Microbial Involvement in the Bioremediation of Total Petroleum Hydrocarbon Polluted Soils: Challenges and Perspectives
Abstract
:1. Introduction
2. The Soil Scenario
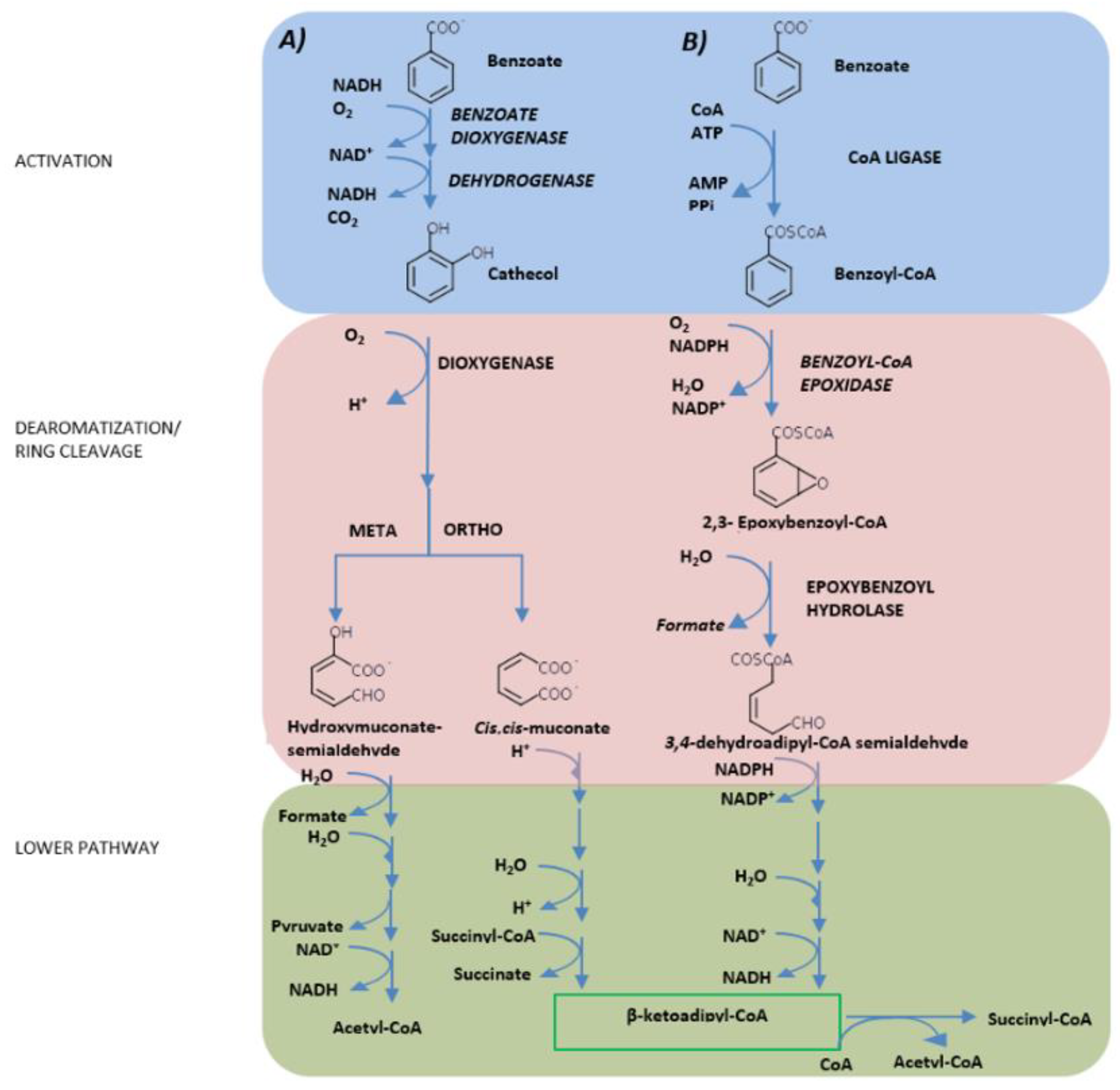
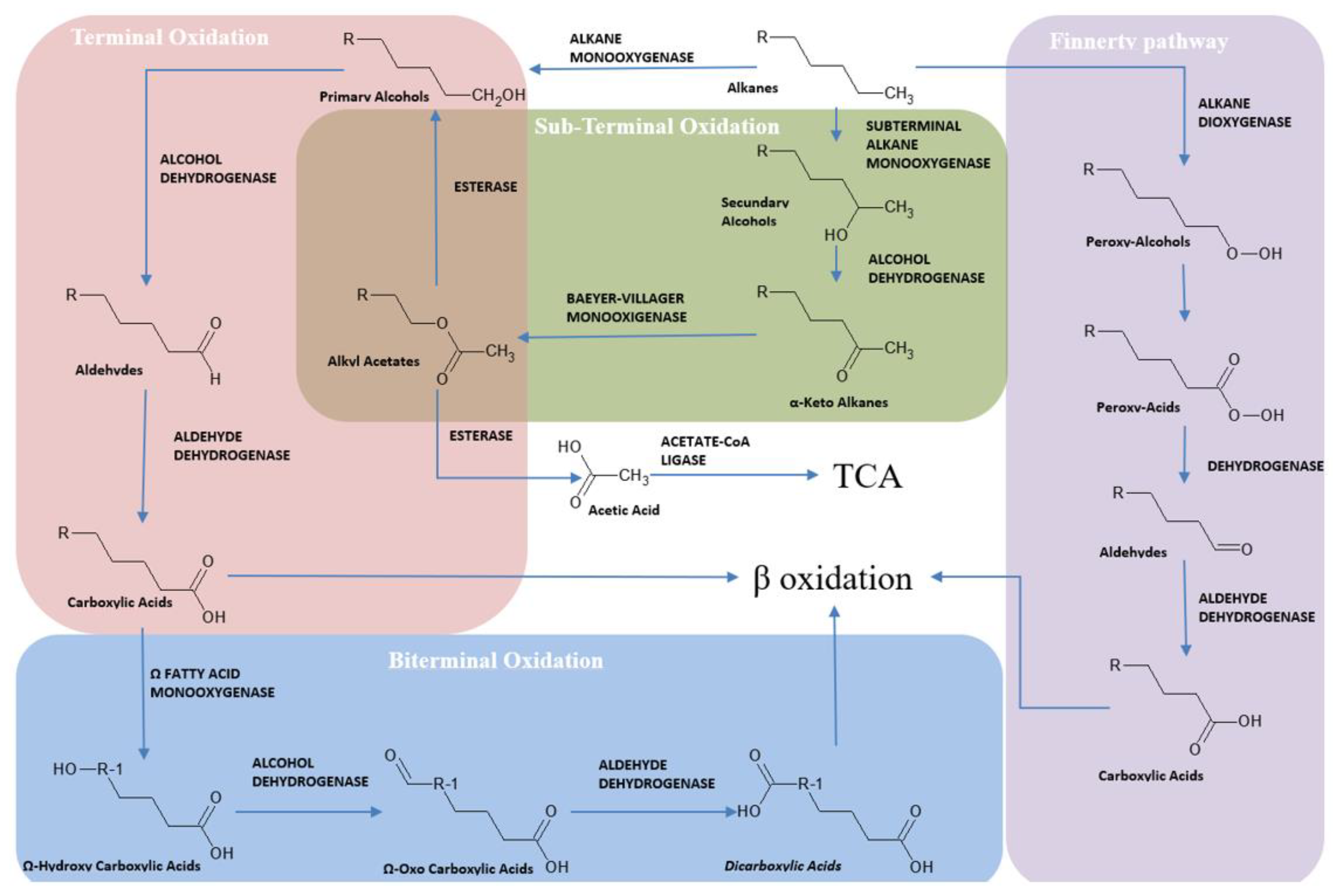
3. Hydrocarburoclastic Bacterial Pathways
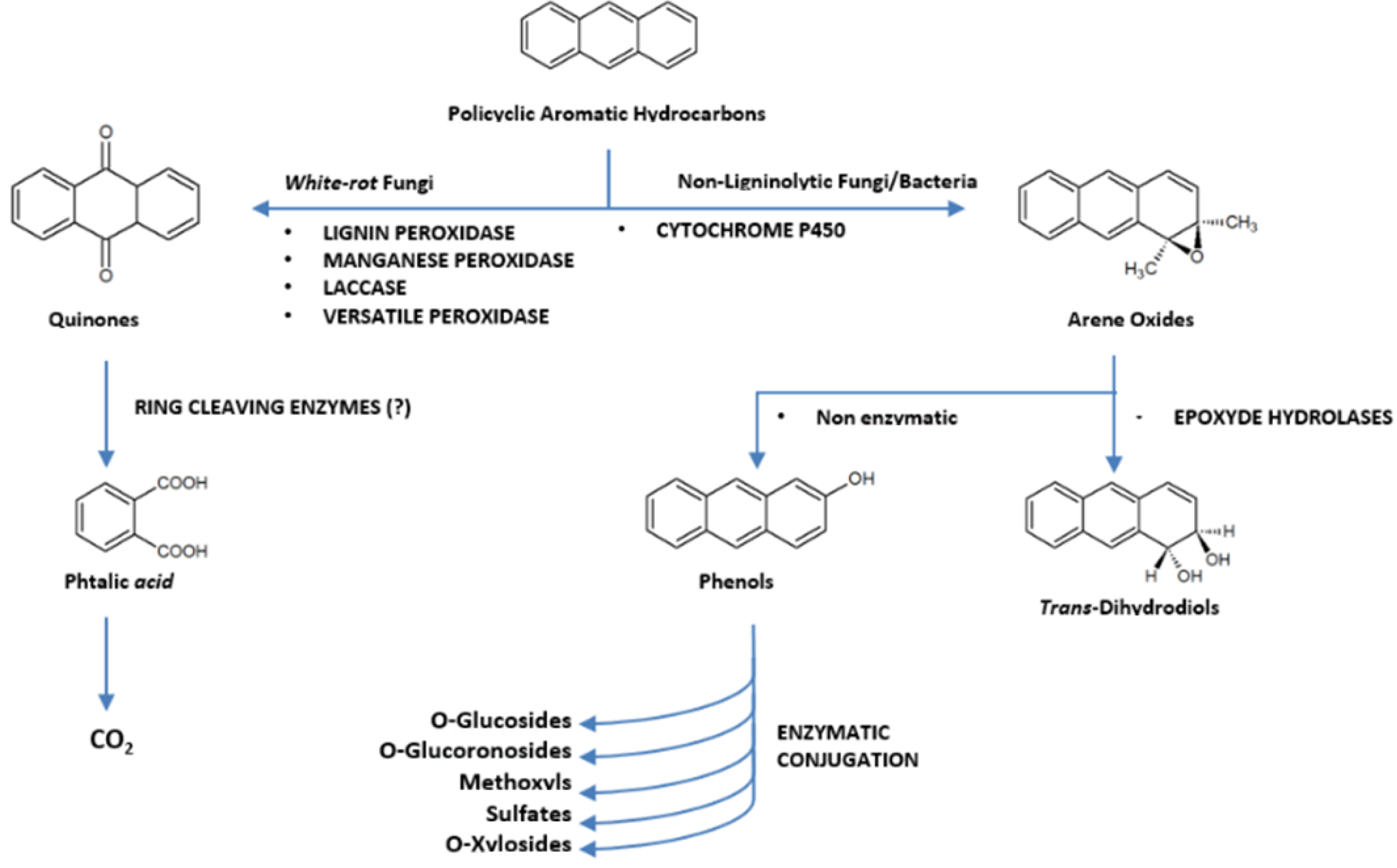
4. Fungal Pathways
5. Microbial Interactions
6. Plasticity of Microbial Metabolic Profiles
7. Taxonomical and Functional Metagenomics for Bioremediation
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Available online: www.ibisworld.com/global/market-size/global-oil-gas-exploration-production/ (accessed on 1 March 2022).
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A Comprehensive Review of Aliphatic Hydrocarbon Biodegradation by Bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef] [PubMed]
- Meckenstock, R.U.; Boll, M.; Mouttaki, H.; Koelschbach, J.S.; Cunha Tarouco, P.; Weyrauch, P.; Dong, X.; Himmelberg, A.M. Anaerobic Degradation of Benzene and Polycyclic Aromatic Hydrocarbons. Microb. Physiol. 2016, 26, 92–118. [Google Scholar] [CrossRef] [PubMed]
- Harayama, S.; Kasai, Y.; Hara, A. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 2004, 15, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Sharma, R.; Singh, K.; Sharma, A. Application of bioremediation technology in the environment contaminated with petroleum hydrocarbon. Ann. Microbiol. 2013, 63, 417–431. [Google Scholar] [CrossRef]
- Du, W.; Wan, Y.; Zhong, N.; Fei, J.; Zhang, Z.; Chen, L.; Hao, J. Status quo of soil petroleum contamination and evolution of bioremediation. Pet. Sci. 2011, 8, 502–514. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.I.; Biswas, B.; Smith, E.; Naidu, R.; Megharaj, M. Toxicity assessment of fresh and weathered petroleum hydrocarbons in contaminated soil- a review. Chemosphere 2018, 212, 755–767. [Google Scholar] [CrossRef]
- Ollivier, B.; Magot, M. Petroleum Microbiology; Wiley Online Library: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef] [Green Version]
- Hatzinger, P.B.; Alexander, M. Effect of Aging of Chemicals in Soil on Their Biodegradability and Extractability. Environ. Sci. Technol. 2002, 29, 537–545. [Google Scholar] [CrossRef]
- Piatt, J.J.; Brusseau, M.L. Rate-limited sorption of hydrophobic organic compounds by soils with well-characterized organic matter. Environ. Sci. Technol. 1998, 32, 1604. [Google Scholar] [CrossRef]
- Reid, B.J.; Jones, K.C.; Semple, K.T. Bioavailability of persistent organic pollutants in soils and sediments-a perspective on mechanisms 1608, consequences and assessment. Environ. Pollut. 2000, 108, 103–112. [Google Scholar] [CrossRef]
- Nam, K.; Alexander, M. Role of nonaporosity and hydrophobicity in sequestration and bioavailability: Tests with model solids. Environ. Sci. Technol. 1998, 32, 71–74. [Google Scholar] [CrossRef]
- Pignatello, J.J.; Xing, B. Mechanisms of Slow Sorption of Organic Chemicals to Natural Particles. Environ. Sci. Technol. 1995, 30, 1–11. [Google Scholar] [CrossRef]
- Ball, W.P.; Roberts, P.V. Long-Term Sorption of Halogenated Organic Chemicals by Aquifer Material. 1. Equilibrium. Environ. Sci. Technol. 1991, 25, 1223. [Google Scholar] [CrossRef]
- Ball, W.P.; Roberts, P.V. Long-Term Sorption of Halogenated Organic Chemicals by Aquifer Material. 2. Intraparticle Diffusion. Environ. Sci. Technol. 1991, 25, 1237. [Google Scholar] [CrossRef]
- Mader, B.T.; Uwe-Goss, K.; Eisenreich, S.J. Sorption of nonionic, hydrophobic organic chemicals to mineral surfaces. Environ. Sci. Technol. 1997, 31, 1079. [Google Scholar] [CrossRef]
- Trindade, P.V.O.; Sobral, L.G.; Rizzo, A.C.L.; Leite, S.G.F.; Soriano, A.U. Bioremediation of a weathered and a recently oil-contaminated soils from Brazil: A comparison study. Chemosphere 2005, 58, 515–522. [Google Scholar] [CrossRef]
- Maletić, S.; Dalmacija, B.; Rončević, S.; Agbaba, J.; Petrović, O. Degradation kinetics of an aged hydrocarbon-contaminated soil. Water Air Soil Pollut. 2009, 202, 149–159. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, H. Microbial Consortia Are Needed to Degrade Soil Pollutants. Microorganisms 2022, 24, 261. [Google Scholar] [CrossRef]
- Minna Laine, M.; Jørgensen, K.S. Straw compost and bioremediated soil as inocula for the bioremediation of chlorophenol-contaminated soil. Appl. Environ. Microbiol. 1996, 62, 1507. [Google Scholar] [CrossRef] [Green Version]
- Kästner, M.; Streibich, S.; Beyrer, M.; Richnow, H.H.; Fritsche, W. Formation of bound residues during microbial degradation of [14C]anthracene in soil. Appl. Environ. Microbiol. 1999, 65, 1834. [Google Scholar] [CrossRef] [Green Version]
- Boethling, R.S.; Alexander, M. Effect of Concentration of Organic Chemicals on Their Biodegradation by Natural Microbial Communities. Appl. Environ. Microbiol. 1979, 37, 1211–1216. [Google Scholar] [CrossRef] [Green Version]
- Fu, C.; Pfanstiel, S.; Gao, C.; Yan, X.; Govind, R.; Tabak, H.H. Studies on Contaminant Biodegradation in Slurry 1211, Wafer, and Compacted Soil Tube Reactors. Environ. Sci. Technol. 1996, 30, 743–750. [Google Scholar] [CrossRef]
- Park, J.H.; Zhao, X.; Voice, T.C. Biodegradation of Non-desorbable Naphthalene in Soils. Environ. Sci. Technol. 2001, 35, 2734. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.H.; Lee, M.W.; Park, J.M. Biodegradation of phenanthrene in soil-slurry systems with different mass transfer regimes and soil contents. J. Biotechnol. 2004, 110, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Voice, T.C. Assessment of bioavailability using a multicolumn system. Environ. Sci. Technol. 2000, 34, 1506. [Google Scholar] [CrossRef]
- Zhao, X.; Szafranski, M.J.; Maraqa, M.A.; Voice, T.C. Sorption and bioavailability of carbon tetrachloride in a low organic content sandy soil. Environ. Toxicol. Chem. 1999, 18, 1755. [Google Scholar] [CrossRef]
- Harms, H.; Zehnder, A.J.B. Bioavailability of sorbed 3-chlorodibenzofuran. Appl. Environ. Microbiol. 1762, 61, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Pignatello, J.J.; Martinson, M.M.; Steiert, J.G.; Carlson, R.E.; Crawford2, R.L. Biodegradation and Photolysis of Pentachlorophenol in Artificial Freshwater Streams. Appl. Environ. Microbiol. 1983, 46, 1024–1031. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds—From one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef]
- Fetzner, S. Ring-cleaving dioxygenases with a cupin fold. Appl. Environ. Microbiol. 2012, 78, 2505. [Google Scholar] [CrossRef] [Green Version]
- Steiner, R.A.; Janssen, H.J.; Roversi, P.; Oakley, A.J.; Fetzner, S. Structural basis for cofactor-independent dioxygenation of N-heteroaromatic compounds at the α/β-hydrolase fold. Proc. Natl. Acad. Sci. USA 2010, 107, 657. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Liu, X.; Yang, W.; Xu, F.; Wang, W.; Feng, L.; Bartlam, M.; Wang, L.; Rao, Z. Crystal Structure of Long-Chain Alkane Monooxygenase (LadA) in Complex with Coenzyme FMN: Unveiling the Long-Chain Alkane Hydroxylase. J. Mol. Biol. 2008, 376, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Watkinson, R.J.; Morgan, P. Physiology of aliphatic hydrocarbon-degrading microorganisms. Physiol. Biodegrad. Microorg. 1991, 1, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Kester, A.S.; Foster, J.W. Diterminal oxidation of long-chain alkanes by bacteria. J. Bacteriol. 1963, 85, 859–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coon, M.J. Omega Oxygenases: Nonheme-iron enzymes and P450 cytochromes. Biochem. Biophys. Res. Commun. 2005, 338, 378–385. [Google Scholar] [CrossRef]
- Forney, F.W.; Markovetz, A.J. Subterminal Oxidation of Aliphatic Hydrocarbons. J. Bacteriol. 1970, 102, 281. [Google Scholar] [CrossRef] [Green Version]
- Finnerty, W. Lipids of Acinetobacter. In Proceedings of the World Conference on Biotechnology for the Fats and Oil Industry; Applewhite, T.H., Ed.; Amer Oil Chemists Society: Urbana, IL, USA, 1988; pp. 184–188. [Google Scholar]
- Maeng, J.H.O.; Sakai, Y.; Tani, Y.; Kato, N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J. Bacteriol. 1996, 178, 3695. [Google Scholar] [CrossRef] [Green Version]
- Cameotra, S.S.; Makkar, R.S. Biosurfactant-enhanced bioremediation of hydrophobic pollutants. Pure Appl. Chem. 2010, 82, 97–116. [Google Scholar] [CrossRef]
- Costa, S.G.V.A.O.; Nitschke, M.; Lépine, F.; Déziel, E.; Contiero, J. Structure, properties and applications of rhamnolipids produced by Pseudomonas aeruginosa L2-1 from cassava wastewater. Process Biochem. 2010, 45, 1511. [Google Scholar] [CrossRef]
- Harkins, W.D.; Jordan, H.F. A method for the determination of surface and interfacial tension from the maximum pull on a ring. J. Am. Chem. Soc. 2002, 52, 1751. [Google Scholar] [CrossRef]
- Banat, I.M.; Satpute, S.K.; Cameotra, S.S.; Patil, R.; Nyayanit, N.V. Cost effective technologies and renewable substrates for biosurfactants’ production. Front. Microbiol. 1772, 5, 697. [Google Scholar] [CrossRef]
- Ganesh, A.; Lin, J. Diesel degradation and biosurfactant production by Gram-positive isolates. Afr. J. Biotechnol. 2011, 8, 5847. [Google Scholar] [CrossRef]
- Abbasian, F.; Lockington, R.; Megharaj, M.; Naidu, R. A Review on the Genetics of Aliphatic and Aromatic Hydrocarbon Degradation. Appl. Biochem. Biotechnol. 2016, 178, 224–250. [Google Scholar] [CrossRef] [PubMed]
- Becarelli, S.; Chicca, I.; la China, S.; Siracusa, G.; Bardi, A.; Gullo, M.; Petroni, G.; Levin, D.B.; Di Gregorio, S. A New Ciboria sp. for Soil Mycoremediation and the Bacterial Contribution to the Depletion of Total Petroleum Hydrocarbons. Front. Microbiol. 2021, 12, 647373. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Leon, V.; de Sisto Materano, A.; Ilzins, O.A. Enhancement of oil degradation by co-culture of hydrocarbon degrading and biosurfactant producing bacteria. Pol. J. Microbiol. 2006, 55, 139–146. [Google Scholar]
- Di Gregorio, S.; Becarelli, S.; Siracusa, G.; Ruffini Castiglione, M.; Petroni, G.; Masini, G.; Gentini, A.; de Lima e Silva, M.R.; Lorenzi, R. Pleurotus ostreatus spent mushroom substrate for the degradation of polycyclic aromatic hydrocarbons: The case study of a pilot dynamic biopile for the decontamination of a historically contaminated soil. J. Chem. Technol. Biotechnol. 2016, 91, 1654. [Google Scholar] [CrossRef]
- Chandankere, R.; Yao, J.; Cai, M.; Masakorala, K.; Jain, A.K.; Choi, M.M.F. Properties and characterization of biosurfactant in crude oil biodegradation by bacterium Bacillus methylotrophicus USTBa. Fuel 2014, 122, 140–148. [Google Scholar] [CrossRef]
- Worrich, A.; Stryhanyuk, H.; Musat, N.; König, S.; Banitz, T.; Centler, F.; Frank, K.; Thullner, M.; Harms, H.; Richnow, H.H.; et al. Mycelium-mediated transfer of water and nutrients stimulates bacterial activity in dry and oligotrophic environments. Nat. Commun. 2017, 8, 1. [Google Scholar] [CrossRef]
- González-Abradelo, D.; Pérez-Llano, Y.; Peidro-Guzmán, H.; del Sánchez-Carbente, M.R.; Folch-Mallol, J.L.; Aranda, E.; Vaidyanathan, V.K.; Cabana, H.; Gunde-Cimerman, N.; Batista-García, R.A. First demonstration that ascomycetous halophilic fungi (Aspergillus sydowii and Aspergillus destruens) are useful in xenobiotic mycoremediation under high salinity conditions. Bioresour. Technol. 2019, 279, 287–296. [Google Scholar] [CrossRef]
- Peidro-Guzmán, H.; Pérez-Llano, Y.; González-Abradelo, D.; Fernández-López, M.G.; Dávila-Ramos, S.; Aranda, E.; Hernández, D.R.O.; García, A.O.; Lira-Ruan, V.; Pliego, O.R.; et al. Transcriptomic analysis of polyaromatic hydrocarbon degradation by the halophilic fungus Aspergillus sydowii at hypersaline conditions. Environ. Microbiol. 2021, 23, 3435. [Google Scholar] [CrossRef]
- Aranda, E.; Godoy, P.; Reina, R.; Badia-Fabregat, M.; Rosell, M.; Marco-Urrea, E.; García-Romera, I. Isolation of Ascomycota fungi with capability to transform PAHs: Insights into the biodegradation mechanisms of Penicillium oxalicum. Int. Biodeterior. Biodegrad. 2017, 122, 141–150. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; de Hoog, G.S.; Summerbell, R.C. Fungal Communities in Hydrocarbon Degradation. In Microbial Communities Utilizing Hydrocarbons and Lipids: Members, Metagenomics and Ecophysiology, Handbook of Hydrocarbon and Lipid Microbiology; McGenity, T., Ed.; Springer: Cham, Switzerland, 2019; pp. 1–36. [Google Scholar] [CrossRef]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Marco-Urrea, E.; García-Romera, I.; Aranda, E. Potential of non-ligninolytic fungi in bioremediation of chlorinated and polycyclic aromatic hydrocarbons. New Biotechnol. 2015, 32, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P. Increase of laccase activity during interspecific interactions of white-rot fungi. FEMS Microbiol. Ecol. 2004, 50, 245–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, K.R.; Chand, S.; Gostomski, P.A.; Boyd-Wilson, K.S.H.; Ford, C.; Walter, M. Fungal Inoculum Properties and Its Effect on Growth and Enzyme Activity of Trametes versicolor in Soil. Biotechnol. Prog. 2005, 21, 377–385. [Google Scholar] [CrossRef]
- Wong, D.W.S. Structure and action mechanism of ligninolytic enzymes. Appl. Biochem. Biotechnol. 2009, 157, 174–209. [Google Scholar] [CrossRef]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef] [Green Version]
- Aranda, E. Promising approaches towards biotransformation of polycyclic aromatic hydrocarbons with Ascomycota fungi. Curr. Opin. Biotechnol. 2016, 38, 1–8. [Google Scholar] [CrossRef]
- Covino, S.; D’Annibale, A.; Stazi, S.R.; Cajthaml, T.; Čvančarová, M.; Stella, T.; Petruccioli, M. Assessment of degradation potential of aliphatic hydrocarbons by autochthonous filamentous fungi from a historically polluted clay soil. Sci. Total Environ. 2015, 505, 545–554. [Google Scholar] [CrossRef]
- Becarelli, S.; Chicca, I.; Siracusa, G.; la China, S.; Gentini, A.; Lorenzi, R.; Munz, G.; Petroni, G.; Levin, D.B.; Di Gregorio, S. Hydrocarbonoclastic Ascomycetes to enhance co-composting of total petroleum hydrocarbon (TPH) contaminated dredged sediments and lignocellulosic matrices. New Biotechnol. 2019, 50, 27–36. [Google Scholar] [CrossRef]
- Siracusa, G.; Yuan, Q.; Chicca, I.; Bardi, A.; Spennati, F.; Becarelli, S.; Levin, D.B.; Munz, G.; Petroni, G.; Di Gregorio, S. Mycoremediation of Old and Intermediate Landfill Leachates with an Ascomycete Fungal Isolate, Lambertella sp. Water 2020, 12, 800. [Google Scholar] [CrossRef] [Green Version]
- Zavarzina, A.G.; Lisov, A.A.; Zavarzin, A.A.; Leontievsky, A.A. Fungal Oxidoreductases and Humification in Forest Soils. In Soil Enzymology. Soil Biology; Shukla, G., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 22, pp. 207–228. [Google Scholar] [CrossRef]
- Loss, E.M.O.; Lee, M.K.; Wu, M.Y.; Martien, J.; Chen, W.; Amador-Noguez, D.; Jefcoate, C.; Remucal, C.; Jung, S.; Kim, S.C.; et al. Cytochrome P450 monooxygenase-mediated metabolic utilization of benzo[a]pyrene by aspergillus species. mBio 2019, 10, e00558-19. [Google Scholar] [CrossRef]
- Verdin, A.; Lounès-Hadj Sahraoui, A.; Newsam, R.; Robinson, G.; Durand, R. Polycyclic aromatic hydrocarbons storage by Fusarium solani in intracellular lipid vesicles. Environ. Pollut. 1987, 133, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Fayeulle, A.; Veignie, E.; Slomianny, C.; Dewailly, E.; Munch, J.C.; Rafin, C. Energy-dependent uptake of benzo[a]pyrene and its cytoskeleton-dependent intracellular transport by the telluric fungus Fusarium solani. Environ. Sci. Pollut. Res. Int. 2014, 21, 3515. [Google Scholar] [CrossRef]
- Meulenberg, R.; Rijnaarts, H.H.M.; Doddema, H.J.; Field, J.A. Partially oxidized polycyclic aromatic hydrocarbons show an increased bioavailability and biodegradability. FEMS Microbiol. Lett. 1997, 152, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Capotorti, G.; Cesti, P.; Lombardi, A.; Guglielmetti, G. Formation of sulfate conjugates metabolites in the degradation of phenanthrene, anthracene, pyrene and benzo[a]pyrene by the ascomycete Aspergillus terreus. Polycycl. Aromat. Compd. 2005, 25, 197–213. [Google Scholar] [CrossRef]
- Chen, W.; Lee, M.K.; Jefcoate, C.; Kim, S.C.; Chen, F.; Yu, J.H. Fungal cytochrome p450 monooxygenases: Their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol. Evol. 2014, 6, 1620. [Google Scholar] [CrossRef] [Green Version]
- Sepic, E.; Leskovsek, H.; Trier, C. Aerobic bacterial degradation of selected polyaromatic compounds and n-alkanes found in petroleum. J. Chromatogr. 1995, 697, 515–523. [Google Scholar] [CrossRef]
- Koma, D.; Hasumi, F.; Yamamoto, E.; Ohta, T.; Chung, S.Y.; Kubo, M. Biodegradation of long-chain n-paraffins from waste oil of car engine by Acinetobacter sp. J. Biosci. Bioeng. 2001, 91, 94–96. [Google Scholar] [CrossRef]
- van Hamme, J.D.; Singh, A.; Ward, O.P. Recent Advances in Petroleum Microbiology. Microbiol. Mol. Biol. Rev. 2003, 67, 503–549. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.; Li, H.; Bao, M.; Sun, P. Metabolic pathway for a new strain Pseudomonas synxantha LSH-7′: From chemotaxis to uptake of n-hexadecane. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Li, X.W.; Liu, Z.P. Microbial biodegradation of petroleum hydrocarbon. Acta Microbiol. Siica. 2002, 42, 764–767. [Google Scholar]
- Das, N.; Chandran, P. Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview. Biotechnol. Res. Int. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warmink, J.A.; Nazir, R.; van Elsas, J.D. Universal and species-specific bacterial “fungiphiles” in the mycospheres of different basidiomycetous fungi. Environ. Microbiol. 2009, 11, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, S.; Henault, C.; Aamor, A.; Bdioui, N.; Bloor, J.M.G.; Maire, V.; Mary, B.; Revaillot, S.; Maron, P.A. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol. Biochem. 2011, 43, 86–96. [Google Scholar] [CrossRef]
- Blagodatskaya, E.V.; Blagodatsky, S.A.; Anderson, T.H.; Kuzyakov, Y. Priming effects in Chernozem induced by glucose and N in relation to microbial growth strategies. Appl. Soil Ecol. 2007, 1, 95–105. [Google Scholar] [CrossRef]
- Milcu, A.; Heim, A.; Ellis, R.J.; Scheu, S.; Manning, P. Identification of General Patterns of Nutrient and Labile Carbon Control on Soil Carbon Dynamics Across a Successional Gradient. Ecosystems 2011, 14, 710–719. [Google Scholar] [CrossRef]
- Purahong, W.; Wubet, T.; Lentendu, G.; Schloter, M.; Pecyna, M.J.; Kapturska, D.; Hofrichter, M.; Krüger, D.; Buscot, F. Life in leaf litter: Novel insights into community dynamics of bacteria and fungi during litter decomposition. Mol. Ecol. 2016, 25, 4059. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mediators of mutualistic microbe–microbe interactions. Nat. Prod. Rep. 2018, 35, 303–308. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.; Brookes, P.C.; Xu, J.; Gilbert, J.A. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016, 10, 1891. [Google Scholar] [CrossRef]
- Stopnisek, N.; Zuhlke, D.; Carlier, A.; Barberan, A.; Fierer, N.; Becher, D.; Riedel, K.; Eberl, L.; Weisskopf, L. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 2015, 10, 253–264. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Senbayram, M.; Blagodatsky, S.; Myachina, O.; Dittert, K.; Lin, X.; Blagodatskaya, E.; Kuzyakov, Y. Soil C and N availability determine the priming effect: Microbial N mining and stoichiometric decomposition theories. Glob. Chang. Biol. 2014, 20, 2356. [Google Scholar] [CrossRef] [PubMed]
- Ech Tláskal, V.; Brabcová, V.; Etrovský Tv Jomura, M.; López-Mondéjar, R.; Oliveira Monteiro, L.M.; Saraiva, J.P.; Human, Z.R.; Cajthaml, T.; Nunes Da Rocha, U.; Baldrian, P. Complementary Roles of Wood-Inhabiting Fungi and Bacteria Facilitate Deadwood Decomposition. MSystems 2021, 6, e01078-20. [Google Scholar] [CrossRef] [PubMed]
- Banitz, T.; Fetzer, I.; Johst, K.; Wick, L.Y.; Harms, H.; Frank, K. Assessing biodegradation benefits from dispersal networks. Ecol. Model. 2011, 222, 2552. [Google Scholar] [CrossRef]
- Tecon, R.; Or, D. Bacterial flagellar motility on hydrated rough surfaces controlled by aqueous film thickness and connectedness. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Worrich, A.; König, S.; Miltner, A.; Banitz, T.; Centler, F.; Frank, K.; Thullner, M.; Harms, H.; Kästner, M.; Wick, L.Y. Mycelium-like networks increase bacterial dispersal, growth, and biodegradation in a model ecosystem at various water potentials. Appl. Environ. Microbiol. 2016, 82, 2902. [Google Scholar] [CrossRef] [Green Version]
- Furuno, S.; Foss, S.; Wild, E.; Jones, K.C.; Semple, K.T.; Harms, H.; Wick, L.Y. Mycelia promote active transport and spatial dispersion of polycyclic aromatic hydrocarbons. Environ. Sci. Technol. 2012, 46, 5463. [Google Scholar] [CrossRef]
- Schamfuß, S.; Neu, T.R.; van der Meer, J.R.; Tecon, R.; Harms, H.; Wick, L.Y. Impact of mycelia on the accessibility of fluorene to PAH-degrading bacteria. Environ. Sci. Technol. 2013, 47, 6908. [Google Scholar] [CrossRef]
- Furuno, S.; Päzolt, K.; Rabe, K.; Neu, T.R.; Harms, H.; Wick, L.Y. Fungal mycelia allow chemotactic dispersal of polycyclic aromatic hydrocarbon-degrading bacteria in water-unsaturated systems. Environ. Microbiol. 2010, 12, 1391. [Google Scholar] [CrossRef]
- Rudnick, M.B.; van Veen, J.A.; de Boer, W. Oxalic acid: A signal molecule for fungus-feeding bacteria of the genus Collimonas? Environ. Microbiol. Rep. 2015, 7, 709–714. [Google Scholar] [CrossRef]
- Ul Haq, I.; Oliveira da Rocha Calixto, R.; Yang, P.; Maria Pires dos Santos, G.; Barreto-Bergter, E.; Dirk van Elsas, J. Chemotaxis and adherence to fungal surfaces are key components of the behavioral response of Burkholderia terrae BS001 to two selected soil fungi. FEMS Microbiol. Ecol. 2016, 92, fiw164. [Google Scholar] [CrossRef] [Green Version]
- Mangwani, N.; Dash, H.R.; Chauhan, A.; Das, S. Bacterial quorum sensing: Functional features and potential applications in biotechnology. J. Mol. Microbiol. Biotechnol. 2012, 22, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.C.; Zhong, J.J. Regulation of aromatics biodegradation by rhl quorum sensing system through induction of catechol meta-cleavage pathway. Bioresour. Technol. 2013, 136, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Chicca, I.; Becarelli, S.; Dartiahl, C.; la China, S.; de Kievit, T.; Petroni, G.; Di Gregorio, S.; Levin, D.B. Degradation of BTEX mixture by a new Pseudomonas putida strain: Role of the quorum sensing in the modulation of the upper BTEX oxidative pathway. Environ. Sci. Pollut. Res. Int. 2020, 27, 36203–36214. [Google Scholar] [CrossRef] [PubMed]
- Wongsuk, T.; Pumeesat, P.; Luplertlop, N. Fungal quorum sensing molecules: Role in fungal morphogenesis and pathogenicity. J. Basic Microbiol. 2016, 56, 440–447. [Google Scholar] [CrossRef]
- Stanley, C.E.; Stöckli, M.; van Swaay, D.; Sabotič, J.; Kallio, P.T.; Künzler, M.; Demello, A.J.; Aebi, M. Probing bacterial–fungal interactions at the single cell level. Integr. Biol. 2014, 6, 935–945. [Google Scholar] [CrossRef] [Green Version]
- Sztajer, H.; Szafranski, S.P.; Tomasch, J.; Reck, M.; Nimtz, M.; Rohde, M.; Wagner-Döbler, I. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014, 8, 2256. [Google Scholar] [CrossRef] [Green Version]
- Dixon, E.F.; Hall, R.A. Noisy neighbourhoods: Quorum sensing in fungal–polymicrobial infections. Cell. Microbiol. 2015, 17, 1431. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, A.K.; Hogan, D.A. Candida albicans: Molecular interactions with Pseudomonas aeruginosa and Staphylococcus aureus. Fungal Biol. Rev. 2014, 28, 85–96. [Google Scholar] [CrossRef]
- Medaura, M.C.; Guivernau, M.; Moreno-Ventas, X.; Prenafeta-Boldú, F.X.; Viñas, M. Bioaugmentation of Native Fungi, an Efficient Strategy for the Bioremediation of an Aged Industrially Polluted Soil With Heavy Hydrocarbons. Front. Microbiol. 2021, 12, 713. [Google Scholar] [CrossRef]
- Chen, C.; Li, T. Bacterial dye-decolorizing peroxidases: Biochemical properties and biotechnological opportunities. Phys. Sci. Rev. 2016, 1. [Google Scholar] [CrossRef]
- Chen, C.; Shrestha, R.; Jia, K.; Gao, P.F.; Geisbrecht, B.V.; Bossmann, S.H.; Shi, J.; Li, P. Characterization of Dye-decolorizing Peroxidase (DyP) from Thermomonospora curvata reveals unique catalytic properties of A-type DyPs. J. Biol. Chem. 2015, 290, 23447–23463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, K.; Gong, G.; Woo, H.M.; Kim, Y.; Um, Y. A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and β-ether lignin dimer. Sci. Rep. 2015, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bosma, T.N.P.; Middeldorp, P.J.M.; Schraa, G.; Zehnder, A.J.B. Mass Transfer Limitation of Biotransformation: Quantifying Bioavailability. Environ. Sci. Technol. 1996, 31, 248–252. [Google Scholar] [CrossRef]
- Semple, K.T.; Reid, B.J.; Fermor, T.R. Impact of composting strategies on the treatment of soils contaminated with organic pollutants. Environ. Pollut. 2001, 112, 269–283. [Google Scholar] [CrossRef]
- Tran, H.T.; Lin, C.; Bui, X.T.; Ngo, H.H.; Cheruiyot, N.K.; Hoang, H.G.; Vu, C.T. Aerobic composting remediation of petroleum hydrocarbon-contaminated soil. Current and future perspectives. Sci. Total Environ. 2021, 753, 14. [Google Scholar] [CrossRef]
- Huang, Y.; Pan, H.; Wang, Q.; Ge, Y.; Liu, W.; Christie, P. Enrichment of the soil microbial community in the bioremediation of a petroleum-contaminated soil amended with rice straw or sawdust. Chemosphere 2019, 224, 265–271. [Google Scholar] [CrossRef]
- Hawkes, C.v.; Kivlin, S.N.; Rocca, J.D.; Huguet, V.; Thomsen, M.A.; Suttle, K.B. Fungal community responses to precipitation. Glob. Change Biol. 2011, 17, 1637. [Google Scholar] [CrossRef]
- Jiao, S.; Chen, W.; Wei, G. Resilience and Assemblage of Soil Microbiome in Response to Chemical Contamination Combined with Plant Growth. Appl. Environ. Microbiol. 2019, 85, e02523-18. [Google Scholar] [CrossRef] [Green Version]
- Lǎzǎroaie, M.M. Multiple Responses of Gram-Positive and Gram-Negative Bacteria to Mixture of Hydrocarbons. Braz. J. Microbiol. 2010, 41, 649. [Google Scholar] [CrossRef]
- Chicca, I.; Becarelli, S.; Bernabei, G.; Siracusa, G.; Di Gregorio, S. Innovative Culturomic Approaches and Predictive Functional Metagenomic Analysis: The Isolation of Hydrocarbonoclastic Bacteria with Plant Growth Promoting Capacity. Water 2022, 14, 142. [Google Scholar] [CrossRef]
- Ofaim, S.; Ofek-Lalzar, M.; Sela, N.; Jinag, J.; Kashi, Y.; Minz, D.; Freilich, S. Analysis of microbial functions in the rhizosphere using a metabolic-network based framework for metagenomics interpretation. Front. Microbiol. 2017, 8, 1606. [Google Scholar] [CrossRef] [PubMed]
- Chemerys, A.; Pelletier, E.; Cruaud, C.; Martin, F.; Violet, F.; Jouanneau, Y. Characterization of Novel Polycyclic Aromatic Hydrocarbon Dioxygenases from the Bacterial Metagenomic DNA of a Contaminated Soil. Appl. Environ. Microbiol. 2014, 80, 6591–6600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Röling, W.F.M. Maths on microbes: Adding microbial ecophysiology to metagenomics. Microb. Biotechnol. 2015, 8, 21–22. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Castrejón-Godínez, M.L.; Salazar-Bustamante, E.; Gama-Martínez, Y.; Sánchez-Salinas, E.; Mussali-Galante, P.; Tovar-Sánchez, E.; Ortiz-Hernández, M.L. Omics Approaches to Pesticide Biodegradation. Curr. Microbiol. 2020, 77, 545–563. [Google Scholar] [CrossRef]
- Plewniak, F.; Crognale, S.; Rossetti, S.; Bertin, P.N. A genomic outlook on bioremediation: The case of arsenic removal. Front. Microbiol. 2018, 9, 820. [Google Scholar] [CrossRef] [Green Version]
- Bilal, T.; Malik, B.; Hakeem, K.R. Metagenomic analysis of uncultured microorganisms and their enzymatic attributes. J. Microbiol. Methods 2018, 155, 65–69. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Biniecka, P.; Bondarczuk, K.; Piotrowska-Seget, Z. Metagenomic Functional Profiling Reveals Differences in Bacterial Composition and Function During Bioaugmentation of Aged Petroleum-Contaminated Soil. Front. Microbiol. 2020, 11, 2106. [Google Scholar] [CrossRef]
- Yergeau, E.; Sanschagrin, S.; Beaumier, D.; Greer, C.W. Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high arctic soils. PLoS ONE 2012, 7, e30058. [Google Scholar] [CrossRef]
- Jacquiod, S.; Demanèche, S.; Franqueville, L.; Ausec, L.; Xu, Z.; Delmont, T.O.; Dunon, V.; Cagnon, C.; Mandic-Mulec, I.; Vogel, T.M.; et al. Characterization of new bacterial catabolic genes and mobile genetic elements by high throughput genetic screening of a soil metagenomic library. J. Biotechnol. 2014, 190, 18–29. [Google Scholar] [CrossRef]
- El Hadidi, M.; Ruscheweyh, H.J.; Huson, D. Improved metagenome analysis using MEGAN5. In Proceedings of the Joint 21st Annual International Conference on Intelligent Systems for Molecular Biology (ISMB) and 12th European Conference on Computational Biology (ECCB), Berlin, Germany, 21–23 July 2013. [Google Scholar]
- Seshadri, R.; Kravitz, S.A.; Smarr, L.; Gilna, P.; Frazier, M. CAMERA: A Community Resource for Metagenomics. PLoS Biol. 2007, 5, e75. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Chen, J.; Li, W.; Altintas, I.; Lin, A.; Peltier, S.; Stocks, K.; Allen, E.E.; Ellisman, M.; Grethe, J.; et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: The CAMERA resource. Nucleic Acids Res. 2011, 39, D546–D551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Chettri, B.; Langpoklakpam, J.S.; Basak, P.; Prasad, A.; Mukherjee, A.K.; Bhattacharyya, M.; Singh, A.K.; Chattopadhyay, D. Bioinformatic Approaches Including Predictive Metagenomic Profiling Reveal Characteristics of Bacterial Response to Petroleum Hydrocarbon Contamination in Diverse Environments. Sci. Rep. 2017, 7, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, P.; Ning, Z.; Ning, Z.; Liu, Y.; He, Z.; He, Z.; Shi, J.; Niu, M. Diagnosing bioremediation of crude oil-contaminated soil and related geochemical processes at the field scale through microbial community and functional genes. Ann. Microbiol. 2020, 70, 1–15. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chicca, I.; Becarelli, S.; Di Gregorio, S. Microbial Involvement in the Bioremediation of Total Petroleum Hydrocarbon Polluted Soils: Challenges and Perspectives. Environments 2022, 9, 52. https://doi.org/10.3390/environments9040052
Chicca I, Becarelli S, Di Gregorio S. Microbial Involvement in the Bioremediation of Total Petroleum Hydrocarbon Polluted Soils: Challenges and Perspectives. Environments. 2022; 9(4):52. https://doi.org/10.3390/environments9040052
Chicago/Turabian StyleChicca, Ilaria, Simone Becarelli, and Simona Di Gregorio. 2022. "Microbial Involvement in the Bioremediation of Total Petroleum Hydrocarbon Polluted Soils: Challenges and Perspectives" Environments 9, no. 4: 52. https://doi.org/10.3390/environments9040052
APA StyleChicca, I., Becarelli, S., & Di Gregorio, S. (2022). Microbial Involvement in the Bioremediation of Total Petroleum Hydrocarbon Polluted Soils: Challenges and Perspectives. Environments, 9(4), 52. https://doi.org/10.3390/environments9040052






