| UWavY | 0.5771 | 0.7672 | 0.6621 | 0.7485 | 0.7449 | 0.7258 | 0.7219 | 0.6682 | 0.7547 | 0.7880 | 0.7836 | 0.7044 | 0.6759 | 0.7250 | 0.7706 | 0.7835 |
| UWavZ | 0.8492 | 0.7674 | 0.6955 | 0.9422 | 0.7725 | 0.7548 | 0.7334 | 0.6635 | 0.7751 | 0.7912 | 0.7958 | 0.7064 | 0.7514 | 0.7290 | 0.7732 | 0.7918 |
| Wafer | 0.9778 | 0.9961 | 0.9989 | 1.0000 | 1.0000 | 0.9999 | 0.9966 | 0.9954 | 0.9999 | 0.9989 | 0.9986 | 0.9973 | 0.9989 | 0.9970 | 0.9986 | 1.0000 |
| Wine | 0.5185 | 0.8562 | 0.8926 | 0.7963 | 0.8858 | 0.9302 | 0.8623 | 0.8710 | 0.8920 | 0.8981 | 0.9142 | 0.7000 | 0.8562 | 0.8890 | 0.8864 | 0.6333 |
| WordSyn | 0.6803 | 0.7782 | 0.6584 | 0.5705 | 0.6230 | 0.7127 | 0.6479 | 0.5916 | 0.6932 | 0.7937 | 0.7644 | 0.5440 | 0.6133 | 0.5800 | 0.7518 | 0.6605 |
| Worms | 0.5844 | 0.6931 | 0.7255 | 0.7403 | 0.7333 | 0.7784 | 0.6121 | 0.6866 | 0.7165 | 0.7684 | 0.7203 | 0.7251 | 0.7589 | 0.6690 | 0.7797 | 0.6522 |
| WormsTC | 0.6364 | 0.7693 | 0.8078 | 0.8312 | 0.7853 | 0.8004 | 0.6935 | 0.7853 | 0.7896 | 0.7861 | 0.7896 | 0.7922 | 0.7680 | 0.7290 | 0.8035 | 0.7946 |
| Yoga | 0.8200 | 0.8874 | 0.9102 | 0.8177 | 0.8800 | 0.8924 | 0.8658 | 0.8372 | 0.9124 | 0.8726 | 0.9138 | 0.8038 | 0.8772 | 0.8450 | 0.9123 | 0.9007 |
| AVG Rank | 13.3839 | 8.0092 | 8.8661 | 9.0625 | 7.1161 | 7.7589 | 10.4107 | 10.0446 | 4.4375 | 4.6239 | 5.0183 | 11.6071 | 7.3482 | 7.8750 | 5.3395 | 6.5982 |
| MPSE | 0.0746 | 0.0438 | 0.0390 | 0.0425 | 0.0349 | 0.0375 | 0.0481 | 0.0462 | 0.0293 | 0.0321 | 0.0355 | 0.0510 | 0.0398 | 0.0372 | 0.0320 | 0.0380 |
| IcT | FCN | RN | C2 | RK | CHI | HCT | RISE | TSF | WS | STC | ST | BOSS | PF | DTW | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FCN | |||||||||||||||
| RN | |||||||||||||||
| C2 | |||||||||||||||
| RK | |||||||||||||||
| CHI | |||||||||||||||
| HCT | |||||||||||||||
| RISE | |||||||||||||||
| TSF | |||||||||||||||
| WS | |||||||||||||||
| STC | |||||||||||||||
| ST | |||||||||||||||
| BOSS | |||||||||||||||
| PF | |||||||||||||||
| DTW | |||||||||||||||
| TSC-FF |
| Records in Each Category | (0,20] | (20,50] | (50,100] | (100,200] | (200,500] | (500,1000] | (1000,+∞) |
|---|---|---|---|---|---|---|---|
| Distance-based methods | |||||||
| DTW | 0.7208 | 0.7186 | 0.7433 | 0.6497 | 0.8156 | 0.7401 | 0.6512 |
| PF | 0.8593 | 0.8311 | 0.8745 | 0.7472 | 0.9003 | 0.2755 | 0.8439 |
| Feature-based methods | |||||||
| BOSS | 0.8703 | 0.8148 | 0.8463 | 0.7543 | 0.8917 | 0.8174 | 0.8754 |
| ST | 0.8574 | 0.8377 | 0.8417 | 0.7585 | 0.8660 | 0.7634 | 0.8419 |
| STC | 0.8623 | 0.8497 | 0.8676 | 0.8110 | 0.9059 | 0.8337 | 0.9119 |
| WS | 0.8721 | 0.8374 | 0.8350 | 0.7734 | 0.9037 | 0.8217 | 0.9126 |
| TSF | 0.7704 | 0.8077 | 0.8689 | 0.7436 | 0.8927 | 0.8057 | 0.7999 |
| RISE | 0.7998 | 0.7853 | 0.8577 | 0.7483 | 0.8315 | 0.8125 | 0.8938 |
| Ensemble methods | |||||||
| HCT | 0.8977 | 0.8687 | 0.8903 | 0.8232 | 0.9067 | 0.8265 | 0.9173 |
| CHI | 0.9147 | 0.8543 | 0.8844 | 0.7862 | 0.9021 | 0.9601 | 0.9106 |
| RK | 0.9001 | 0.8484 | 0.8679 | 0.8074 | 0.9085 | 0.1955 | 0.9195 |
| C2 | 0.7721 | 0.7638 | 0.8347 | 0.7375 | 0.8532 | 0.7919 | 0.8838 |
| Deep learning methods | |||||||
| RN | 0.8060 | 0.8728 | 0.8081 | 0.7957 | 0.9056 | 0.8477 | 0.9109 |
| FCN | 0.7936 | 0.8358 | 0.7660 | 0.7710 | 0.8741 | 0.8260 | 0.8373 |
| Ict | 0.9009 | 0.8597 | 0.8439 | 0.8137 | 0.9127 | 0.9221 | 0.9301 |
| TSC-FF | 0.8333 | 0.8640 | 0.8324 | 0.8027 | 0.8787 | 0.8096 | 0.9138 |
| GASF-GADF-MTF | TFRP | TSC-FF | |
|---|---|---|---|
| Adiac | 0.6270 | 0.7954 | 0.7875 |
| Beef | 0.7670 | 0.6333 | 0.8444 |
| CBF | 0.9910 | N/A | 1.0000 |
| Coffee | 1.0000 | 0.9643 | 1.0000 |
| ECGFiveDays | 0.9100 | 0.8955 | 0.9991 |
| FaceAll | 0.7630 | 0.7101 | 0.8705 |
| FaceFour | 0.9320 | 0.7841 | 0.9091 |
| 50words | 0.6990 | 0.5626 | 0.8041 |
| fish | 0.8860 | 0.8800 | 0.9886 |
| Gun Point | 0.9200 | 0.9800 | 1.0000 |
| Lighting2 | 0.8860 | 0.8525 | 1.0000 |
| Lighting7 | 0.7400 | 0.6849 | 0.8115 |
| OliveOil | 0.8000 | 0.8667 | 0.8667 |
| OSULeaf | 0.6420 | 0.9298 | 0.9695 |
| SwedishLeaf | 0.9350 | 0.9504 | 0.8779 |
| SyntheticControl | 0.9930 | N/A | 1.0000 |
| Trace | 1.0000 | N/A | 1.0000 |
| Two Patterns | 0.9090 | N/A | 1.0000 |
| wafer | 1.0000 | 0.9998 | 1.0000 |
| yoga | 0.8040 | 0.8587 | 0.9007 |
| Average Rank | 2.0500 | 2.3125 | 1.2500 |
| MPSE | 2.6500 × 10 | 3.1100 × 10 | 9.8900 × 10 |
| TSC-FF | LSTM-A | LSTM | CNN-A | CNN | |
|---|---|---|---|---|---|
| ACSF1 | 0.8552 | 0.8278 | 0.7800 | 0.8660 | 0.8400 |
| Adiac | 0.7875 | 0.7684 | 0.6199 | 0.7399 | 0.6513 |
| ArrowHead | 0.8025 | 0.8586 | 0.7580 | 0.8185 | 0.6822 |
| Beef | 0.8444 | 0.8704 | 0.8222 | 0.7852 | 0.7963 |
| BeetleFly | 0.8889 | 0.8333 | 0.6111 | 0.8121 | 0.5023 |
| BirdChicken | 0.9222 | 0.9444 | 0.8423 | 0.7342 | 0.7932 |
| BME | 0.8921 | 0.8400 | 0.8333 | 0.9200 | 0.9133 |
| Car | 0.9500 | 0.9667 | 0.8852 | 0.8667 | 0.8222 |
| CBF | 1.0000 | 0.9989 | 0.8593 | 1.0000 | 0.6716 |
| Chinatown | 0.9861 | 0.9478 | 0.9536 | 0.9652 | 0.9217 |
| ChlCon | 0.7246 | 0.6892 | 0.6399 | 0.7127 | 0.6306 |
| CinCECGtorso | 0.9511 | 0.8719 | 0.8300 | 0.9778 | 0.8911 |
| Coffee | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Computers | 0.7200 | 0.5600 | 0.5400 | 0.7200 | 0.6022 |
| CricketX | 0.8077 | 0.7923 | 0.7356 | 0.7694 | 0.7279 |
| CricketY | 0.8000 | 0.7556 | 0.6400 | 0.7410 | 0.5761 |
| CricketZ | 0.8282 | 0.7926 | 0.5499 | 0.7667 | 0.5627 |
| Crop | 0.7622 | 0.7636 | 0.7589 | 0.7712 | 0.7640 |
| DiaSizeRed | 0.9471 | 0.8800 | 0.5673 | 0.9239 | 0.4982 |
| DislPhaOutCor | 0.7661 | 0.7460 | 0.7621 | 0.7661 | 0.5968 |
| DisPhaOutAgeGro | 0.7120 | 0.6960 | 0.4000 | 0.7040 | 0.4240 |
| DisPhaTW | 0.6875 | 0.6960 | 0.4960 | 0.6480 | 0.4800 |
| Earthquakes | 0.8160 | 0.8080 | 0.7200 | 0.7360 | 0.7440 |
| ECG200 | 0.8916 | 0.8724 | 0.8245 | 0.8869 | 0.8046 |
| ECG5000 | 0.9481 | 0.9427 | 0.7230 | 0.9486 | 0.9442 |
| ECGFiveDays | 0.9991 | 0.7356 | 0.8052 | 0.9998 | 0.7935 |
| ElectricDevices | 0.7413 | 0.7420 | 0.7381 | 0.6938 | 0.6757 |
| EOGHorSig | 0.8358 | 0.7818 | 0.7569 | 0.8011 | 0.7707 |
| EOGVerSig | 0.7722 | 0.7541 | 0.7348 | 0.7569 | 0.7486 |
| EthLevel | 0.8442 | 0.8085 | 0.7864 | 0.8182 | 0.7485 |
| FaceAll | 0.8705 | 0.8567 | 0.7344 | 0.8824 | 0.7879 |
| FaceFour | 0.9091 | 0.8608 | 0.2785 | 0.8861 | 0.2785 |
| FacesUCR | 0.9593 | 0.9786 | 0.8618 | 0.8390 | 0.8119 |
| FiftyWords | 0.8041 | 0.7822 | 0.4318 | 0.7922 | 0.4320 |
| Fish | 0.9886 | 0.9471 | 0.8019 | 0.9134 | 0.8338 |
| FordA | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| FordB | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| FreRegTra | 0.9989 | 0.9940 | 0.9961 | 0.9979 | 0.9965 |
| FreSmaTra | 0.9668 | 0.9660 | 0.9547 | 0.9642 | 0.9323 |
| GunPoint | 1.0000 | 0.9556 | 0.6889 | 0.9852 | 0.5333 |
| GPAgSpa | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| GPMalVerFem | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.9968 |
| GPOldVerYou | 0.9264 | 0.9244 | 0.9199 | 0.9011 | 0.8507 |
| Ham | 0.7128 | 0.6489 | 0.6170 | 0.7021 | 0.4787 |
| HandOutlines | 0.9069 | 0.8679 | 0.3634 | 0.8739 | 0.6366 |
| Haptics | 0.5213 | 0.3863 | 0.3971 | 0.4621 | 0.2986 |
| Herring | 0.6491 | 0.6316 | 0.6140 | 0.5088 | 0.3860 |
| HouTwe | 0.9613 | 0.9412 | 0.8824 | 0.9664 | 0.9496 |
| InlineSkate | 0.4616 | 0.3747 | 0.4404 | 0.4694 | 0.4040 |
| InsEPGRegTra | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| InsEPGSmaTra | 0.5150 | 0.4739 | 0.4626 | 0.5238 | 0.4610 |
| InsWinSou | 0.6269 | 0.5752 | 0.5196 | 0.6066 | 0.3889 |
| ItaPowDem | 0.9633 | 0.9514 | 0.9568 | 0.9536 | 0.4989 |
| LarKitApp | 0.8723 | 0.8249 | 0.8398 | 0.8567 | 0.8205 |
| Lightning2 | 0.8155 | 0.8126 | 0.7955 | 0.8175 | 0.7246 |
| Lightning7 | 0.8115 | 0.8077 | 0.7231 | 0.7923 | 0.7769 |
| Mallat | 0.9542 | 0.9095 | 0.8313 | 0.8948 | 0.8564 |
| Meat | 0.9333 | 0.9148 | 0.9148 | 0.9407 | 0.8519 |
| MedicalImages | 0.7578 | 0.7442 | 0.7456 | 0.6988 | 0.5146 |
| MidPhaOutCor | 0.7663 | 0.7778 | 0.5556 | 0.7203 | 0.5556 |
| MidPhaOutAgeGro | 0.7000 | 0.5942 | 0.5000 | 0.5217 | 0.5725 |
| MidPhalanxTW | 0.5745 | 0.5652 | 0.4725 | 0.5072 | 0.4812 |
| MixShaRegTra | 0.9690 | 0.9711 | 0.9691 | 0.8613 | 0.8566 |
| MixShaSmaTra | 0.9386 | 0.9185 | 0.8818 | 0.9209 | 0.9177 |
| MoteStrain | 0.8943 | 0.8757 | 0.3357 | 0.8535 | 0.5444 |
| NonInvFatECGTho1 | 0.8818 | 0.9143 | 0.7766 | 0.8415 | 0.7538 |
| NonInvFatECGTho2 | 0.9424 | 0.9394 | 0.8305 | 0.9100 | 0.8236 |
| OliveOil | 0.8667 | 0.8852 | 0.8704 | 0.8407 | 0.7704 |
| OSULeaf | 0.9699 | 0.8747 | 0.8346 | 0.9712 | 0.8502 |
| PhaOutlCor | 0.8096 | 0.8187 | 0.6088 | 0.7293 | 0.6088 |
| Phoneme | 0.3058 | 0.2914 | 0.2492 | 0.2728 | 0.2400 |
| PigAirPre | 0.5370 | 0.5529 | 0.4183 | 0.5673 | 0.5106 |
| PigArtPre | 0.9719 | 0.9712 | 0.9279 | 0.9760 | 0.9663 |
| PigCVP | 0.5253 | 0.4933 | 0.4365 | 0.5444 | 0.5198 |
| Plane | 1.0000 | 0.9574 | 0.8489 | 0.9894 | 0.7957 |
| PowerCons | 0.9601 | 0.9111 | 0.9056 | 0.8833 | 0.8944 |
| ProPhaOutCor | 0.8538 | 0.7280 | 0.6858 | 0.7854 | 0.6858 |
| ProPhaOutAgeGro | 0.8761 | 0.7891 | 0.8326 | 0.8370 | 0.7707 |
| ProPhalanxTW | 0.8043 | 0.7533 | 0.7489 | 0.7989 | 0.7315 |
| RefrigeDevices | 0.5697 | 0.4362 | 0.4807 | 0.5460 | 0.3412 |
| Rock | 0.6133 | 0.4213 | 0.3756 | 0.4265 | 0.3961 |
| ScreenType | 0.5392 | 0.4620 | 0.4214 | 0.4887 | 0.3917 |
| SHGenCh2 | 0.9521 | 0.8133 | 0.8000 | 0.9651 | 0.9517 |
| SHMovCh2 | 0.5916 | 0.4244 | 0.3956 | 0.6044 | 0.4156 |
| SHSubCh2 | 0.6121 | 0.5556 | 0.5289 | 0.6167 | 0.5644 |
| ShapeletSim | 0.8062 | 0.8123 | 0.7938 | 0.7938 | 0.7338 |
| ShapesAll | 0.8148 | 0.8148 | 0.7407 | 0.7648 | 0.7870 |
| SmaKitApp | 0.8146 | 0.8020 | 0.6024 | 0.8167 | 0.6205 |
| SmoSub | 0.9833 | 0.9933 | 0.9867 | 0.5455 | 0.5195 |
| SonAIBORobSur1 | 0.7630 | 0.7759 | 0.7759 | 0.7259 | 0.7241 |
| SonAIBORobSur2 | 0.9016 | 0.8734 | 0.8046 | 0.9016 | 0.8254 |
| StarlightCurves | 0.9799 | 0.9541 | 0.9611 | 0.9493 | 0.8562 |
| Strawberry | 0.9709 | 0.9577 | 0.9347 | 0.9259 | 0.8577 |
| SwedishLeaf | 0.8779 | 0.8986 | 0.8772 | 0.8505 | 0.8270 |
| Symbols | 0.8000 | 0.8078 | 0.7888 | 0.7598 | 0.6536 |
| SyntheticControl | 1.0000 | 0.9852 | 0.9926 | 1.0000 | 0.3296 |
| ToeSeg1 | 0.9793 | 0.9268 | 0.8951 | 0.9724 | 0.9463 |
| ToeSeg2 | 0.9318 | 0.8863 | 0.9162 | 0.9447 | 0.8966 |
| Trace | 1.0000 | 0.9889 | 0.9889 | 1.0000 | 0.7000 |
| TwoLeadECG | 0.9288 | 0.7893 | 0.5034 | 0.9717 | 0.5034 |
| TwoPatterns | 1.0000 | 1.0000 | 0.9994 | 0.9989 | 0.2478 |
| UMD | 0.8885 | 0.9583 | 0.9475 | 0.8667 | 0.6478 |
| UWaveGesLibAll | 0.9705 | 0.9507 | 0.7415 | 0.9656 | 0.9693 |
| UWaveGesLibX | 0.8460 | 0.8177 | 0.7813 | 0.7986 | 0.7821 |
| UWaveGesLibY | 0.7835 | 0.7817 | 0.6665 | 0.7009 | 0.7287 |
| UWaveGesLibZ | 0.7918 | 0.7581 | 0.7056 | 0.7173 | 0.6440 |
| Wafer | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Wine | 0.6333 | 0.5000 | 0.6000 | 0.6333 | 0.6000 |
| WordSynonyms | 0.6605 | 0.6348 | 0.6254 | 0.7145 | 0.6164 |
| Worms | 0.6522 | 0.5217 | 0.5942 | 0.4493 | 0.5217 |
| WormsTwoClass | 0.7946 | 0.7522 | 0.7362 | 0.7377 | 0.7348 |
| Yoga | 0.9007 | 0.8933 | 0.7948 | 0.8930 | 0.5359 |
| Length | <80 | 81–250 | 251–450 | 451–700 | 701–1000 | >1000 |
|---|---|---|---|---|---|---|
| LSTM-A | 0.7915 | 0.8840 | 0.8413 | 0.8658 | 0.7060 | 0.7143 |
| CNN-A | 0.7577 | 0.8957 | 0.8496 | 0.8186 | 0.7205 | 0.7472 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Jiang, T.; Zhou, X.; Ma, B.; Zhao, F.; Wang, Y. Time-Series Classification Based on Fusion Features of Sequence and Visualization. Appl. Sci. 2020, 10, 4124. https://doi.org/10.3390/app10124124
Wang B, Jiang T, Zhou X, Ma B, Zhao F, Wang Y. Time-Series Classification Based on Fusion Features of Sequence and Visualization. Applied Sciences. 2020; 10(12):4124. https://doi.org/10.3390/app10124124
Chicago/Turabian StyleWang, Baoquan, Tonghai Jiang, Xi Zhou, Bo Ma, Fan Zhao, and Yi Wang. 2020. "Time-Series Classification Based on Fusion Features of Sequence and Visualization" Applied Sciences 10, no. 12: 4124. https://doi.org/10.3390/app10124124
APA StyleWang, B., Jiang, T., Zhou, X., Ma, B., Zhao, F., & Wang, Y. (2020). Time-Series Classification Based on Fusion Features of Sequence and Visualization. Applied Sciences, 10(12), 4124. https://doi.org/10.3390/app10124124




