Abstract
Phytoplankton community composition has been utilized for water quality assessments of various freshwater sources, but studies are lacking on agricultural irrigation ponds. This work evaluated the performance of the random forest algorithm in estimating phytoplankton community structure from in situ water quality measurements at two agricultural ponds. Sampling was performed between 2017 and 2019 and measurements of three phytoplankton groups (green algae, diatoms, and cyanobacteria) and three sets of water quality parameters (physicochemical, organic constituents, and nutrients) were obtained to train and test mathematical models. Models predicting green algae populations had superior performance to the diatom and cyanobacteria models. Spatial models revealed that water in the ponds’ interior sections had lower root mean square errors (RMSEs) compared to nearshore waters. Furthermore, model performance did not change when input datasets were compounded. Models based on physicochemical parameters, which can be obtained in real time, outperformed models based on organic constituent and nutrient parameters. However, the use of nutrient parameters improved model performance when examining cyanobacteria data at the ordinal level. Overall, the random forest algorithm was useful for predicting major phytoplankton taxonomic groups in agricultural irrigation ponds, and this may help resource managers mitigate the use of cyanobacteria bloom-laden waters in agricultural applications.
1. Introduction
Phytoplankton community composition and abundance is often used in assessments of recreational, aquaculture, and drinking water quality. Long-term monitoring studies conducted in marine and estuarine waters used for aquaculture activities [1,2] and in freshwater lakes and reservoirs used to provide drinking water and recreational areas [3,4,5] have demonstrated distinctive relationships between certain phytoplankton community constituents and water temperature, salinity, and nutrient concentrations. However, long-term phytoplankton community composition studies in small-bodied agricultural irrigation waters to examine similar relationships are lacking.
The examination of water for phytoplankton community composition and abundance is a time-consuming activity that relies on the expertise of well-trained phytoplankton taxonomists or automated technologies, such as flow cytometry, that may be cost-prohibitive to many water quality management programs [6,7,8]. Satellite imagery has proven useful for monitoring phytoplankton community structure in large lakes (>24,000 acres, [9]) but does not yet have the spatial scale needed to remotely observe smaller bodies of water that are increasingly being used in agricultural irrigation applications [10]. Hence, alternative techniques are being explored to examine the relationships between more easily measured water quality parameters (i.e., temperature, chlorophyll-a, and specific conductance) and phytoplankton community composition and abundance. The presence of such relationships makes the use of regression analysis feasible for predicting phytoplankton community structure and concentrations using measured water quality parameters. Regression analyses have been used to predict the occurrence of bloom-forming cyanobacteria in shallow lakes [11,12], green algae in reservoirs [13], and diatoms in estuaries, rivers, and lakes [14,15], as well as to evaluate overall irrigation water quality [16,17], but these two parameters have not been examined as distinct input variables within the same mathematical model.
Regression analyses were used to successfully predict the composition of phytoplankton communities in a drinking water reservoir near Beijing, China, that had an area of greater than 44,000 acres [18]. However, as noted by Cheruvelil et al., [19], scale and regionalization are important factors to consider when conducting water quality assessments and applying water quality standards. Models such as those reported by Zeng et al. [18] are only beginning to be constructed for the freshwater reaches of the Chesapeake Bay watershed [20,21], yet even these efforts do not reflect water specifically designated for agricultural uses. Recently, machine learning provided several versatile techniques to establish models suitable to create ‘phytoplankton–water quality’ relationships. The machine learning algorithm of random forests was specifically chosen for its ability to elucidate nonlinear relationships between input variables and because of its built-in mechanism to limit potential overfitting of the model. The objective of this work was to evaluate the performance of the random forest algorithm in estimating the phytoplankton community structure from in situ water quality measurements of different complexities obtained during three years of spatially intensive observations at two 1-acre agricultural irrigation ponds.
Phytoplankton community structure has long been used to assess trophic changes in aquatic systems [22] with shifts from green algae-dominated communities to cyanobacteria-dominated communities indicating eutrophic conditions [23,24]. Equally as important is the influence various phytoplankton groups can have on water chemistry [25,26], especially carbon cycling [27]. For this study three phytoplankton groups were considered critical to assess in relationship to water quality parameters due to their abundance within local freshwater phytoplankton populations. Previous studies by Parson and Parker [28] and Marshall [29,30] demonstrated that between 70–80% of regional freshwater lake phytoplankton community structure was composed of green algae (Chlorophytes), diatoms (Bacillariophytes), and cyanobacteria (Cyanophytes). Due to the harmful and potentially toxic effects of cyanobacteria blooms on human and environmental health, the detection, prediction, and modeling of these blooms has become a focus for resource managers [31,32,33]. Additionally, there is growing concern about the risk that cyanotoxins may pose to the agriculture industry through the transfer of cyanotoxins from irrigation waters to crops and livestock, particularly as climate change increases the occurrence and toxicity of cyanobacteria blooms [34,35,36]. Other concerns include both toxic and non-toxic cyanobacteria blooms altering carbon cycling, alkalizing waters, and increasing turbidity, thus further degrading water quality [37,38]. Thus, additional analyses with the random forest algorithm were conducted to determine if there were correlations between water quality parameters and the cyanobacteria orders Chroococcales and Nostocales, as these orders contain many pelagic, toxigenic species that are of particular concern in surface waters [39].
2. Materials and Methods
2.1. Experimental Design and Sample Collection
Phytoplankton and water quality sampling was conducted every two weeks at two 1-acre ponds on working farms in Maryland during the 2017 and 2018 growing seasons (May–October). Pond 1 (Figure 1-P1) located in Germantown, Maryland is a man-made embankment pond with in-flow from a co-located pond; 23 stations were routinely sampled in this pond. Pond 2 (Figure 1-P2) located in Wye Mills, Maryland (University of Maryland Wye Research Center) is an excavated pond with in-flow from an ephemeral creek; 34 stations were routinely sampled in this pond. Phytoplankton samples and water quality measurements were made at all stations in Pond 1. Phytoplankton samples at Pond 2 were collected at fewer locations, consisting of odd-numbered nearshore locations and all interior sampling locations (22 stations), whereas water quality measurements were made at all stations. Full site descriptions are provided in Smith et al. [40]. In situ measurements were taken along with a water sample for laboratory processing at each sampling location. A YSI Exo-2 sonde (Yellow Springs Instruments, Yellow Spring, OH, USA) was used to measure temperature (TEMP), dissolved oxygen (DO), specific conductance (SPC), pH, fluorescent dissolved organic matter (FDOM), and turbidity (NTU). As a proxy for phytoplankton density, both chlorophyll-a (CHL) and phycocyanin (Phyco) were measured with the YSI Exo-2 sonde as demonstrated by Brient et al., [41] and Song et al., [42]. Water samples were measured for colored dissolved organic matter (CDOM) using a Turner Designs AquaFluor fluorometer (Turner Designs, San Jose, CA, USA). Identification and enumeration of phytoplankton was performed using a modified Utermöhl method as described in Marshall and Alden [43], with taxa identified according to Komárek [39], John et al. [44] and Bellinger and Sigee [45]. For full details of sampling methodologies see Smith et al. [46]. Water quality sampling methods and phytoplankton analyses for the 2019 sampling year were the same as those performed in 2017 and 2018 but occurred on a less routine schedule. In Pond 1 there were six sampling dates in 2017, six sampling dates in 2018, and three sampling dates in 2019. In Pond 2 there were five sampling dates in 2017, six sampling dates in 2018, and two sampling dates in 2019. Sampling dates are presented in Supplementary Table S1.

Figure 1.
Sampling locations for both Pond 1 (P1) and Pond 2 (P2). Sampling location number is shown inside the circle. Yellow circles indicate interior water sampling locations and orange circles indicate nearshore sampling locations.
Field work conducted in 2017 and 2018 yielded 518 phytoplankton samples, in situ measurements, and laboratory-based water quality measurements (Supplementary Table S1). For the purpose of the random forest analysis, phytoplankton data was examined at the taxonomic group level (diatoms, green algae, and cyanobacteria). While other taxa (i.e., dinoflagellates) were observed with microscopy analyses, the low spatial and temporal occurrence and abundance of these taxa over the course of the study precluded examination with the random forest analysis. The data collected in 2019 were used as a blind dataset to test the random forest model.
2.2. Modelling with the Random Forest Algorithm
The machine learning random forest algorithm designed by Liaw and Wiener [47] was used to predict phytoplankton functional group concentrations and the most influential parameters for each group. The default number of trees run for each model was 500. The input data was split into training and testing datasets with the default value of 70% training and 30% testing. The result is more accurate outputs which are better suited for prediction models. To alleviate the potential overfitting of the random forest models, the ‘mtry’ parameter was used, which limits the number of variables sampled at each split of a tree. The default ‘mtry’ value was selected which is calculated as where P is the total number of input variables.
Random forest models with various inputs and outputs were developed in this study. Three input datasets (A, B, and C, Table 1) were used for each of the three output datasets of phytoplankton groups (diatoms, green algae, and cyanobacteria). When considering model performance on a finer taxonomic scale, the cyanobacteria orders Nostocales and Chroococcales were used since cyanobacteria blooms during the study period comprised organisms within these orders. The input set A included physicochemical parameters, i.e., TEMP, pH, DO, NTU, and SPC. In 2018, photosynthetic active radiation (PAR) was added to input set A. The input set B included parameters related to organic constituents, i.e., CHL, Phyco, and FDOM. In 2018, CDOM was added to input set B. Input set C included nutrients and macro elements, i.e., potassium, calcium, magnesium, ammonium, nitrate, and phosphate, which were only measured in 2018. For 2017 and 2017 + 2018 data sets, the random forest model was developed with input set A, input set B, and combined input sets A and B (AB). For the 2018 dataset, random forest models were developed with input set A, input set B, input set C, and combinations of these input sets (AB, AC, BC, and ABC). Models were run individually for 2017 and 2018 to allow for the inclusion of additional measurements added in 2018. These measurements were excluded when running multiyear models to keep datasets balanced. All random forest computations were completed in Rstudio (Rstudio Team, Boston, MA, USA) using the ‘randomForest’ package.

Table 1.
Constituents of three input sets used for the random forest analysis.
2.3. Performance Metrics
Pearson correlations were used to assess the strength of linear relationships between measured water quality parameters. All Pearson correlations were computed in Rstudio. Moderate correlations were defined as correlations with an r value between 0.5 and 0.7 and strong correlations were defined as having an r value greater than 0.7.
To evaluate the model’s prediction capabilities, the root-mean-squared errors (RMSEs) were computed with the predicted and measured values as:
where logCi,meas and logCi,predict are measured and predicted concentrations for the ith dataset and N is the total number of datasets. The RMSE values were computed for training and testing datasets for each of the individual regression trees of the random forest models and then averaged. If the independent data was available (the 2019 validation dataset), the RMSE vales were computed from that validation dataset with predictions of the trained random forest models. T-tests were used to determine significant differences in accuracy metric results, and a p value of 0.05 was selected.
The Williams-Kloot test [48] was utilized to compare the performance of pairs of random forest models obtained with different inputs for estimating phytoplankton concentrations. The test consists of computing the slope of the inward regression using the following equation:
where is the measured concentration, is the predicted concentration from model 1, and is the predicted concentration from model 2. If λ was positive and significantly different from zero, then the performance of model 2 was considered better than the performance of model 1. If λ was negative and significantly different from zero, then the performance of model 1 was considered the better of the two models. A p-value of 0.05 was selected to determine significance in the Williams-Kloot test applications.
The ratio of coefficients of variance (CVs) were also calculated to compare the variation of interior locations with the variation of nearshore sampling locations for phytoplankton functional groups and water quality parameters. The equation for calculating the ratio of CVs for each parameter is as follows:
where σ is the standard deviation and μ is the mean of the interior (i) parameters or nearshore (n) parameters.
The input variable importance was quantified by the Mean Decrease Accuracy (%IncMSE) as implemented in the Rstudio randomForest package. The %IncMSE reflects the loss of model accuracy when a variable is scrambled, i.e., its values are randomly rearranged. The model decreases of accuracy were computed for each tree in the forest and the percentage of decrease of accuracy was averaged over all trees to get the mean value. A higher %IncMSE value indicated that a variable had a greater effect on the model accuracy and was therefore a more influential variable.
3. Results
3.1. Data Summary
The most dominant and commonly occurring phytoplankton taxa were all representatives of eutrophic, shallow, small water bodies per the functional group classifications of Reynolds et al. [22]. Diatom concentrations for both years ranged from 4.19 to 7.59 log cells·L−1 and from 4.19 to 7.77 log cells·L−1 in Pond 1 (Aulacoseira spp.) and Pond 2 (Aulacoseira spp. and Cyclotella spp.), respectively. In Pond 1, cyanobacteria (Microcystis spp.) ranged from 4.19 to 7.95 log cells·L−1, and green algae (Coelastrum spp. and Scenedesmus spp.) ranged from 5.49 to 8.08 log cells·L−1 for both years. In Pond 2, cyanobacteria (Aphanizomenon spp., Dolichospermum spp., and Microcystis spp.) and green algae (Closterium spp. and Scenedesmus spp.) ranged from 4.67 to 8.69 log cells·L−1 and from 4.89 to 8.18 log cells·L−1, respectively. In 2017 and 2018, green algae had the highest average cell concentrations, followed by cyanobacteria and then diatoms in Pond 1. At Pond 2, cyanobacteria had the lowest average concentrations of the phytoplankton groups in 2017, whereas in 2018, diatoms had the lowest average concentrations. Water quality, weather, and phytoplankton data are presented in Supplemental Table S2 and Supplemental Figure S1.
In 2017, a total of eight physicochemical parameters and organic constituents commonly used to assess water quality were measured in the field and used in sets A and B for training and testing the random forest algorithm. An additional 11 physicochemical, organic constituents, and nutrient/macro element parameters were added in 2018 and applied across input sets A, B, and C training and testing datasets. Average TEMP, DO, SPC, pH, NTU, and Phyco did not differ from 2017 to 2018 in both Pond 1 and Pond 2. CHL averages doubled from 2017 to 2018 at Pond 2. While CHL concentrations were low at Pond 1 for both 2017 and 2018, there was a decrease in 2018. This may be the result of routine algicide application to Pond 1 during the study period.
The strength of the linear relationship between the water quality parameters measured for this study was assessed with Pearson correlation statistics (Supplemental Table S3). Only DO-pH and NTU-Phyco exhibited moderate or strong correlations in both ponds in both years. SPC-FDOM and TEMP-SPC had moderate or strong correlations in both ponds in 2017, but not in 2018. Phyco-CHL was correlated in both ponds in 2017, but this correlation was only found in Pond 2 in 2018. Moderate correlations between pH-FDOM, DO-FDOM, and NTU-CHL were found only in one pond over both years. A strong correlation between TEMP-FDOM was observed only in Pond 2 in 2017.
3.2. Performance of Models
3.2.1. Model Accuracy
The RMSEs that characterized the random forest model performance are shown in Figure 2 for each phytoplankton group. The differences in RMSE between ponds were relatively small. However, in almost all instances, RMSE values for Pond 2 were larger than those for Pond 1 for all three phytoplankton groups (green algae, diatoms, and cyanobacteria) and all three time periods (2017, 2018, and 2017 + 2018). It is also worth noting that the ranges of log phytoplankton concentrations, computed from minimum and maximum values in Supplemental Table S2, were also slightly greater in Pond 2 than in Pond 1 for all three phytoplankton groups and all three time periods. The smallest and the largest RMSEs for the combined year data were found for green algae and cyanobacteria, respectively. RMSE values for diatoms were in most cases an intermediate value. An exception to this was in 2018, in Pond 2; here diatom RMSEs were larger than the cyanobacteria values. RMSEs of the 2018 model were lower than RMSE of 2017 model. Mean values of measured parameters in the 2018 dataset were also lower than those from 2017 (Supplemental Table S2). RMSE values of the combined dataset 2017 + 2018 were smaller than the RMSE values of 2018. Creating the combined year dataset improved the robustness of the random forest models.
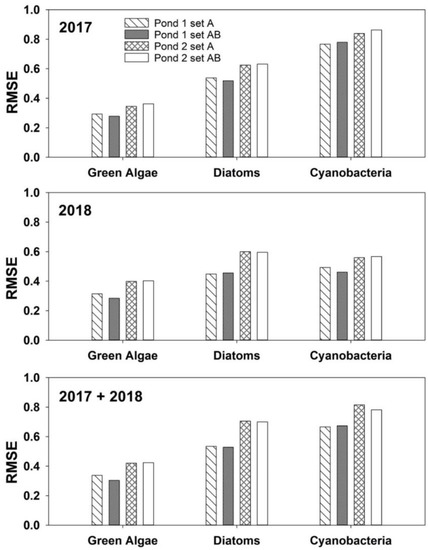
Figure 2.
Root-mean-squared errors (RMSEs) of the random forest models for green algae, diatoms, and cyanobacteria for Ponds 1 and 2, with data from 2017, 2018, and 2017 + 2018.
The small differences in RMSE between random forest models using input set A and input set AB implied that there may be not a significant difference between model performance. All Williams-Kloot tests yielded positive λ values indicating that modeling with set AB as the input may be superior to the model created with set A as the input. The Williams-Kloot test showed that there was a significant difference between models for green algae in Pond 1 (p < 0.001) and cyanobacteria in Pond 2 (p = 0.010), but not for green algae in Pond 2, cyanobacteria in Pond 1, or diatoms in either pond.
The RMSEs for the random forest model performance of cyanobacteria orders, Nostocales and Chroococcales, are shown in Table 2. Values of RMSE for the Nostocales order were larger in Pond 1 compared to Pond 2 for all years. Overall, RMSEs for Chroococcales were larger for Pond 2 compared to Pond 1 for 2018, while Pond 1 had larger RMSE values for Chroococcales 2017 and 2017 + 2018. The RMSEs for both Nostocales and Chroococcales were the lowest in 2018 when compared to 2017 and combined years 2017 + 2018. In all instances, the model error for Chroococcales decreased with the addition of additional input datasets, whereas the model error for Nostocales only decreased with increased datasets for 2018 in Pond 1 and 2018 as well as combined years (2017 + 2018) for Pond 2.

Table 2.
Random forest accuracy RMSE for cyanobacteria orders.
3.2.2. Model Validation
Models developed with the 2017 and 2018 datasets were tested using data collected in 2019. The results are shown in Figure 3. When using the random forest model on blind 2019 data, the RMSE results did not mirror what was predicted during model development using the 2017 and 2018 data. The RMSE values for green algae 2019 predictions were larger than the values for the 2017 and 2018 datasets using any combination of input sets A and B. For 2019, cyanobacteria continued to produce the higher RMSE values, whereas diatoms presented the lowest RMSE values. Pond 2 continued to have higher RMSEs for diatoms than Pond 1. Cyanobacteria in Pond 1 displayed higher RMSE values in 2019, whereas for years 2017 and 2018, Pond 2 typically had higher cyanobacteria RMSE values. Overall, green algae RMSEs were much higher for the 2019 data compared to the 2017 and 2018 dataset. In all instances, RMSE values were lower when the model was run with set AB parameters. The Williams-Kloot test determined that the AB model was superior to the A model. The AB model performance was significantly different (p < 0.05) for all groups and both ponds, except for Pond 2 diatoms (p = 0.84).
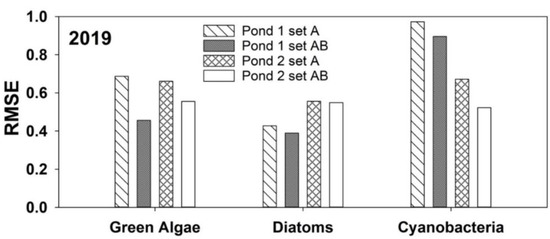
Figure 3.
Root-mean-squared errors (RMSEs) of the random forest models for green algae, diatoms, and cyanobacteria for Ponds 1 and 2, with blind data from 2019.
3.3. Spatial Patterns of Random Forest Model Performances
Spatial distribution of the individual location errors with data from 2017 + 2018 and set A parameters is shown in Supplemental Table S4. There was a pattern of lower RMSE values for interior locations compared with nearshore locations in each pond. For all three groups of phytoplankton within both ponds, the lowest RMSE values were found in the interior of the ponds, except the outflow area of Pond 2 (location 23). The average RMSE values were larger, and the performance of the models was reduced at nearshore locations compared to interior locations for all phytoplankton groups at Pond 2. Separation of nearshore locations from interior locations revealed that in Pond 2, the probability (t-test) of the average RMSE being the same over nearshore and interior locations was very low (p < 0.01) for green algae and cyanobacteria. The probability of RMSE values being the same for nearshore and interior locations for diatoms in Pond 2 was greater but still small (p < 0.1). In Pond 1, no substantial differences in average RMSE for nearshore and interior locations were found for green algae (p > 0.5), and only moderate differences were found for diatoms and cyanobacteria (p < 0.1). The percentage of sampling dates in which the CV was larger for nearshore locations compared to interior locations can be found in Supplemental Table S5. For diatoms and cyanobacteria, more than 54.6% of the sampling dates had higher CVs for nearshore samples compared to interior samples for both ponds. For green algae, Pond 2 (63.6%) had a higher percentage of dates with nearshore variability being higher than Pond 1 (41.7%). Most of water quality measurements had high CVs at nearshore locations with most being greater than 75% of the sampling dates. The exception to this is both SPC (63.6% of dates) and Phyco (72.7% of dates) in Pond 2.
Spatial distribution of the individual location errors with data from 2017 + 2018 and set AB parameters is shown in Supplemental Table S4. Similar to the spatial distributions of errors of the model using set A parameters, set AB parameters show a similar pattern of interior locations mostly containing the lowest RMSE values. This was true for cyanobacteria and diatoms at Pond 1 and green algae and cyanobacteria at Pond 2. A t-test of set AB showed that no differences were found in the average RMSEs for interior and nearshore locations for green algae (p > 0.05) at Pond 1. Diatom RMSEs at both ponds exhibited moderate (p < 0.1) differences between interior and nearshore locations. Significant (p < 0.05) differences between nearshore and interior RMSEs were found for cyanobacteria at both ponds and green algae at Pond 2.
3.4. Importance of Variables-Predictors
The top three most important predictors for each dataset and model for are shown in Table 2 and Table 3. The larger the value of %IncMSE indicates that the variable has the most effect on the model accuracy. The three variables with the highest %IncMSE were the most important predictors for each model. SPC and TEMP were the most influential predictors; found in 63% of all cases when using input set A and in 46% of all cases when using input set AB. NTU was seen in 7% and FDOM was seen in 11% of all cases where input set A and input set AB were used, respectively. Predictors from set A continued to have high importance (total of 61%) when the input set AB was used.

Table 3.
Most influential predictors and the increase in accuracy (%)for that variable for Pond 1 as determined using the random forest algorithm.
There was no significant difference between the ponds when considering the top three most influential predictors when input set A was used. Using input set A, SPC was the most influential predictor, with nine occurrences for each pond. TEMP was in the top three most influential predictors nine times for Pond 1 and seven times in Pond 2. There was a greater difference between the ponds when input set AB was used. SPC was the most influential predictor three times for Pond 1 and nine times for Pond 2. TEMP was among the most influential predictors eight times for Pond 1 and five times for Pond 2. The influence of CHL was more prominent for Pond 1 (four times) than for Pond 2 (once). Similarly, the FDOM was more prominent for Pond 1 (five times) than for the Pond 2 (once). Overall, with the AB input set, predictors of the input set A were less influential in Pond 1 (52% of all occurrences) than in Pond 2 (78% of all occurrences).
There were clear differences among the phytoplankton groups. NTU was the most influential predictor when assessing cyanobacteria, yet CHL was not. For green algae, TEMP and SPC were the most frequent influential predictor with the input set A. When the organic constituent-related inputs were included as part of set AB, TEMP and FDOM became the most frequent influential predictors. CDOM was found as an influential predictor only for diatoms. Diatoms in Pond 2 had the same most influential predictors with inputs sets A and AB. The same was true for diatoms with combined 2017 + 2018 data in Pond 1.
The top three most influential predictors were different in 2017 and 2018 in most cases, with green algae in Pond 1 being an exception. The 2017 + 2018 dataset, in some cases, led to the influential predictors being the same as individual years modeled separately (e.g., green algae in Pond 2 with the input set A; diatoms in Pond 1 with the input set A; green algae with the input set AB in Pond 1; and cyanobacteria in Pond 2 with input sets A and AB). The nutrient-related variables available in 2018 are grouped in input set C. These proved to be most important when all available input variables (input set ABC) were used as input for green algae and diatoms (Table 3 and Table 4), but not cyanobacteria. Because nutrient data was only collected in 2018, it was excluded from the 2017 + 2018 dataset to avoid unequal weighting across all parameters.

Table 4.
Most influential predictors and increase in mean square error for that variable for Pond 2 as determined using the random forest algorithm.
The top three most important predictors for each data set of the cyanobacteria orders (Nostocales and Chroococcales) are presented in Table 5 and Table 6. Temperature and SPC were among the most frequent influential predictors. When comparing the overlap between Nostocales and Chroococcales for each input dataset, the top three most influential predictors were similar 67% and 51% of the time for Pond 1 and Pond 2, respectively. Comparing the top three important variables between ponds for each order reveals that variables are similar across both ponds in 54% of instances for Nostocales and 44% for Chroococcales. In 2018, when all input datasets were included, nutrients were some of the most important predictors for both orders at Pond 2 and for Chroococcales at Pond 1.

Table 5.
Most influential predictors for cyanobacteria orders Nostocales and Chroococcales, in Pond 1 as determined using the random forest algorithm.

Table 6.
Most influential predictors for cyanobacteria orders, Nostocales and Chroococcales, in Pond 2 as determined using the random forest algorithm.
3.5. Sensitivity to Inputs
The mean decrease of accuracy (%IncMSE) is shown below each variable in Table 3 and Table 4. For Pond 1 in 2017 and 2018, %IncMSE values were low for green algae and diatoms (<15), and slightly higher for cyanobacteria (>15). The combination of years (2017 + 2018) produced higher increases of mean square error, indicating that multiyear data allowed for predictors to be more influential. For Pond 2, the sensitivity to the important variables tended to be higher than in Pond 1. The values of %IncMSE in Pond 2 were less than 30 for green algae and diatoms for both years. Cyanobacteria had a larger (>30) increase in mean square error values, with the highest value being 143, indicating cyanobacteria predictions were more sensitive to the influential predictors than the predictions for green algae and diatoms. Multiyear data tended to increase the %IncMSE values, causing greater sensitivity to influential predictors.
4. Discussion
Earlier work by Smith et al. [40,46] demonstrated the correlation between several basic water quality parameters and cyanobacteria populations, as well as the temporal stability of phytoplankton populations within these ponds. Here, the relationship between more complex water quality parameters and phytoplankton groups were examined with machine learning. Phytoplankton group concentrations in the two agricultural irrigation ponds in this study did not vary greatly, nor were the community compositions significantly different, both representing communities of eutrophic, shallow, small-bodied waters. Average diatom and green algae concentrations were similar between years and the two ponds. Despite the routine application of the algicide copper sulfate during the study, phytoplankton concentrations in Pond 1 were comparable to those reported in regional [29,30] and global lakes [49,50]. Pond 2 had recurrent cyanobacteria blooms during the study, making the phytoplankton concentrations more comparable to those reported in small lakes by Lee et al. [51] and in local waters by Tango and Butler [52]. Pond 2 phytoplankton concentrations were slightly higher than Pond 1 concentrations and can potentially be explained by routine algicide use in Pond 1. All three phytoplankton populations in Pond 1 were greater in 2017 than 2018, whereas the opposite was true for Pond 2, except for diatom concentrations, which were slightly higher in 2017 than 2018.
Root-mean-square errors (RMSEs), a metric used to evaluate model performance, for the 2017, 2018, and 2017 + 2018 models (sets A and AB) varied depending on phytoplankton group. Green algae models tended to have the best performance, followed by diatoms, and then cyanobacteria. In a review by Shimoda and Arhonditsis [53], green algae were found to have the least error of the three phytoplankton groups similar to the results in this study. Cyanobacteria models had higher RMSEs than green algae models in both our findings and those reviewed by Shimoda and Arhonditsis [53]. This could be explained by the natural spatial and temporal variability of cyanobacteria blooms making accurate population predictions more challenging [46,54]. While various types of models were used in the review by Shimoda and Arhonditsis [53], the RMSEs from this work indicate that the random forest model is a superior model for predicting green algae when compared to the diatom and cyanobacteria models. In the work of Di Maggio et al. [55] where the same three functional groups were studied, cyanobacteria were found to have the least accurate model performance during peak biomass periods. However, Thomas et al. [56] noted that cyanobacteria were more predictable than diatoms and green algae across many time scales in an alpine lake. Both ecosystem type and available input variables appear to affect the comparative performance of the random forest algorithm in predictions of phytoplankton functional groups. The robustness of the model during the growing season is characterized by the RMSE values presented in this paper since these RMSEs are averages over the datasets used for training and testing by the random forest algorithm. Since this study only focused on assessing the accuracy of the prediction model in agricultural irrigation ponds during the growing season (May–October) (when waters were used for irrigation purposes and when cyanobacteria biomass, and subsequently risks from cyanotoxins, was expected to be greatest in this region [52,57]), to better assess this model’s performance in comparison to similar models, additional training and validation needs to be done using data collected outside of the growing season and in varying waterbody types. In this study, sampling was conducted during periods of time between rainfall events, when irrigation is more likely to take place due to crop production demands, elevated temperatures, and reduced soil moisture [58]. To better equip this model for prediction during all weather conditions and all seasons, additional sampling and training of the model would be necessary.
Model performance did not differ drastically between years. The exception to this is for cyanobacteria predictions wherein RMSE values decreased substantially from 2017 to 2018, indicating better performance of the 2018 models. In Pond 1, models predicting diatoms and cyanobacteria performed better in 2018 compared to 2017. Similarly, in Pond 2, better model performance in 2018 were seen for cyanobacteria predictions and, to a lesser extent, for diatom predictions. The combined 2017 + 2018 datasets had higher RMSE values than when using just the 2018 dataset, but lower than when only the 2017 dataset was used. For all three groups and both sets of parameters (A and AB), 2018 had the best model performance as indicated by the lowest RMSEs. Thomas et al. [56] found that multiyear datasets were able to produce reasonable performance and attributed it to the model having more data points to train the machine learning algorithm with. Our individual years had fewer data points than the combined year models. While 2018 had the lowest RMSE values of the three data sets, the use of 2017 + 2018 caused a decrease in RMSE values for 2017. Furthermore, it was determined that the prediction of the 2019 data was not as accurate as the prediction of the 2017 and 2018 years. Additional monitoring would help to determine if the model performance of future years is comparable to the accuracy represented in the 2017 and 2018 evaluations.
The addition of organic constituent-related input parameters did not improve model performance overall. While some aspects of the model saw a small increase in performance, others saw a small decrease, and no general pattern could be defined. This follows many other studies that showed the use of inputs, similar to this study’s set A parameters (DO, pH, NTU, and TEMP), tended to be most important and produced the best prediction results [59,60,61]. According to Rigosi et al. [62], a model based on water quality physical parameters often has superior performance, and this was attributed to the high level of complexity found in biological processes. Likewise, while the nutrient and macro element parameters in input set C were highly influential when evaluating the 2018 data, the difference in model performance across phytoplankton groups may be due to the complex and interrelated way each phytoplankton group utilizes different nutrients and macro elements [63,64], and subsequently interacts with other organisms [65], which was not captured with just one year of data. The presence of short blooms of both nitrogen-fixing and non-nitrogen-fixing cyanobacteria in the study area [46], which can utilize different forms of nitrogen and impact the overall nitrogen budget [66,67], also may not have been equitably represented in this dataset. When just the potentially toxigenic cyanobacteria, represented in the dataset as Chroococcales and Nostocales, were examined alone, inclusion of nutrient parameters in the training and testing dataset did improve model performance and warrants further consideration. However, the ability to use the random forest algorithm to predict phytoplankton groups using only set A inputs is beneficial for a wide range of resource monitoring applications, including the differentiation of discolored water caused by cyanobacteria, including subsurface bloom species like Raphidiopsis raciborskii [68], from discolored water caused by chlorophytes and euglenophytes, both of which are known to occur in the study area [29,30,52,69]. Set A input parameters are often the least expensive and easiest parameters to collect, thus predictions can be quickly and easily performed and provide a guideline to expanding resource monitoring efforts should cyanobacteria blooms be predicted.
Overall, spatial distributions of RMSE values differed based on phytoplankton group. Green algae had the lowest spatial average RMSEs (P1 = 0.278, P2 = 0.356); cyanobacteria had the highest spatial average RMSEs (P1 = 0.567, P2 = 0.679); and average RMSEs for diatoms were in between (P1 = 0.446, P2 = 0.578) for both ponds and models. This indicates that the set A and AB models were the most accurate in predicting the spatial green algae concentrations for the 2017 + 2018 dataset. In general, interior waters tended to exhibit the lowest RMSE values in both ponds and for models, with both input sets A and AB showing that the random forest algorithm predicted interior concentrations of green algae best, followed by diatoms and cyanobacteria. In a prior study on the temporal and spatial variability of phytoplankton functional groups within these two agricultural irrigation ponds, it was established that interior waters tended to be less variable than nearshore waters [46]. This stability allows the model to better predict the phytoplankton community structure in those locations. Variations in phytoplankton concentrations tended to be greater in nearshore samples when compared to interior waters using an assessment of CV. In over 50% of the sampling dates, CVs were higher for nearshore samples except for green algae in Pond 1. Similarly, water quality CVs in both ponds were almost always higher for nearshore locations, with most nearshore variability being higher in 75% or more of the sampling dates. This pattern was also observed in the study by Awada et al. [70] for marine waters; the model developed by these authors performed best in open water locations of the Gulf of Sirte and had poorer agreement between measured and simulated concentrations of chlorophyll-a along the shoreline. In Lake Taihu, locations closer to the shoreline tended to have higher simulation errors than central lake locations [60]. However, in a study on Lake Okeechobee, the random forest algorithm had better model results at nearshore locations as opposed to pelagic locations, and Zhang et al. [71] attributed this to poor phytoplankton growth in the pelagic zones caused by wind-driven sediment resuspension.
For all three phytoplankton groups, there was almost no change in RMSE values from models run using set A parameters to models run using set AB parameters, indicating that the additional parameters did not impact the predictive abilities of the random forests. The ability of the random forest model to predict phytoplankton community structure or chlorophyll-a concentrations accurately on set A parameters (TEMP, pH, NTU, and SPC) alone has been noted in several other studies [18,61,72]. Whereas other studies [73,74] found that biological parameters (biological oxygen demand; chlorophyll-a concentrations) were more important for phytoplankton prediction models, biological oxygen demand was not measured in this study. It should, however, be considered for future modeling efforts as it is known to be spatially and temporally variable in lake waters [75,76] and can be positively correlated with potentially toxigenic cyanobacteria species [77] and overall phytoplankton biomass [76], both of which are of concern to agricultural resource managers looking to meet water quality standards.
Overall, this study found that the most important variables tended to be set A parameters (TEMP, pH, NTU, and SPC) for both ponds. TEMP was determined to be the most recurrent parameter in the top three most influential parameters for all groups and both ponds. This is comparable to numerous other random forest models used for phytoplankton prediction [33,61,72,74,78]. Other set A parameters which were also reported in the top three most influential parameters, but to a lesser degree than TEMP, in this study were SPC, NTU, pH, and DO. SPC appeared to be the most influential predictor in input set A. A possible reason for this could be the correlation between SPC and nutrient ion concentrations in agricultural waters [79] and the intercoupled relationship between specific nutrient forms and concentrations and phytoplankton groups [80]. Correlations between water quality parameters reflected both commonalities of the water quality-forming processes in the studied ponds and the specifics of ponds. The strength of correlations between inputs depended on the ponds and years. This indicates the importance of processes not well-characterized by the available input variables. However, one cannot exclude the presence of confounding factors, i.e., factors affecting both input and target variables. Alleviating the effect of confounding variables is efficient if the relationships between the target variable and independent variables are found from designed experiments when the confounding variable is known and can be measured. This is not the case in the present work. More needs to be learned about the functioning of phytoplankton communities in small agricultural irrigation ponds now and how future community structure may be shaped by climate change and increased anthropogenic forces to adequately discover and monitor compounding variables.
The presence of correlations between independent variables in this work illustrates the common multicollinearity problem. Whereas the accuracy of the random forest models typically is not affected by multicollinearity [81], the causality conclusions, including the ranks of correlated variables in the lists of important inputs, can be affected [82]. We realize that the possible effect of multicollinearity was not fully addressed in this work. Multiple methods of variable elimination are suggested to reduce the input variable list and to characterize the effects of the input reduction on variable importance determinations [83,84]. Applying these methods to low-dimensional data can improve the model’s reliability as more data are available per model coefficient [85]. However, this may uncontrollably change conclusions on the relative importance of input variables [86]. Comparing the efficiency of the input reduction methods presents an interesting research avenue. In the present study, we made the first step in this type of investigation by analyzing correlations between inputs. For example, the expected strong correlations in both ponds in 2017 and 2018 were found between DO and pH (Supplemental Table S3). The probable cause for this is high phytoplankton biomass undergoing photosynthesis, which causes pH to increase due to CO2 consumption and O2 release. Thus, we cannot exclude the effect of this correlation on the occurrence and position of DO and pH in lists of important inputs (Table 3 and Table 4).
The only instance when set A parameters were not the most influential parameters was in 2018 when nutrients (input set C) were measured and used as inputs. Nutrients being the most influential or important parameters is in line with numerous assessments of phytoplankton community structure using random forest algorithms. Total nitrogen (TN), total phosphate (TP), nitrate, and nitrite were identified as the most important predictors in the phytoplankton models used in Lake Okeechobee [71] and Lake Taihu [76]. However, these studies took place in lakes considerably larger than the ponds studied here. Small waterbodies (<12 acres), which are increasingly being used in agricultural practices in the Mid-Atlantic region, often have a greater biodiversity than larger bodies of water and can experience more anthropogenic and climatic stress [87,88,89], highlighting the need to refine models to local conditions. Since nutrients were only measured in 2018, these parameters were not included in the 2017 or combined-year models. The modeling robustness of nutrient parameters, when compared to set A parameters, has yet to be determined for ponded agricultural waters. When we examined the use of nutrient parameters to predict potentially toxic cyanobacteria species in the orders Chroococcales and Nostocales, inclusion of nutrient parameters did improve model performance. Due to the small number of water samples tested for cyanotoxins during this study (data not shown), we cannot correlate this model’s performance with cyanotoxin production. However, this preliminary assessment showed improved performance when nutrient parameters were incorporated into this model tuned for small, ponded water systems with recurrent cyanobacteria blooms, and can be used as a baseline for our future monitoring and modeling efforts as there currently are no prediction models available that can differentiate between toxigenic and non-toxigenic cyanobacteria blooms [33,90], even though it is known through field and laboratory studies that TN and TP enrichment can stimulate the production of cyanotoxins in numerous species [37,91].
In a review of predictive and forecasting models for cyanobacteria by Rousso et al. [33], it was found that parameters similar to this study’s input set A (TEMP, DO, and pH) were reported as the most influential predictors in 38.5% of publications surveyed. Nutrients were reported as the most influential parameters in 30.5% of the total publications surveyed. One of the least influential predictors reported (6% of publications) was similar to the parameters included input set B (FDOM, CHL, and Phyco), which is comparable to the findings in this study. As noted in a cyanobacteria research forecast by Burford et al. [91], future modeling efforts should incorporate CO2 dynamics that will reflect future climate scenarios, temporally relevant weather patterns, and the intricate relationship cyanobacteria have with the food web, all factors which ultimately will influence agricultural irrigation water quality. This study only focused on between-rain events when irrigation water from these agriculture ponds was used most frequently. However, rain events, specifically those which cause surface run-off, will ultimately influence nutrient concentrations within surface waters used for irrigation. Defining the relationship between water quality parameters and cyanobacteria blooms under numerous weather conditions with an easy-to-use model would aid local resource managers charged with safeguarding irrigation water quality and mitigating the risks posed to the agriculture industry from cyanotoxins.
Physicochemical parameters being the most important predictors for the three major phytoplankton groups is beneficial for water quality management. Enumeration of phytoplankton is time intensive, requires highly-trained staff, and/or expensive infrastructure [8,18,32], whereas parameters such as temperature, dissolved oxygen, pH, conductivity, and turbidity can be easily and affordably measured in real time with an in situ sensor. The quick acquisition and input of these parameters into a modeling application allows for the prediction of major phytoplankton groups by machine learning algorithms to be performed by a broader group of individuals that could lead to more timely alerts of potentially harmful phytoplankton species.
5. Conclusions
The prediction of phytoplankton groups in two agricultural irrigation ponds was feasible with machine learning methodology via the random forest algorithm. Random forest predictions for green algae were more accurate compared with predictions for diatoms and cyanobacteria. The RMSE values of the model obtained with two years of data were in between the RMSE values obtained with data from individual years. Interior sampling locations had a lower model error than nearshore sampling locations. Minimal differences in model performance were seen when organic constituent concentrations were added as predictors. Models using physicochemical parameters (input set A) were the models with the best performance. Modeling of potentially toxic cyanobacteria was improved with the inclusion of nitrogen and phosphorus data. However, commonly measured water quality variables such as temperature, pH, dissolved oxygen, specific conductance, and turbidity were the most frequent influential predictors. The use of these easy-to-measure parameters evaluated with regionally tuned models could allow water quality managers to potentially bypass time intensive and expensive monitoring procedures for those which can be obtained easily, affordably, and in real time, offering improved resource management and health safeguards.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/environments9110142/s1, Figure S1: Weather data for Pond 1 and Pond 2 for 2017 and 2018; Table S1: Time series data of water quality parameters for Pond 1 and Pond 2 for 2017, 2018, and 2019; Table S2: Summary statistics for all measured parameters and phytoplankton functional groups for 2017 and 2018; Table S3: Pearson correlation coefficients between water quality parameters in 2017 and 2018; Table S4: Spatial root-mean-squared errors (RMSEs) calculated by sampling location for 2017 + 2018 calculated for green algae, diatoms, and cyanobacteria using set A and AB parameters in Pond 1 and Pond 2; Table S5: Percent of dates in which the coefficient of variation (CV) was larger for nearshore locations.
Author Contributions
Conceptualization, M.D.S. and Y.P.; methodology, J.E.S., J.L.W. and M.D.S.; formal analysis, J.E.S. and Y.P.; investigation, J.E.S. and M.D.S.; resources, M.D.S. and Y.P.; data curation, J.E.S. and Y.P.; writing—original draft preparation, J.E.S.; writing—review and editing, J.L.W., R.L.H. and Y.P.; supervision, R.L.H. and Y.P.; project administration, Y.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data shown in the text and supplementary document are currently under a 2-year hold as part of a doctoral dissertation. As of May 2024, all data will be publicly available. Requests for data before then can be made to the corresponding author, who will work with requestees in order to share the maximum allowable content.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Marić, D.; Kraus, R.; Godrijan, J.; Supić, N.; Djakovac, T.; Precali, R. Phytoplankton Response to Climatic and Anthropogenic Influences in the North-Eastern Adriatic during the Last Four Decades. Estuar. Coast. Shelf Sci. 2012, 115, 98–112. [Google Scholar] [CrossRef]
- Marshall, H.G.; Lane, M.F.; Nesius, K.K.; Burchardt, L. Assessment and Significance of Phytoplankton Species Composition within Chesapeake Bay and Virginia Tributaries through a Long-Term Monitoring Program. Environ. Monit. Assess. 2009, 150, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qin, B.; Teubner, K.; Dokulil, M.T. Long-Term Dynamics of Phytoplankton Assemblages: Microcystis-Domination in Lake Taihu, a Large Shallow Lake in China. J. Plankton Res. 2003, 25, 445–453. [Google Scholar] [CrossRef]
- Wynne, T.T.; Stumpf, R.P. Spatial and Temporal Patterns in the Seasonal Distribution of Toxic Cyanobacteria in Western Lake Erie from 2002–2014. Toxins 2015, 7, 1649–1663. [Google Scholar] [CrossRef]
- Znachor, P.; Nedoma, J.; Hejzlar, J.; Seďa, J.; Komárková, J.; Kolář, V.; Mrkvička, T.; Boukal, D.S. Changing Environmental Conditions Underpin Long-Term Patterns of Phytoplankton in a Freshwater Reservoir. Sci. Total Environ. 2020, 710, 135626. [Google Scholar] [CrossRef]
- Bergkemper, V.; Weisse, T. Do Current European Lake Monitoring Programmes Reliably Estimate Phytoplankton Community Changes? Hydrobiologia 2018, 824, 143–162. [Google Scholar] [CrossRef]
- Clayton, S.; Gibala-Smith, L.; Mogatas, K.; Flores-Vargas, C.; Marciniak, K.; Wigginton, M.; Mulholland, M.R. Imaging Technologies Build Capacity and Accessibility in Phytoplankton Species Identification Expertise for Research and Monitoring: Lessons Learned During the COVID-19 Pandemic. Front. Microbiol. 2022, 13, 823109. [Google Scholar] [CrossRef]
- Lawton, L.; Marsalek, B.; Padisak, J.; Chorus, I. Determination of Cyanobacteria in the Laboratory. In Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring and Management; Chorus, I., Bartram, J., Eds.; E & FN Spon: London, UK; New York, NY, USA, 1999; ISBN 978-0-419-23930-7. [Google Scholar]
- Ho, J.C.; Michalak, A.M.; Pahlevan, N. Widespread Global Increase in Intense Lake Phytoplankton Blooms since the 1980s. Nature 2019, 574, 667–670. [Google Scholar] [CrossRef]
- López-Felices, B.; Aznar-Sánchez, J.A.; Velasco-Muñoz, J.F.; Piquer-Rodríguez, M. Contribution of Irrigation Ponds to the Sustainability of Agriculture. A Review of Worldwide Research. Sustainability 2020, 12, 5425. [Google Scholar] [CrossRef]
- Descy, J.-P.; Leprieur, F.; Pirlot, S.; Leporcq, B.; Van Wichelen, J.; Peretyatko, A.; Teissier, S.; Codd, G.A.; Triest, L.; Vyverman, W.; et al. Identifying the Factors Determining Blooms of Cyanobacteria in a Set of Shallow Lakes. Ecol. Inform. 2016, 34, 129–138. [Google Scholar] [CrossRef]
- Rao, K.; Zhang, X.; Wang, M.; Liu, J.; Guo, W.; Huang, G.; Xu, J. The Relative Importance of Environmental Factors in Predicting Phytoplankton Shifting and Cyanobacteria Abundance in Regulated Shallow Lakes. Environ. Pollut. 2021, 286, 117555. [Google Scholar] [CrossRef] [PubMed]
- Fornarelli, R.; Galelli, S.; Castelletti, A.; Antenucci, J.P.; Marti, C.L. An Empirical Modeling Approach to Predict and Understand Phytoplankton Dynamics in a Reservoir Affected by Interbasin Water Transfers. Water Resour. Res. 2013, 49, 3626–3641. [Google Scholar] [CrossRef]
- Gayoso, A. Long-Term Phytoplankton Studies in the Bahía Blanca Estuary, Argentina. ICES J. Mar. Sci. 1998, 55, 655–660. [Google Scholar] [CrossRef][Green Version]
- Schönfelder, I.; Gelbrecht, J.; Schönfelder, J.; Steinberg, C.E.W. Relationships between Littoral Diatoms and Their Chemical Environment in Northeastern German Lakes and Rivers. J. Phycol. 2002, 38, 66–89. [Google Scholar] [CrossRef]
- Mokhtar, A.; Elbeltagi, A.; Gyasi-Agyei, Y.; Al-Ansari, N.; Abdel-Fattah, M.K. Prediction of Irrigation Water Quality Indices Based on Machine Learning and Regression Models. Appl. Water Sci. 2022, 12, 76. [Google Scholar] [CrossRef]
- Yıldız, S.; Karakuş, C.B. Estimation of Irrigation Water Quality Index with Development of an Optimum Model: A Case Study. Environ. Dev. Sustain. 2020, 22, 4771–4786. [Google Scholar] [CrossRef]
- Zeng, Q.; Liu, Y.; Zhao, H.; Sun, M.; Li, X. Comparison of Models for Predicting the Changes in Phytoplankton Community Composition in the Receiving Water System of an Inter-Basin Water Transfer Project. Environ. Pollut. 2017, 223, 676–684. [Google Scholar] [CrossRef]
- Cheruvelil, K.S.; Soranno, P.A.; Bremigan, M.T.; Wagner, T.; Martin, S.L. Grouping Lakes for Water Quality Assessment and Monitoring: The Roles of Regionalization and Spatial Scale. Environ. Manag. 2008, 41, 425–440. [Google Scholar] [CrossRef]
- Maloney, K.O.; Smith, Z.M.; Buchanan, C.; Nagel, A.; Young, J.A. Predicting Biological Conditions for Small Headwater Streams in the Chesapeake Bay Watershed. Freshw. Sci. 2018, 37, 795–809. [Google Scholar] [CrossRef]
- Zhang, Q.; Blomquist, J.D.; Moyer, D.L.; Chanat, J.G. Estimation Bias in Water-Quality Constituent Concentrations and Fluxes: A Synthesis for Chesapeake Bay Rivers and Streams. Front. Ecol. Evol. 2019, 7, 109. [Google Scholar] [CrossRef]
- Reynolds, C.S.; Huszar, V.; Kruk, C.; Naselli-Flores, L.; Melo, S. Towards a Functional Classification of the Freshwater Phytoplankton. J. Plankton Res. 2002, 24, 417–428. [Google Scholar] [CrossRef]
- Duarte, C.M.; Agustí, S.; Canjield, D.E., Jr. Patterns in Phytoplankton Community Structure in Florida Lakes. Limnol. Oceanogr. 1992, 37, 155–161. [Google Scholar] [CrossRef]
- Watson, S.B.; McCauley, E.; Downing, J.A. Patterns in Phytoplankton Taxonomic Composition across Temperate Lakes of Differing Nutrient Status. Limnol. Oceanogr. 1997, 42, 487–495. [Google Scholar] [CrossRef]
- Heini, A.; Puustinen, I.; Tikka, M.; Jokiniemi, A.; Leppäranta, M.; Arvola, L. Strong Dependence between Phytoplankton and Water Chemistry in a Large Temperate Lake: Spatial and Temporal Perspective. Hydrobiologia 2014, 731, 139–150. [Google Scholar] [CrossRef]
- Hu, S.; Liu, H.; Zhao, W.; Shi, T.; Hu, Z.; Li, Q.; Wu, G. Comparison of Machine Learning Techniques in Inferring Phytoplankton Size Classes. Remote Sens. 2018, 10, 191. [Google Scholar] [CrossRef]
- Halsey, K.H.; Giovannoni, S.J.; Graus, M.; Zhao, Y.; Landry, Z.; Thrash, J.C.; Vergin, K.L.; de Gouw, J. Biological Cycling of Volatile Organic Carbon by Phytoplankton and Bacterioplankton. Limnol. Oceanogr. 2017, 62, 2650–2661. [Google Scholar] [CrossRef]
- Parson, M.J.; Parker, B.C. Algal Flora in Mountain Lake, Virginia: Past and Present. Castanea 1989, 54, 79–86. [Google Scholar]
- Marshall, H.G. Phytoplankton in Virginia Lakes and Reservoirs. Va. J. Sci. 2013, 64, 3–15. [Google Scholar] [CrossRef]
- Marshall, H.G. Phytoplankton in Virginia Lakes and Reservoirs: Part II. Va. J. Sci. 2014, 65, 3–8. [Google Scholar] [CrossRef]
- Stumpf, R.P.; Johnson, L.T.; Wynne, T.T.; Baker, D.B. Forecasting Annual Cyanobacterial Bloom Biomass to Inform Management Decisions in Lake Erie. J. Great Lakes Res. 2016, 42, 1174–1183. [Google Scholar] [CrossRef]
- Stauffer, B.A.; Bowers, H.A.; Buckley, E.; Davis, T.W.; Johengen, T.H.; Kudela, R.; McManus, M.A.; Purcell, H.; Smith, G.J.; Vander Woude, A.; et al. Considerations in Harmful Algal Bloom Research and Monitoring: Perspectives from a Consensus-Building Workshop and Technology Testing. Front. Mar. Sci. 2019, 6, 399. [Google Scholar] [CrossRef]
- Rousso, B.Z.; Bertone, E.; Stewart, R.; Hamilton, D.P. A Systematic Literature Review of Forecasting and Predictive Models for Cyanobacteria Blooms in Freshwater Lakes. Water Res. 2020, 182, 115959. [Google Scholar] [CrossRef] [PubMed]
- Wood, R. Acute Animal and Human Poisonings from Cyanotoxin Exposure—A Review of the Literature. Environ. Int. 2016, 91, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jiang, X.; Manubolu, M.; Riedl, K.; Ludsin, S.A.; Martin, J.F.; Lee, J. Fresh Produce and Their Soils Accumulate Cyanotoxins from Irrigation Water: Implications for Public Health and Food Security. Food Res. Int. 2017, 102, 234–245. [Google Scholar] [CrossRef]
- Weralupitiya, C.; Wanigatunge, R.P.; Gunawardana, D.; Vithanage, M.; Magana-Arachchi, D. Cyanotoxins Uptake and Accumulation in Crops: Phytotoxicity and Implications on Human Health. Toxicon 2022, 211, 21–35. [Google Scholar] [CrossRef]
- Aguilera, A.; Aubriot, L.; Echenique, R.O.; Salerno, G.L.; Brena, B.M.; Pírez, M.; Bonilla, S. Synergistic Effects of Nutrients and Light Favor Nostocales over Non-Heterocystous Cyanobacteria. Hydrobiologia 2017, 794, 241–255. [Google Scholar] [CrossRef]
- Verspagen, J.M.H.; de Waal, D.B.V.; Finke, J.F.; Visser, P.M.; Donk, E.V.; Huisman, J. Rising CO2 Levels Will Intensify Phytoplankton Blooms in Eutrophic and Hypertrophic Lakes. PLoS ONE 2014, 9, e104325. [Google Scholar] [CrossRef]
- Komárek, J. Review of the Cyanobacterial Genera Implying Planktic Species after Recent Taxonomic Revisions According to Polyphasic Methods: State as of 2014. Hydrobiologia 2016, 764, 259–270. [Google Scholar] [CrossRef]
- Smith, J.E.; Stocker, M.D.; Wolny, J.L.; Hill, R.L.; Pachepsky, Y.A. Intraseasonal Variation of Phycocyanin Concentrations and Environmental Covariates in Two Agricultural Irrigation Ponds in Maryland, USA. Environ. Monit. Assess. 2020, 192, 706. [Google Scholar] [CrossRef]
- Brient, L.; Lengronne, M.; Bertrand, E.; Rolland, D.; Sipel, A.; Steinmann, D.; Baudin, I.; Legeas, M.; Rouzic, B.L.; Bormans, M. A Phycocyanin Probe as a Tool for Monitoring Cyanobacteria in Freshwater Bodies. J. Environ. Monit. 2008, 10, 248–255. [Google Scholar] [CrossRef]
- Song, K.; Li, L.; Tedesco, L.; Clercin, N.; Hall, B.; Li, S.; Shi, K.; Liu, D.; Sun, Y. Remote Estimation of Phycocyanin (PC) for Inland Waters Coupled with YSI PC Fluorescence Probe. Environ. Sci. Pollut. Res. 2013, 20, 5330–5340. [Google Scholar] [CrossRef] [PubMed]
- Marshall, H.G.; Alden, R.W. A Comparison of Phytoplankton Assemblages and Environmental Relationships in Three Estuarine Rivers of the Lower Chesapeake Bay. Estuaries 1990, 13, 287–300. [Google Scholar] [CrossRef]
- John, D.M.; Whitton, B.A.; Brook, A.J. The Freshwater Algal Flora of the British Isles: An Identification Guide to Freshwater and Terrestrial Algae; Cambridge University Press: London, UK, 2011; 724p, ISBN 978-0-521-19375-7. [Google Scholar]
- Bellinger, E.D.; Sigee, D.C. Freshwater Algae, 1st ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Smith, J.E.; Wolny, J.L.; Stocker, M.D.; Hill, R.L.; Pachepsky, Y.A. Temporal Stability of Phytoplankton Functional Groups within Two Agricultural Irrigation Ponds in Maryland, USA. Front. Water 2021, 3, 14. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Williams, E.J.; Kloot, N. Interpolation in a Series of Correlated Observations. Aust. J. Appl. Sci. 1953, 4, 1–17. [Google Scholar]
- Dembowska, E.A.; Mieszczankin, T.; Napiórkowski, P. Changes of the Phytoplankton Community as Symptoms of Deterioration of Water Quality in a Shallow Lake. Environ. Monit. Assess. 2018, 190, 95. [Google Scholar] [CrossRef]
- Jia, J.; Gao, Y.; Song, X.; Chen, S. Characteristics of Phytoplankton Community and Water Net Primary Productivity Response to the Nutrient Status of the Poyang Lake and Gan River, China. Ecohydrology 2019, 12, e2136. [Google Scholar] [CrossRef]
- Lee, T.A.; Rollwagen-Bollens, G.; Bollens, S.M. The Influence of Water Quality Variables on Cyanobacterial Blooms and Phytoplankton Community Composition in a Shallow Temperate Lake. Environ. Monit. Assess. 2015, 187, 315. [Google Scholar] [CrossRef]
- Tango, P.J.; Butler, W. Cyanotoxins in Tidal Waters of Chesapeake Bay. Northeast. Nat. 2008, 15, 403–416. [Google Scholar] [CrossRef]
- Shimoda, Y.; Arhonditsis, G.B. Phytoplankton Functional Type Modelling: Running before We Can Walk? A Critical Evaluation of the Current State of Knowledge. Ecol. Model. 2016, 320, 29–43. [Google Scholar] [CrossRef]
- Beversdorf, L.J.; Weirich, C.A.; Bartlett, S.L.; Miller, T.R. Variable Cyanobacterial Toxin and Metabolite Profiles across Six Eutrophic Lakes of Differing Physiochemical Characteristics. Toxins 2017, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Di Maggio, J.; Fernández, C.; Parodi, E.R.; Diaz, M.S.; Estrada, V. Modeling Phytoplankton Community in Reservoirs. A Comparison between Taxonomic and Functional Groups-Based Models. J. Environ. Manag. 2016, 165, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.K.; Fontana, S.; Reyes, M.; Kehoe, M.; Pomati, F. The Predictability of a Lake Phytoplankton Community, from Hours to Years. Ecol. Lett. 2017, 21, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Marshall, H.G.; Burchardt, L.; Lacouture, R. A Review of Phytoplankton Composition within Chesapeake Bay and Its Tidal Estuaries. J. Plankton Res. 2005, 27, 1083–1102. [Google Scholar] [CrossRef]
- Paul, M.; Negahban-Azar, M.; Shirmohammadi, A.; Montas, H. Developing a Multicriteria Decision Analysis Framework to Evaluate Reclaimed Wastewater Use for Agricultural Irrigation: The Case Study of Maryland. Hydrology 2021, 8, 4. [Google Scholar] [CrossRef]
- Fragoso, C.R.; Marques, D.M.L.M.; Collischonn, W.; Tucci, C.E.M.; van Nes, E.H. Modelling Spatial Heterogeneity of Phytoplankton in Lake Mangueira, a Large Shallow Subtropical Lake in South Brazil. Ecol. Model. 2008, 219, 125–137. [Google Scholar] [CrossRef]
- Huang, J.; Gao, J.; Hörmann, G.; Fohrer, N. Modeling the Effects of Environmental Variables on Short-Term Spatial Changes in Phytoplankton Biomass in a Large Shallow Lake, Lake Taihu. Environ. Earth Sci. 2014, 72, 3609–3621. [Google Scholar] [CrossRef]
- Liu, X.; Feng, J.; Wang, Y. Chlorophyll a Predictability and Relative Importance of Factors Governing Lake Phytoplankton at Different Timescales. Sci. Total Environ. 2019, 648, 472–480. [Google Scholar] [CrossRef]
- Rigosi, A.; Fleenor, W.; Rueda, F. State-of-the-Art and Recent Progress in Phytoplankton Succession Modelling. Environ. Rev. 2010, 18, 423–440. [Google Scholar] [CrossRef]
- Bradshaw, C.; Kautsky, U.; Kumblad, L. Ecological Stoichiometry and Multi-Element Transfer in a Coastal Ecosystem. Ecosystems 2012, 15, 591–603. [Google Scholar] [CrossRef]
- Finkel, Z.V.; Beardall, J.; Flynn, K.J.; Quigg, A.; Rees, T.A.V.; Raven, J.A. Phytoplankton in a Changing World: Cell Size and Elemental Stoichiometry. J. Plankton Res. 2010, 32, 119–137. [Google Scholar] [CrossRef]
- Guedes, I.A.; Rachid, C.T.C.C.; Rangel, L.M.; Silva, L.H.S.; Bisch, P.M.; Azevedo, S.M.F.O.; Pacheco, A.B.F. Close Link Between Harmful Cyanobacterial Dominance and Associated Bacterioplankton in a Tropical Eutrophic Reservoir. Front. Microbiol. 2018, 9, 424. [Google Scholar] [CrossRef] [PubMed]
- Agawin, N.S.R.; Rabouille, S.; Veldhuis, M.J.W.; Servatius, L.; Hol, S.; van Overzee, H.M.J.; Huisman, J. Competition and Facilitation between Unicellular Nitrogen-Fixing Cyanobacteria and Non—Nitrogen-Fixing Phytoplankton Species. Limnol. Oceanogr. 2007, 52, 2233–2248. [Google Scholar] [CrossRef]
- Newell, S.E.; Davis, T.W.; Johengen, T.H.; Gossiaux, D.; Burtner, A.; Palladino, D.; McCarthy, M.J. Reduced Forms of Nitrogen Are a Driver of Non-Nitrogen-Fixing Harmful Cyanobacterial Blooms and Toxicity in Lake Erie. Harmful Algae 2019, 81, 86–93. [Google Scholar] [CrossRef]
- Chapman, A.D.; Schelske, C.L. Recent Appearance of Cylindrospermopsis (Cyanobacteria) in Five Hypereutrophic Florida Lakes. J. Phycol. 1997, 33, 191–195. [Google Scholar] [CrossRef]
- Burchardt, L.; Marshall, H. Algal Composition and Abundance in the Neuston Surface Micro Layer From a Lake and Pond in Virginia (U.S.A.). J. Limnol. 2003, 62, 139–142. [Google Scholar] [CrossRef]
- Awada, H.; Aronica, S.; Bonanno, A.; Basilone, G.; Zgozi, S.W.; Giacalone, G.; Fontana, I.; Genovese, S.; Ferreri, R.; Mazzola, S.; et al. A Novel Method to Simulate the 3D Chlorophyll Distribution in Marine Oligotrophic Waters. Commun. Nonlinear Sci. Numer. Simul. 2021, 103, 106000. [Google Scholar] [CrossRef]
- Zhang, J.; Zhi, M.; Zhang, Y. Combined Generalized Additive Model and Random Forest to Evaluate the Influence of Environmental Factors on Phytoplankton Biomass in a Large Eutrophic Lake. Ecol. Indic. 2021, 130, 108082. [Google Scholar] [CrossRef]
- Derot, J.; Yajima, H.; Schmitt, F.G. Benefits of Machine Learning and Sampling Frequency on Phytoplankton Bloom Forecasts in Coastal Areas. Ecol. Inform. 2020, 60, 101174. [Google Scholar] [CrossRef]
- Cheng, Y.; Bhoot, V.N.; Kumbier, K.; Sison-Mangus, M.P.; Brown, J.B.; Kudela, R.; Newcomer, M.E. A Novel Random Forest Approach to Revealing Interactions and Controls on Chlorophyll Concentration and Bacterial Communities during Coastal Phytoplankton Blooms. Sci. Rep. 2021, 11, 19944. [Google Scholar] [CrossRef]
- Yajima, H.; Derot, J. Application of the Random Forest Model for Chlorophyll-a Forecasts in Fresh and Brackish Water Bodies in Japan, Using Multivariate Long-Term Databases. J. Hydroinformatics 2017, 20, 206–220. [Google Scholar] [CrossRef]
- Carpenter, S.R.; Gurevitch, A.; Adams, M.S. Factors Causing Elevated Biological Oxygen Demand in the Littoral Zone of Lake Wingra, Wisconsin. Hydrobiologia 1979, 67, 3–9. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Y.; He, G.; Han, J.; Wang, T. Exploration of Relationships between Phytoplankton Biomass and Related Environmental Variables Using Multivariate Statistic Analysis in a Eutrophic Shallow Lake: A 5-Year Study. J. Environ. Sci. 2007, 19, 920–927. [Google Scholar] [CrossRef]
- Karadžić, V.; Simić, G.S.; Natić, D.; Ržaničanin, A.; Ćirić, M.; Gačić, Z. Changes in the Phytoplankton Community and Dominance of Cylindrospermopsis raciborskii (Wolosz.) Subba Raju in a Temperate Lowland River (Ponjavica, Serbia). Hydrobiologia 2013, 711, 43–60. [Google Scholar] [CrossRef]
- Kehoe, M.J.; Chun, K.P.; Baulch, H.M. Who Smells? Forecasting Taste and Odor in a Drinking Water Reservoir. Environ. Sci. Technol. 2015, 49, 10984–10992. [Google Scholar] [CrossRef] [PubMed]
- Taboada-Castro, M.M.; Diéguez-Villar, A.; Taboada-Castro, M.T. Transfer of Nutrients and Major Ions of an Agricultural Catchment to Runoff Waters: Analysis of Their Spatial Distribution. In Proceedings of the Conserving Soil and Water for Society: Sharing Solutions, Brisbane, Australia, 4–8 July 2004; pp. 1–4. [Google Scholar]
- Varol, M. Phytoplankton Functional Groups in a Monomictic Reservoir: Seasonal Succession, Ecological Preferences, and Relationships with Environmental Variables. Environ. Sci. Pollut. Res. 2019, 26, 20439–20453. [Google Scholar] [CrossRef] [PubMed]
- Lindner, T.; Puck, J.; Verbeke, A. Beyond Addressing Multicollinearity: Robust Quantitative Analysis and Machine Learning in International Business Research. J. Int. Bus. Stud. 2022, 53, 1307–1314. [Google Scholar] [CrossRef]
- Darst, B.F.; Malecki, K.C.; Engelman, C.D. Using Recursive Feature Elimination in Random Forest to Account for Correlated Variables in High Dimensional Data. BMC Genet. 2018, 19, 65. [Google Scholar] [CrossRef]
- Bøvelstad, H.M.; Nygård, S.; Størvold, H.L.; Aldrin, M.; Borgan, Ø.; Frigessi, A.; Lingjærde, O.C. Predicting Survival from Microarray Data—A Comparative Study. Bioinformatics 2007, 23, 2080–2087. [Google Scholar] [CrossRef]
- Sauerbrei, W.; Boulesteix, A.-L.; Binder, H. Stability Investigations of Multivariable Regression Models Derived from Low- and High-Dimensional Data. J. Biopharm. Stat. 2011, 21, 1206–1231. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Y.; Wallace, J.; Campbell, E. Estimating Annual Runoff in Response to Forest Change: A Statistical Method Based on Random Forest. J. Hydrol. 2020, 589, 125168. [Google Scholar] [CrossRef]
- Ransom, C.J.; Kitchen, N.R.; Camberato, J.J.; Carter, P.R.; Ferguson, R.B.; Fernández, F.G.; Franzen, D.W.; Laboski, C.A.M.; Myers, D.B.; Nafziger, E.D.; et al. Statistical and Machine Learning Methods Evaluated for Incorporating Soil and Weather into Corn Nitrogen Recommendations. Comput. Electron. Agric. 2019, 164, 104872. [Google Scholar] [CrossRef]
- Brönmark, C.; Hansson, L.-A. Environmental Issues in Lakes and Ponds: Current State and Perspectives. Environ. Conserv. 2002, 29, 290–307. [Google Scholar] [CrossRef]
- Chopyk, J.; Allard, S.; Nasko, D.J.; Bui, A.; Mongodin, E.F.; Sapkota, A.R. Agricultural Freshwater Pond Supports Diverse and Dynamic Bacterial and Viral Populations. Front. Microbiol. 2018, 9, 792. [Google Scholar] [CrossRef] [PubMed]
- Merem, E.C.; Isokpehi, R.; Foster, D.; Wesley, J.; Nwagboso, E.; Romorno, C.; Richardson, C. Using Geo-Information Systems in Assessing Water Quality in the Mid-Atlantic Region Agricultural Watershed of Maryland. Int. J. Ecosyst. 2012, 2, 112–139. [Google Scholar] [CrossRef]
- Binding, C.E.; Pizzolato, L.; Zeng, C. EOLakeWatch; Delivering a Comprehensive Suite of Remote Sensing Algal Bloom Indices for Enhanced Monitoring of Canadian Eutrophic Lakes. Ecol. Indic. 2021, 121, 106999. [Google Scholar] [CrossRef]
- Burford, M.A.; Carey, C.C.; Hamilton, D.P.; Huisman, J.; Paerl, H.W.; Wood, S.A.; Wulff, A. Perspective: Advancing the Research Agenda for Improving Understanding of Cyanobacteria in a Future of Global Change. Harmful Algae 2020, 91, 101601. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).