Missing Value Imputation in Stature Estimation by Learning Algorithms Using Anthropometric Data: A Comparative Study
Abstract
1. Introduction
2. Background
2.1. Human Biological Information
2.2. Imputation Method for Handling Missing Values
2.3. Machine Learning Classifier
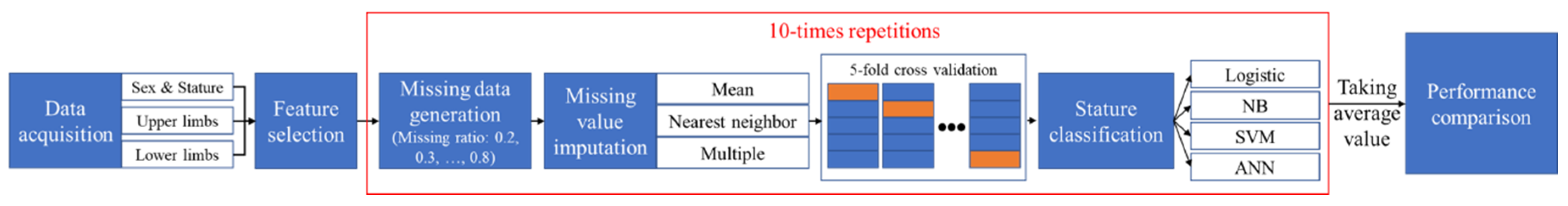
3. Method
3.1. Participants
3.2. Measurements
3.3. Experimental Procedure
4. Results
4.1. Stature Classification: Upper Limb
4.2. Stature Classification: Lower Limb
4.3. Stature Classification: Both
5. Discussion and Future Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Attallah, N.L.; Marshall, W.A. The estimation of stature from anthropometric and photogrammetric measurements of the limbs. Med. Sci. Law 1986, 26, 53–59. [Google Scholar] [CrossRef]
- Awais, M.; Naeem, F.; Rasool, N.; Mahmood, S. Identification of sex from footprint dimensions using machine learning: A study on population of Punjab in Pakistan. Egypt. J. Forensic Sci. 2018, 8, 72. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.S.; Lee, U.Y.; Park, D.K.; Jeong, Y.K.; Lee, N.S.; Han, S.Y.; Han, S.H. Stature estimation from partial measurements and maximum length of lower limb bones in Koreans. Aust. J. Forensic Sci. 2014, 46, 330–338. [Google Scholar] [CrossRef]
- Mahakizadeh, S.; Moghani-Ghoroghi, F.; Moshkdanian, G.; Mokhtari, T.; Hassanzadeh, G. The determination of correlation between stature and upper limb and hand measurements in Iranian adults. Forensic Sci. Int. 2016, 260, 27–30. [Google Scholar] [CrossRef]
- Nor, F.M.; Abdullah, N.; Mustapa, A.M.; Wen, L.Q.; Faisal, N.A.; Nazari, D.A.A.A. Estimation of stature by using lower limb dimensions in the Malaysian population. J. Forensic Leg. Med. 2013, 20, 947–952. [Google Scholar] [CrossRef]
- Bidmos, M.A.; Adebesin, A.A.; Mazengenya, P.; Olateju, O.I.; Adegboye, O. Estimation of sex from metatarsals using discriminant function and logistic regression analyses. Aust. J. Forensic Sci. 2020, 1–14. [Google Scholar] [CrossRef]
- Ahmed, A.A. Estimation of stature using lower limb measurements in Sudanese Arabs. J. Forensic Leg. Med. 2013, 20, 483–488. [Google Scholar] [CrossRef]
- Bhavna, N.S.; Nath, S. Use of lower limb measurements in reconstructing stature among Shia Muslims. Internet J. Biol. Anthropol. 2009, 2, 86–97. [Google Scholar]
- Moshkdanian, G.; Mahaki Zadeh, S.; Moghani Ghoroghi, F.; Mokhtari, T.; Hassanzadeh, G. Estimation of stature from the anthropometric measurement of lower limb in Iranian adults. Anat. Sci. J. 2014, 11, 149–154. [Google Scholar]
- Kim, W.; Kim, Y.M.; Yun, M.H. Estimation of stature from hand and foot dimensions in a Korean population. J. Forensic Leg. Med. 2018, 55, 87–92. [Google Scholar] [CrossRef]
- Ahmed, A.A. Estimation of stature from the upper limb measurements of Sudanese adults. Forensic Sci. Int. 2013, 228, 178-e1. [Google Scholar] [CrossRef]
- Muñoz, J.I.; Liñares-Iglesias, M.; Suárez-Peñaranda, J.M.; Mayo, M.; Miguéns, X.; Rodríguez-Calvo, M.S.; Concheiro, L. Stature estimation from radiographically determined long bone length in a Spanish population sample. J. Forensic Sci. 2001, 46, 363–366. [Google Scholar] [CrossRef]
- Ruff, C.B.; Holt, B.M.; Niskanen, M.; Sladék, V.; Berner, M.; Garofalo, E.; Garvin, H.M.; Hora, M.; Maijanen, H.; Niinimäki, S.; et al. Stature and body mass estimation from skeletal remains in the European Holocene. Am. J. Phys. Anthropol. 2012, 148, 601–617. [Google Scholar] [CrossRef]
- Czibula, G.; Ionescu, V.S.; Miholca, D.L.; Mircea, I.G. Machine learning-based approaches for predicting stature from archaeological skeletal remains using long bone lengths. J. Archaeol. Sci. 2016, 69, 85–99. [Google Scholar] [CrossRef]
- Miguel-Hurtado, O.; Guest, R.; Stevenage, S.V.; Neil, G.J.; Black, S. Comparing machine learning classifiers and linear/logistic regression to explore the relationship between Hand dimensions and demographic characteristics. PLoS ONE 2016, 11, e0165521. [Google Scholar] [CrossRef]
- Rativa, D.; Fernandes, B.J.; Roque, A. Height and Weight Estimation From Anthropometric Measurements Using Machine Learning Regressions. IEEE J. Transl. Eng. Health Med. 2018, 6, 1–9. [Google Scholar] [CrossRef]
- Ortiz, A.G.; Costa, C.; Silva, R.H.A.; Biazevic, M.G.H.; Michel-Crosato, E. Sex estimation: Anatomical references on panoramic radiographs using Machine Learning. Forensic Imaging 2020, 200356. [Google Scholar] [CrossRef]
- Arun Kumar, A.; Soodeen-Lalloo, A.K. Estimation of stature from fragmented human remains. Anthropology 2013, 1, 2. [Google Scholar]
- Pablos, A.; Gómez-Olivencia, A.; García-Pérez, A.; Martínez, I.; Lorenzo, C.; Arsuaga, J.L. From toe to head: Use of robust regression methods in stature estimation based on foot remains. Forensic Sci. Int. 2013, 226, 299-e1. [Google Scholar] [CrossRef]
- Duyar, I.; Pelin, C. Body height estimation based on tibia length in different stature groups. Am. J. Phys. Anthropol. Off. Publ. Am. Assoc. Phys. Anthropol. 2003, 122, 23–27. [Google Scholar] [CrossRef]
- Chikhalkar, B.G.; Mangaonkar, A.A.; Nanandkar, S.D.; Peddawad, R.G. Estimation of stature from measurements of long bones, hand and foot dimensions. J. Indian Acad. Forensic Med. 2010, 32, 329–333. [Google Scholar]
- Mahakkanukrauh, P.; Khanpetch, P.; Prasitwattanseree, S.; Vichairat, K.; Case, D.T. Stature estimation from long bone lengths in a Thai population. Forensic Sci. Int. 2011, 210, 279-e1. [Google Scholar] [CrossRef] [PubMed]
- Abrahamyan, D.O.; Gazarian, A.; Braillon, P.M. Estimation of stature and length of limb segments in children and adolescents from whole-body dual-energy X-ray absorptiometry scans. Pediatric Radiol. 2008, 38, 311–315. [Google Scholar] [CrossRef]
- Kim, W. A comparative study on the statistical modelling for the estimation of stature in Korean adults using hand measurements. Anthropol. Anz. Ber. Uber Die Biol. Anthropol. Lit. 2019, 76, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Akhlaghi, M.; Hajibeygi, M.; Zamani, N.; Moradi, B. Estimation of stature from upper limb anthropometry in Iranian population. J. Forensic Leg. Med. 2012, 19, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Little, R.J. Regression with missing X’s: A review. J. Am. Stat. Assoc. 1992, 87, 1227–1237. [Google Scholar] [CrossRef]
- Sande, I.G. Hot-deck imputation procedures. Incomplete Data Sample Surv. 1983, 3, 339–349. [Google Scholar]
- Andridge, R.R.; Little, R.J. A review of hot deck imputation for survey non-response. Int. Stat. Rev. 2010, 78, 40–64. [Google Scholar] [CrossRef]
- Cotton, C. Functional description of the Generalized Edit and Imputation System. Business Survey Methods Division. Stat. Can. 1991, 59, 447–461. [Google Scholar]
- Kim, J.K.; Fuller, W. Fractional hot deck imputation. Biometrika 2004, 91, 559–578. [Google Scholar] [CrossRef]
- Van Hulse, J.; Khoshgoftaar, T.M. Incomplete-case nearest neighbor imputation in software measurement data. Inf. Sci. 2014, 259, 596–610. [Google Scholar] [CrossRef]
- Rubin, D.B. Multiple imputations in sample surveys-a phenomenological Bayesian approach to nonresponse. Proc. Surv. Res. Methods Sect. Am. Stat. Assoc. 1978, 1, 20–34. [Google Scholar]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B (Methodol.) 1977, 1–38. [Google Scholar] [CrossRef]
- Schafer, J.L. Analysis of Incomplete Multivariate Data; CRC Press: Boca Raton, FL, USA, 1997. [Google Scholar]
- Lin, T.H. A comparison of multiple imputation with EM algorithm and MCMC method for quality of life missing data. Qual. Quant. 2010, 44, 277–287. [Google Scholar] [CrossRef]
- Park, S.; Lee, J.; Son, Y. Predicting market impact costs using nonparametric machine learning models. PLoS ONE 2016, 11, e0150243. [Google Scholar] [CrossRef]
- Kim, Y.M.; Son, Y.; Kim, W.; Jin, B.; Yun, M.H. Classification of children’s sitting postures using machine learning algorithms. Appl. Sci. 2018, 8, 1280. [Google Scholar] [CrossRef]
- Kim, C.; Son, Y.; Youm, S. Chronic Disease Prediction Using Character-Recurrent Neural Network in The Presence of Missing Information. Appl. Sci. 2019, 9, 2170. [Google Scholar] [CrossRef]
- Lee, S.; Lee, Y.S.; Son, Y. Forecasting Daily Temperatures with Different Time Interval Data Using Deep Neural Networks. Appl. Sci. 2020, 10, 1609. [Google Scholar] [CrossRef]
- Son, Y.; Byun, H.; Lee, J. Nonparametric machine learning models for predicting the credit default swaps: An empirical study. Expert Syst. Appl. 2016, 58, 210–220. [Google Scholar] [CrossRef]
- Maron, M.E. Automatic indexing: An experimental inquiry. J. ACM (JACM) 1961, 8, 404–417. [Google Scholar] [CrossRef]
- Rosenblatt, F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Rhiu, I.; Kim, W. Estimation of stature from finger and phalange lengths in a Korean adolescent. J. Physiol. Anthropol. 2019, 38, 13. [Google Scholar] [CrossRef] [PubMed]
- Simmons, K.P.; Istook, C.L. Body measurement techniques: Comparing 3D body-scanning and anthropometric methods for apparel applications. J. Fash. Mark. Manag. 2003, 7, 306–332. [Google Scholar] [CrossRef]
- Son, Y.; Noh, D.J.; Lee, J. Forecasting trends of high-frequency KOSPI200 index data using learning classifiers. Expert Syst. Appl. 2012, 39, 11607–11615. [Google Scholar] [CrossRef]
- Auerbach, B.M. Methods for estimating missing human skeletal element osteometric dimensions employed in the revised fully technique for estimating stature. Am. J. Phys. Anthropol. 2011, 145, 67–80. [Google Scholar] [CrossRef]
- Uhrová, P.; Beňuš, R.; Masnicová, S.; Obertová, Z.; Kramárová, D.; Kyselicová, K.; Dörnhöferová, M.; Bodoriková, S.; Neščáková, E. Estimation of stature using hand and foot dimensions in Slovak adults. Leg. Med. 2015, 17, 92–97. [Google Scholar] [CrossRef]
- Wilson, R.J.; Herrmann, N.P.; Jantz, L.M. Evaluation of stature estimation from the database for forensic anthropology. J. Forensic Sci. 2010, 55, 684–689. [Google Scholar] [CrossRef]
- Feng, L.; Peng, F.; Li, S.; Jiang, L.; Sun, H.; Ji, A.; Zeng, C.; Li, C.; Liu, F. Systematic feature selection improves accuracy of methylation-based forensic age estimation in Han Chinese males. Forensic Sci. Int. Genet. 2018, 35, 38–45. [Google Scholar] [CrossRef]
| Category | Measurement Variable | Definition |
|---|---|---|
| Upper Limb | Upper Arm Length | The distance from lateral shoulder to radial |
| Arm Length | The distance from lateral shoulder to ulnar styloid over radial | |
| Under Arm Length | The distance from axilla to ulnar styloid | |
| Hand Length | The distance between a line connecting radial styloid and dactylion III | |
| Palm Length | The distance between a line connecting radial styloid and base of the middle finger | |
| Hand Breadth | The distance between metacarpal V and metacarpal II | |
| Hand Thickness | The maximum thickness between dorsum of hand and palm at metacarpal III | |
| Inner Grip Circumference | The circumference of grip, shaped as the interphalangeal joint of thumb where it meets the tip of index finger | |
| Hand Circumference | The circumference of the hand over metacarpal V and metacarpal II | |
| Elbow Circumference | The circumference of the elbow at center olecranon with the arm bent 90° | |
| Wrist Circumference | The smallest circumference from the elbow to the knuckles of the hand | |
| Upper Arm Circumference | The circumference of the upper arm around the flexed biceps with the upper arm extended horizontally and the elbow flexed 90° | |
| Lower Limb | Hip Height | The vertical distance between a buttock protrusion and standing surface |
| Waist Height | The vertical distance between a standing surface and side waist band (half the distance between the tenth rib and iliac crest) | |
| Iliac Spine Height | The vertical distance between a standing surface and anterior superior iliac spine | |
| Knee Height | The vertical distance between a standing surface and the tibia | |
| Thigh Vertical Length | The distance between gluteal fold and popliteal fossa | |
| Outside Leg Length | The vertical distance between a standing surface and side waist band (half the distance between the tenth rib and iliac crest) | |
| Foot Breadth | The horizontal length between metatarsophalangeal V and metatarsophalangeal I | |
| Foot Length | The straight length between ptemion and acropodion | |
| Lateral Malleolus Height | The vertical distance between a standing surface and lateral malleolus | |
| Thigh Circumference | The horizontal circumference at gluteal fold | |
| Knee Circumference | The horizontal circumference at mid-patella | |
| Ankle Circumference | the maximum circumference over lateral malleolus and medial malleolus |
| Sex | Method | Imputation | Missing Ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | |||
| Combined sexes | Logistic | Mean | 0.721 | 0.700 | 0.680 | 0.654 | 0.628 | 0.584 | 0.549 |
| Nearest neighbor (NN) | 0.657 | 0.594 | 0.539 | 0.487 | 0.445 | 0.417 | 0.407 | ||
| Multiple | 0.745 | 0.732 | 0.713 | 0.684 | 0.643 | 0.579 | 0.493 | ||
| Naïve Bayes classifier (NB) | Mean | 0.666 | 0.660 | 0.650 | 0.634 | 0.614 | 0.581 | 0.537 | |
| NN | 0.622 | 0.586 | 0.550 | 0.508 | 0.465 | 0.432 | 0.398 | ||
| Multiple | 0.662 | 0.650 | 0.633 | 0.605 | 0.563 | 0.510 | 0.436 | ||
| Support vector machine (SVM) | Mean | 0.749 | 0.734 | 0.717 | 0.696 | 0.671 | 0.638 | 0.595 | |
| NN | 0.698 | 0.647 | 0.597 | 0.539 | 0.491 | 0.452 | 0.445 | ||
| Multiple | 0.756 | 0.741 | 0.719 | 0.688 | 0.644 | 0.582 | 0.493 | ||
| Artificial neural network (ANN) | Mean | 0.734 | 0.714 | 0.697 | 0.670 | 0.642 | 0.609 | 0.565 | |
| NN | 0.692 | 0.642 | 0.586 | 0.526 | 0.476 | 0.439 | 0.432 | ||
| Multiple | 0.753 | 0.738 | 0.717 | 0.687 | 0.644 | 0.582 | 0.495 | ||
| Female | Logistic | Mean | 0.681 | 0.661 | 0.642 | 0.622 | 0.596 | 0.566 | 0.528 |
| NN | 0.625 | 0.576 | 0.529 | 0.486 | 0.457 | 0.432 | 0.420 | ||
| Multiple | 0.699 | 0.686 | 0.663 | 0.634 | 0.591 | 0.535 | 0.466 | ||
| NB | Mean | 0.644 | 0.634 | 0.620 | 0.597 | 0.570 | 0.533 | 0.479 | |
| NN | 0.600 | 0.572 | 0.536 | 0.501 | 0.464 | 0.434 | 0.406 | ||
| Multiple | 0.635 | 0.619 | 0.594 | 0.559 | 0.509 | 0.453 | 0.405 | ||
| SVM | Mean | 0.697 | 0.682 | 0.665 | 0.645 | 0.622 | 0.591 | 0.551 | |
| NN | 0.652 | 0.611 | 0.569 | 0.525 | 0.486 | 0.454 | 0.440 | ||
| Multiple | 0.707 | 0.691 | 0.668 | 0.638 | 0.595 | 0.539 | 0.469 | ||
| ANN | Mean | 0.684 | 0.664 | 0.645 | 0.625 | 0.600 | 0.571 | 0.532 | |
| NN | 0.635 | 0.590 | 0.545 | 0.506 | 0.470 | 0.441 | 0.429 | ||
| Multiple | 0.703 | 0.689 | 0.665 | 0.636 | 0.594 | 0.538 | 0.467 | ||
| Male | Logistic | Mean | 0.729 | 0.710 | 0.686 | 0.662 | 0.637 | 0.605 | 0.562 |
| NN | 0.658 | 0.596 | 0.537 | 0.488 | 0.451 | 0.431 | 0.425 | ||
| Multiple | 0.757 | 0.744 | 0.727 | 0.699 | 0.658 | 0.601 | 0.514 | ||
| NB | Mean | 0.696 | 0.686 | 0.669 | 0.648 | 0.620 | 0.580 | 0.519 | |
| NN | 0.651 | 0.615 | 0.573 | 0.531 | 0.488 | 0.451 | 0.420 | ||
| Multiple | 0.692 | 0.679 | 0.659 | 0.627 | 0.583 | 0.528 | 0.458 | ||
| SVM | Mean | 0.734 | 0.715 | 0.695 | 0.671 | 0.645 | 0.614 | 0.573 | |
| NN | 0.688 | 0.640 | 0.585 | 0.532 | 0.484 | 0.448 | 0.438 | ||
| Multiple | 0.762 | 0.748 | 0.729 | 0.700 | 0.660 | 0.602 | 0.517 | ||
| ANN | Mean | 0.749 | 0.733 | 0.717 | 0.695 | 0.673 | 0.643 | 0.604 | |
| NN | 0.704 | 0.658 | 0.607 | 0.555 | 0.510 | 0.468 | 0.453 | ||
| Multiple | 0.760 | 0.746 | 0.728 | 0.700 | 0.660 | 0.603 | 0.518 | ||
| Sex | Method | Imputation | Missing Ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | |||
| Combined sexes | Logistic | Mean | 0.778 | 0.757 | 0.735 | 0.710 | 0.679 | 0.643 | 0.596 |
| NN | 0.694 | 0.624 | 0.559 | 0.500 | 0.454 | 0.433 | 0.427 | ||
| Multiple | 0.816 | 0.805 | 0.788 | 0.756 | 0.702 | 0.615 | 0.497 | ||
| NB | Mean | 0.784 | 0.772 | 0.757 | 0.737 | 0.709 | 0.668 | 0.613 | |
| NN | 0.709 | 0.655 | 0.599 | 0.543 | 0.493 | 0.457 | 0.428 | ||
| Multiple | 0.800 | 0.790 | 0.770 | 0.734 | 0.668 | 0.570 | 0.451 | ||
| SVM | Mean | 0.822 | 0.811 | 0.796 | 0.777 | 0.752 | 0.716 | 0.668 | |
| NN | 0.755 | 0.700 | 0.638 | 0.573 | 0.519 | 0.486 | 0.493 | ||
| Multiple | 0.837 | 0.824 | 0.803 | 0.767 | 0.706 | 0.617 | 0.501 | ||
| ANN | Mean | 0.811 | 0.798 | 0.777 | 0.761 | 0.732 | 0.689 | 0.636 | |
| NN | 0.770 | 0.722 | 0.657 | 0.579 | 0.511 | 0.475 | 0.477 | ||
| Multiple | 0.835 | 0.822 | 0.802 | 0.767 | 0.707 | 0.617 | 0.498 | ||
| Female | Logistic | Mean | 0.757 | 0.738 | 0.715 | 0.696 | 0.669 | 0.637 | 0.596 |
| NN | 0.690 | 0.624 | 0.568 | 0.520 | 0.485 | 0.462 | 0.456 | ||
| Multiple | 0.791 | 0.784 | 0.768 | 0.740 | 0.690 | 0.612 | 0.513 | ||
| NB | Mean | 0.759 | 0.747 | 0.729 | 0.705 | 0.674 | 0.631 | 0.574 | |
| NN | 0.701 | 0.653 | 0.603 | 0.558 | 0.513 | 0.477 | 0.440 | ||
| Multiple | 0.771 | 0.762 | 0.743 | 0.710 | 0.649 | 0.557 | 0.462 | ||
| SVM | Mean | 0.788 | 0.775 | 0.758 | 0.740 | 0.718 | 0.682 | 0.637 | |
| NN | 0.732 | 0.680 | 0.629 | 0.576 | 0.533 | 0.504 | 0.496 | ||
| Multiple | 0.808 | 0.797 | 0.778 | 0.745 | 0.692 | 0.614 | 0.517 | ||
| ANN | Mean | 0.771 | 0.751 | 0.730 | 0.709 | 0.681 | 0.647 | 0.603 | |
| NN | 0.719 | 0.668 | 0.611 | 0.558 | 0.516 | 0.487 | 0.477 | ||
| Multiple | 0.806 | 0.795 | 0.776 | 0.745 | 0.692 | 0.613 | 0.514 | ||
| Male | Logistic | Mean | 0.770 | 0.749 | 0.726 | 0.703 | 0.674 | 0.639 | 0.594 |
| NN | 0.683 | 0.610 | 0.547 | 0.493 | 0.459 | 0.440 | 0.443 | ||
| Multiple | 0.813 | 0.804 | 0.787 | 0.756 | 0.702 | 0.618 | 0.512 | ||
| NB | Mean | 0.776 | 0.762 | 0.741 | 0.718 | 0.684 | 0.639 | 0.577 | |
| NN | 0.707 | 0.655 | 0.602 | 0.552 | 0.505 | 0.469 | 0.439 | ||
| Multiple | 0.792 | 0.781 | 0.762 | 0.724 | 0.659 | 0.565 | 0.465 | ||
| SVM | Mean | 0.808 | 0.793 | 0.776 | 0.757 | 0.732 | 0.693 | 0.651 | |
| NN | 0.748 | 0.693 | 0.637 | 0.581 | 0.533 | 0.501 | 0.501 | ||
| Multiple | 0.833 | 0.819 | 0.798 | 0.763 | 0.705 | 0.621 | 0.519 | ||
| ANN | Mean | 0.788 | 0.768 | 0.75 | 0.724 | 0.694 | 0.657 | 0.609 | |
| NN | 0.748 | 0.693 | 0.631 | 0.567 | 0.517 | 0.485 | 0.480 | ||
| Multiple | 0.830 | 0.817 | 0.796 | 0.763 | 0.706 | 0.621 | 0.515 | ||
| Sex | Method | Imputation | Missing Ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | |||
| Both | Logistic | Mean | 0.801 | 0.784 | 0.769 | 0.748 | 0.724 | 0.693 | 0.650 |
| NN | 0.717 | 0.648 | 0.578 | 0.515 | 0.465 | 0.424 | 0.407 | ||
| Multiple | 0.829 | 0.821 | 0.811 | 0.791 | 0.755 | 0.694 | 0.594 | ||
| NB | Mean | 0.771 | 0.765 | 0.754 | 0.740 | 0.720 | 0.688 | 0.642 | |
| NN | 0.700 | 0.654 | 0.605 | 0.551 | 0.498 | 0.447 | 0.410 | ||
| Multiple | 0.791 | 0.784 | 0.773 | 0.749 | 0.706 | 0.634 | 0.529 | ||
| SVM | Mean | 0.834 | 0.821 | 0.809 | 0.793 | 0.775 | 0.746 | 0.702 | |
| NN | 0.765 | 0.708 | 0.647 | 0.585 | 0.523 | 0.467 | 0.434 | ||
| Multiple | 0.857 | 0.847 | 0.833 | 0.810 | 0.767 | 0.698 | 0.595 | ||
| ANN | Mean | 0.818 | 0.800 | 0.784 | 0.761 | 0.736 | 0.705 | 0.658 | |
| NN | 0.758 | 0.700 | 0.628 | 0.561 | 0.498 | 0.442 | 0.417 | ||
| Multiple | 0.854 | 0.845 | 0.831 | 0.808 | 0.767 | 0.699 | 0.596 | ||
| Female | Logistic | Mean | 0.768 | 0.753 | 0.734 | 0.718 | 0.696 | 0.667 | 0.625 |
| NN | 0.701 | 0.645 | 0.581 | 0.533 | 0.486 | 0.448 | 0.426 | ||
| Multiple | 0.794 | 0.789 | 0.778 | 0.759 | 0.725 | 0.664 | 0.567 | ||
| NB | Mean | 0.756 | 0.748 | 0.735 | 0.717 | 0.689 | 0.653 | 0.591 | |
| NN | 0.694 | 0.652 | 0.604 | 0.556 | 0.509 | 0.460 | 0.422 | ||
| Multiple | 0.771 | 0.763 | 0.750 | 0.727 | 0.679 | 0.596 | 0.481 | ||
| SVM | Mean | 0.798 | 0.783 | 0.769 | 0.751 | 0.730 | 0.700 | 0.656 | |
| NN | 0.740 | 0.691 | 0.638 | 0.586 | 0.533 | 0.482 | 0.449 | ||
| Multiple | 0.822 | 0.812 | 0.797 | 0.774 | 0.732 | 0.667 | 0.572 | ||
| ANN | Mean | 0.787 | 0.769 | 0.751 | 0.732 | 0.710 | 0.681 | 0.635 | |
| NN | 0.717 | 0.663 | 0.603 | 0.554 | 0.503 | 0.460 | 0.434 | ||
| Multiple | 0.819 | 0.810 | 0.795 | 0.773 | 0.732 | 0.666 | 0.571 | ||
| Male | Logistic | Mean | 0.798 | 0.784 | 0.765 | 0.743 | 0.719 | 0.688 | 0.647 |
| NN | 0.712 | 0.642 | 0.572 | 0.515 | 0.470 | 0.434 | 0.420 | ||
| Multiple | 0.823 | 0.820 | 0.811 | 0.793 | 0.759 | 0.698 | 0.604 | ||
| NB | Mean | 0.787 | 0.776 | 0.762 | 0.743 | 0.717 | 0.678 | 0.615 | |
| NN | 0.713 | 0.670 | 0.618 | 0.570 | 0.516 | 0.466 | 0.424 | ||
| Multiple | 0.803 | 0.796 | 0.782 | 0.757 | 0.711 | 0.633 | 0.526 | ||
| SVM | Mean | 0.829 | 0.816 | 0.798 | 0.782 | 0.761 | 0.732 | 0.689 | |
| NN | 0.769 | 0.717 | 0.659 | 0.601 | 0.543 | 0.488 | 0.453 | ||
| Multiple | 0.857 | 0.847 | 0.832 | 0.810 | 0.768 | 0.702 | 0.609 | ||
| ANN | Mean | 0.817 | 0.799 | 0.779 | 0.758 | 0.732 | 0.701 | 0.655 | |
| NN | 0.742 | 0.687 | 0.625 | 0.561 | 0.507 | 0.455 | 0.431 | ||
| Multiple | 0.853 | 0.844 | 0.829 | 0.807 | 0.768 | 0.702 | 0.607 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, Y.; Kim, W. Missing Value Imputation in Stature Estimation by Learning Algorithms Using Anthropometric Data: A Comparative Study. Appl. Sci. 2020, 10, 5020. https://doi.org/10.3390/app10145020
Son Y, Kim W. Missing Value Imputation in Stature Estimation by Learning Algorithms Using Anthropometric Data: A Comparative Study. Applied Sciences. 2020; 10(14):5020. https://doi.org/10.3390/app10145020
Chicago/Turabian StyleSon, Youngdoo, and Wonjoon Kim. 2020. "Missing Value Imputation in Stature Estimation by Learning Algorithms Using Anthropometric Data: A Comparative Study" Applied Sciences 10, no. 14: 5020. https://doi.org/10.3390/app10145020
APA StyleSon, Y., & Kim, W. (2020). Missing Value Imputation in Stature Estimation by Learning Algorithms Using Anthropometric Data: A Comparative Study. Applied Sciences, 10(14), 5020. https://doi.org/10.3390/app10145020






