Abstract
Microbial communities are vital components of freshwater ecosystems due to their role in nutrient cycling and energy flow; however, the mechanisms driving their variation are still being explored. In aquatic systems, water flow (hydrology) can impact microbial community composition through community connectivity; however, the details of hydrology’s effects on microbial connectivity remain unclear. To address this question, we used 16S rRNA metabarcoding to determine bacterial community composition and connectivity across flow transects in three connected Great Lakes waterbodies with very different water-flow regimes: the Little River (high flow), the Detroit River (moderate flow), and Lake Erie (low flow). Bacterial alpha diversity (Chao1) did not differ among the three locations or sample sites along the transects. Analyses of beta diversity using community dissimilarity matrices identified significant differences among the three locations and among sample sites within locations. Bacterial community connectivity varied among the three locations, with a significant distance–decay relationship observed only in the low-flow location, which is indicative of connectivity driven by spatial proximity. Directional analyses showed that the water-flow direction affected bacterial similarity, consistent with the expected hydrological effects on community connectivity and previous published work. Our results indicate that (1) microbial community composition varies within and among even geographically close sampling locations and (2) the specific water-flow regime appears to affect bacterial community connectivity. Including hydrology in models of bacterial community composition will improve our understanding of the relative roles of selection versus stochastic effects on bacterial community diversity and composition in freshwater ecosystems.
1. Introduction
Microorganisms play a pivotal role in ecosystems where their community composition and activity directly influence biogeochemical cycles and energy flow [1,2]. A major component of the whole ecosystem energy budget is transferred through the microbial community, making them critical, but often overlooked, members of the aquatic food web [3]. Thus, the composition, diversity, and function of aquatic microbial communities can dictate ecosystem-level biogeochemical cycling and, since those biogeochemical processes affect ecosystem function, ecosystem health, and stability. Although much research has been focused on the composition of microbial communities in widely diverse ecosystems, the forces driving microbial community variation (i.e., bacterial community composition) and the mechanisms behind spatial variation in community composition still require further characterization.
Many factors contribute to the variation observed in aquatic bacterial community composition among sites and ecosystems [4]. Bacterial communities can be very specialized, with high levels of spatial and temporal variation, where significant differences can be found even a few meters apart in the water column [5]. While the factors driving bacterial community variation are not always resolved, they generally include both biotic and abiotic factors [4,6]. Furthermore, the ongoing discussion of the relative contributions of environmental selection versus ecological drift (neutral factors) indicates that both processes are important, and their relative contribution is context dependent [7,8]. One widely recognized factor contributing to variation in the composition of bacterial communities (especially aquatic ones) is connectivity, or bacterial dispersal among communities [9,10]. Indeed, microbial connectivity is widely recognized as an important driver of community structure across a diverse array of habitats, e.g., Arctic watersheds [11], floodplain soils [12], the Laurentian Great Lakes [13], agricultural and urban land/waterscapes [14,15], and even deep-sea sediments [16]. Thus, while patterns of microbial connectivity have been reported, there is less known about how it contributes to the mechanisms driving bacterial community composition in aquatic habitats.
In aquatic ecosystems, microbial community composition is affected by key environmental factors, such as water chemistry [17], season [18], pH [19], carbon availability [20], and biotic interactions [4,21], among others. However, the hydrology of the ecosystem can also have profound effects on the structure of microbial communities [13,22,23]. For example, high water velocity was reported to reduce microbial adhesion and the formation of biofilms relative to habitats with lower water velocities [22,24]. Microbial community connectivity fundamentally depends on the rate of microbial dispersal among patches within an environment [25,26]. Thus, high habitat connectivity (e.g., due to high water flow) is expected to result in reduced beta diversity across aquatic landscapes. However, connectivity is also strongly affected by landscape characteristics and structure, as well as the mobility of organisms within the landscape [12,27]. Since most microbes are non-motile [19], they depend on passive means of transportation such as water flow [19,28]. Thus, specific hydrological conditions may be important in determining microbial community connectivity because water flow can facilitate microbial dispersal in aquatic ecosystems [12].
The goal of this study was to characterize bacterial community composition and connectivity within and among aquatic habitats that differed in water-flow regimes. We predicted that higher water-flow habitats will exhibit greater connectivity and, hence, more homogeneous microbial community composition relative to lower-flow sites, since aquatic microbial community connectivity is expected to be affected by water flow. To investigate this, we characterized bacterial community composition using 16S rRNA metabarcoding along flow transects in three physically connected water bodies in the Huron–Erie corridor of the Laurentian Great Lakes. Specifically, we sampled the Little River (a high-flow tributary of the Detroit River), the Detroit River (a moderate-flow tributary of Lake Erie), and Lake Erie (low flow). We tested two hypotheses: (1) bacterial community composition and diversity would vary among the sample locations due to habitat differences associated with the flow (and possibly other abiotic differences) and (2) low-flow locations would exhibit lower connectivity relative to higher-flow locations and, hence, greater bacterial community divergence among sample sites. While hydrology has been previously explored as a factor in microbial community composition (e.g., [11,13]), few studies have addressed water-flow effects on bacterial communities at fine spatial scales. If bacterial community composition is significantly affected by fine spatial scale (<1000 m) hydrological effects, then factors such as dispersal (connectivity) and ecological drift will need to be included in models of microbial ecosystem dynamics. Additionally, such fine-scale effects should inform future lotic versus lentic water-body sampling design for microbial community analyses. Thus, our results address issues in basic microbial ecology as well as applications of metabarcoding for aquatic microbiome monitoring.
2. Materials and Methods
This research was conducted as part of a senior undergraduate course in Evolutionary, Ecological, and Environmental Genetics in March 2019, where students contributed to the design, execution, and analyses of the project and data. The applications of eDNA metabarcoding (e.g., bacterial metabarcoding) are far-reaching, and early practical experiences in these technologies are a critical part of genetics and environmental education.
2.1. Sampling
Water samples were collected on 21–23 March 2019 in three days from three connected water bodies in the Huron–Erie corridor of the Laurentian Great Lakes in Southwestern Ontario, Canada (Figure 1). The three sampling locations varied considerably in flow rates: the Little River, a tributary to the upper Detroit River (“LR”, mean flow rate = 1.2 m/s; Table 1), the Detroit River, a tributary of Lake Erie (“DR”, mean flow rate = 0.67 m/s; Table 1), and Lake Erie (“LE”, mean flow rate = 0.51 m/s; Table 1). While the three sampling locations vary in hydrological flow, they are all within a 50 km radius and connected by water flow along the Huron–Erie corridor (Figure 1). At each location, we sampled water at ∼0.5 m depth along a flow transect, taking two replicate samples per site (500 mL each), and the geographical position was determined using GPS. The sampling gear and water bottles were cleaned and sterilized with a 10% bleach treatment followed by 2–3 rinses with ddH2O in the lab before each sampling event. In Lake Erie, the sample sites were approximately two kilometers apart along the shore transect, but the Detroit River and Little River sampling site distances varied due to access limitations (Figure 1). In the Detroit River, there was a gap between two sampling sites of approximately 6.24 km due to industrial sites preventing access to the river. Overall, sample sites spanned ~6.0 km in Little River, ~19 km in Detroit River, and ~22 km in Lake Erie (Figure 1). Field negative controls were conducted at the first, third, and tenth sampling sites along each transect to test for field contamination. The field negative controls consisted of ddH2O transferred from one sterile bottle to another and were otherwise treated the same as the other samples in the field and the lab. Overall, 60 samples (20 samples per sampling location (LR, DR, and LE)) were collected. The samples were held on ice and filtered (500 mL) within two hours of collection using 0.2 µm 47 mm diameter polycarbonate filters (Isopore™, Millipore, MA, USA). The filters were stored at −20 °C until DNA extractions were performed.
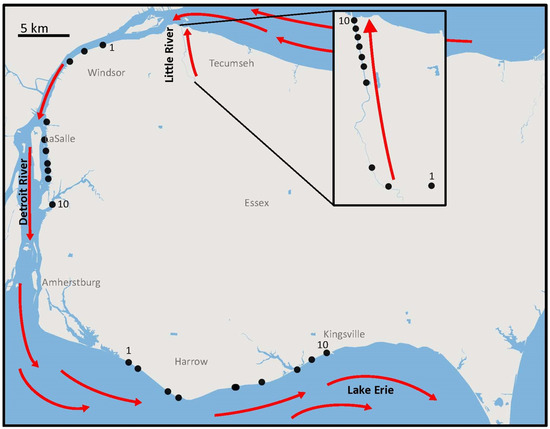
Figure 1.
Map of water collection sites and locations. Map showing the 10 water collection sites at three locations (Little River (inset)), Detroit River, and Lake Erie in the Huron–Erie corridor of the Laurentian Great Lakes. General flow patterns are shown with red arrows and the first and last (upstream to downstream) sample-site numbers are indicated.

Table 1.
Summary of abiotic characteristics of the three sampled locations.
2.2. DNA Extraction and Library Preparation
DNA was extracted from the filters using a sucrose lysis buffer method, following Shahraki et al. [30]. The V5 (787 F-acctgcctgccg-ATTAGATACCCNGGTAG) and V6 (1046 R-acgccaccgagc-CGACAGCCATGCANCACCT) variable regions of the 16S rRNA were selected for PCR amplification as described in Shahraki et al. [30]. Briefly, the first round of PCR amplified the V5–V6 region of the 16S microbial ribosomal RNA. We included a single lab negative control PCR that had ddH2O instead of extracted DNA as the template to test for lab-based contamination. The PCR products (including lab and field negative controls) were cleaned using Sera-Mag Magnetic Beads [30] to remove short strands of DNA, primers, and dNTPs. The cleaned PCR products were used as a template for a second-round short-cycle PCR to ligate the adaptor and sample identity (barcode) sequences for high throughput sequencing (HTS). After the second round of PCR, amplicons were pooled using various volumes based on their band intensity, and the combined PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN, Toronto, ON, Canada). The gel-extracted library was quantified using an Agilent 2100 Bioanalyzer with a High Sensitivity DNA kit (Agilent Technologies, Mississauga, ON, Canada) to determine the library concentration and purity. The library was then diluted to 60 pmol·μL−1 and sequenced on an ION Torrent PGM high throughput sequencer using the Ion PGM™ Sequencing 400 bp chemistry and an Ion 318™ Chip (Thermo Fisher Scientific, Burlington, ON, Canada).
2.3. Data Processing
The FASTQ raw sequence file was analyzed using the Quantitative Insights Into Microbial Ecology (QIIME2-2020.11) platform [31]. The sequence file was demultiplexed using cutadapt demux-single to remove barcodes and primers. The cutadapt trim-single command was also used to identify and remove the adapters [32], and perform a sequence quality check. The DADA2 pipeline [33] was used to denoise the sequences, dereplicate, filter chimeras, and for amplicon sequence variant (ASV) picking. We used 260bp for the read truncation length (p-trunc-len 260), but for all other parameters, default values were used. Taxonomic classification was done using the feature-classifier classify-consensus-blast plugin with the Greengenes (13_8) [34,35] reference database. The table was summarized with the feature-table summarize command. Using the taxa filter table, ASVs related to mitochondria, chloroplasts, eukaryotes (combined: 0.0003% of total sequence reads), and unassigned sequences (0.003% of total sequence reads) were further removed. ASVs that showed up in only one sample or had fewer than 2 reads were removed using the filter-features command in QIIME2, as these ASVs may not represent real biological diversity but rather PCR or sequencing artifacts. For calculating alpha diversity indices, sequence data for each sample were rarefied to 3000 sequences, as most of the samples were at the plateau of their rarefaction curves at this read depth.
2.4. Statistical Analyses
We analyzed the bacterial community data to determine (1) if there are differences in bacterial community composition within and among our three sampling locations (i.e., LR, DR, and LE) and (2) the extent of connectivity among sample sites within each location. These analyses were conducted using both alpha and beta diversity indices.
2.4.1. Community Composition
We estimated Chao 1 (alpha diversity) for each replicate at each site using QIIME2. Linear mixed-effects models (lme4) in R (Version R-4.0.5) were used to test for differences in sample alpha diversity (Chao1) at two hierarchical levels. First, we tested for differences among locations (LR, DR, and LE) using a nested model with the sample site (1–10) nested within the location and replicates nested within the sample site. We then tested for differences among the sample sites within each location separately using three lme4 models with the sample site and replicates (nested within the sample site). To visualize variation in alpha diversity, we combined replicates and plotted Chao1 for each site for each location against their relative spatial distance downstream from the most upstream sample site.
We calculated the Bray–Curtis dissimilarity matrix across sites and replicates for all three locations in QIIME2 and used that for a principal coordinate analysis (PCoA) to assess differences in patterns of species abundance among sites and locations. We plotted the PCoA axes 1 and 2 for the 60 samples (30 sites plus replicates) on a scatterplot to visualize location and site bacterial community variation. We selected PCoA axes 1 and 2 as our beta diversity indices. They explained high levels of variance (22% and 13%, respectively), and their eigenvalues were greater than one (4.0 and 2.3, respectively). Similar to the Chao1 analysis, we used PCoA axes 1 and 2 as dependent variables in a global lme4 model to test for differences in beta diversity PCoA values among locations, among sample sites nested within each location, and with sample replicates nested within each sample site. That analysis was followed by three lme4 analyses, one for each of the three locations testing for the effects of the sample site and replicating nested within the sample site.
Additionally, we used PERMANOVA in R and Bray–Curtis dissimilarity matrices to test for beta diversity differences among locations and sample sites; sampling replicates were nested within the sampling site and sample sites were nested within the locations. We also used three separate PERMANOVAs to test for sample-site effects for each of the three locations separately, with replicates nested within the sampling site. These three sub-models were used to test for location-specific differences in beta diversity among sampling sites.
2.4.2. Community Connectivity
After combining replicate samples at each site, we tested for a distance–decay model of pairwise geographical distance versus dissimilarity in bacterial community composition (i.e., connectivity) using a Mantel test with the Bray–Curtis dissimilarity distance matrix and pairwise Euclidean distance generated using the geographical coordinates of the sample sites at all three locations. Specifically, we tested for three distance–decay relationships (one within each location separately) with pairwise dissimilarity among sample sites within the three locations using a Mantel test and the Bray–Curtis dissimilarity distance matrix and pairwise Euclidean distance.
To assess directional patterns of bacterial community connectivity (i.e., downstream flow), we tested for changes in bacterial community diversity along the sampling transects from upstream to downstream using a regression analysis of the distance downstream versus Chao1 (IBM SPSS Statistics, Vs 29.0.1.0). We performed a similar analysis using mean pairwise Bray–Curtis dissimilarity (from the most upstream site to each downstream site) versus distance downstream to assess patterns of community divergence with distance. We plotted measures of alpha and beta diversity against distance downstream for all three locations.
Finally, we characterized the bacterial taxa contributing to sample-site bacterial community similarity (connectivity). We first square-root transformed the bacterial ASV’s normalized read count data in the software package PRIMER (v.7) and then used the Similarity Percentage (SIMPER) analysis software package in PRIMER (v.7) to determine which ASVs (i.e., taxa) were contributing to patterns of Bray–Curtis similarity among the sample sites [36]. We reanalyzed the data at the family level in SIMPER to identify broad categories of bacteria contributing to differences among sites within the three locations.
3. Results
A total of 3,103,650 sequences were obtained for the 60 experimental samples (30 replicated sites), with a median frequency of 16,630 reads. The nine negative field controls resulted in a total of fewer than 1000 reads with no particularly abundant taxa identified, while the negative lab control generated no sequence reads. Thus, the controls were excluded from our analyses, and we concluded our data were not biased by contamination.
3.1. Bacterial Community Composition
The bacterial communities across all three locations consisted predominantly of the orders Burkholderiales (range = 6% to 77%), Desulfovibrionales (range = 0% to 80%), and Lactobacillales (range = 0% to 30%), although there was considerable variation among the sample-site bacterial community compositions at the order level (Figure 2).
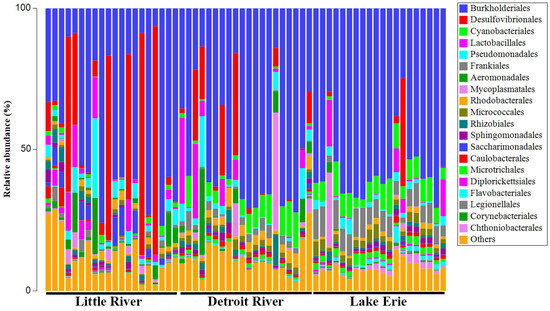
Figure 2.
Bacterial community composition. Relative abundance of bacterial community taxa at the Order taxonomic level for the three sampled locations (LR, DR, and LE) determined by 16S metabarcoding. Each location had 10 sample sites with two replicates at each sample site. The sample site relative abundance bars are arranged in downstream order. The ‘other taxa’ category includes the sum of all bacterial taxa that occurred at less than 5% relative abundance.
Our global linear mixed-effects model showed that alpha diversity (Chao1) did not differ among locations or sampling sites within locations (Table 2). When we tested for sample-site effects on Chao1 within each location separately, no significant effects were found, although the Lake Erie location approached significance (p = 0.071; Table 3).

Table 2.
Sample location and site effects on bacterial community diversity. Results of linear mixed-effects (lme) model analyses for the effects of sample location (LR, DR, and LE) and sample site nested within the location on bacterial community alpha (Chao1) and beta diversity (PCoA 1 and 2 from Bray–Curtis dissimilarity matrix). Significant p-values are in bold.

Table 3.
Sample-site effects on bacterial community diversity. Results of linear mixed-effects (lme) model analyses for the effects of sample site (within each location separately) on bacterial community alpha (Chao1) and beta (PCoA 1 and 2 from Bray–Curtis dissimilarity matrix) diversity. Significant p-values are indicated in bold.
Bacterial community beta diversity was well characterized by the first two PCoA axes from the Bray–Curtis matrix; PCoA 1 and PCoA 2 explained 22% (Eigenvalue = 4.0) and 13% (eigenvalue = 2.3) of the variance in the Bray–Curtis dissimilarity matrix, respectively. The PCoA plot shows LE and LR samples clustered separately; however, the DR samples were variable and overlapped both LE and LR samples (Figure 3). As expected, the global linear mixed-effects model showed highly significant location effects for both PCoA 1 and PCoA 2; however, no significant sample-site (nested within location) effects were found in that analysis (Table 2). When we considered the locations separately, the linear mixed-effects analyses showed significant site effects on PCoA 1 for the DR location (p = 0.042) and on PCoA 2 for the LE location (p = 0.013) but not for any other comparisons (Table 3).
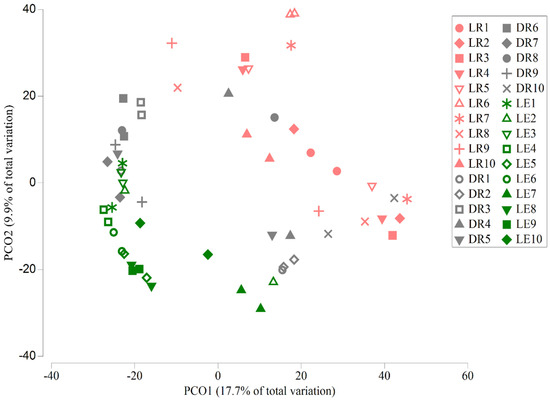
Figure 3.
PCoA plot of bacterial communities from three sample locations. Scatterplot of PCoA axes 1 and 2 for bacterial community Bray–Curtis similarity matrices. Water was sampled from 10 sites (with two replicates) at each of the three locations (LR, DR, and LE); all 60 water sample bacterial communities are shown.
The nested PERMANOVA analysis resulted in significant effects of both location and sample site (nested within location) on bacterial community composition dissimilarity (Bray–Curtis dissimilarity matrix; Table 4). The location effect explained 18% of the variation in Bray–Curtis similarity, while the site nested within the location explained 45% of the variation (Table 4).

Table 4.
Location and site effects on bacterial community beta diversity. Results of PERMANOVA analysis for the effects of sample location (LR, DR, and LE) and sample site nested within location on bacterial community beta diversity (Bray–Curtis dissimilarity matrix). Significant p-values are indicated in bold.
The separate PERMANOVA analyses for the three locations also showed highly significant site effects on bacterial community composition dissimilarity for LE and DR (Bray–Curtis dissimilarity matrix; Table 5). The DR sample-site effect explained 58% of the variation in the pairwise Bray–Curtis dissimilarity, while the LE sample-site effect explained 68% of the variance (Table 5).

Table 5.
Sample-site effects on bacterial community beta diversity. Results of PERMANOVA analyses for the effects of sample site (within each location separately) on bacterial community beta diversity (Bray–Curtis dissimilarity matrix). Significant effect p-values are indicated in bold.
3.2. Bacterial Community Connectivity
We tested for a distance–decay (isolation-by-distance) model of bacterial community dissimilarity using Mantel correlation analyses of pairwise Bray–Curtis community dissimilarity and Euclidean geographic distance matrices (Figure 4). Significant distance–decay relationships reflect connectivity among sample sites, since zero connectivity among bacterial communities would result in community divergence developing independent of distance (hence no distance–decay relationship). The Mantel test across all samples resulted in geographical distance having a significant correlation with bacterial community Bray–Curtis similarity patterns (Mantel test: Rho: 0.176, p < 0.05). When we tested for distance–decay relationships within each location separately, the Mantel correlation analyses returned a significant correlation for LE, but no evidence for distance–decay relationships for either LR or DR (LR Mantel test: Rho = 0.12, p = 0.22; DR Mantel test: Rho = 0.21, p = 0.11; and LE Mantel test: Rho = 0.34, p = 0.007). The significant relationship between Bray–Curtis dissimilarity and geographic distance among the Lake Erie sites is consistent with a distance–decay effect (Figure 4).
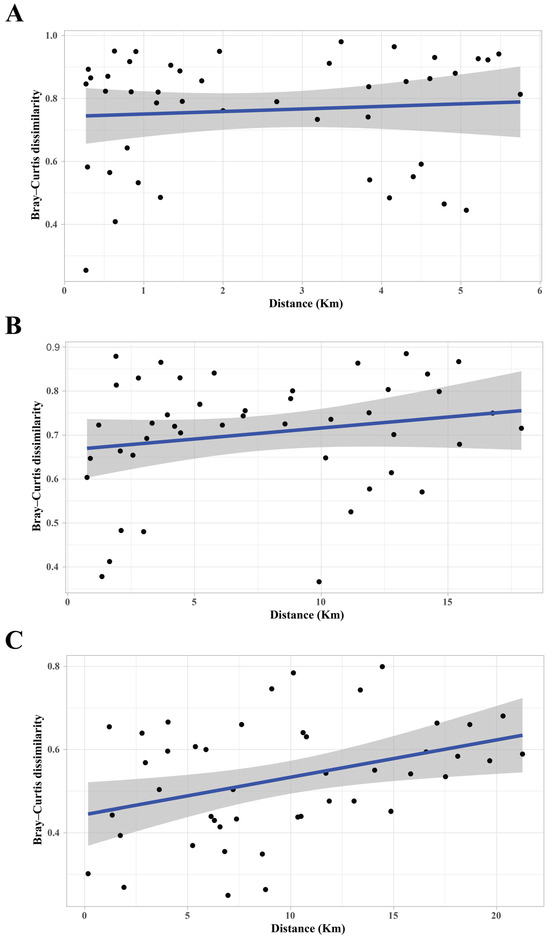
Figure 4.
Distance–decay relationships among sites within three sample locations. Distance–decay plots showing pairwise geographic distance (km) versus bacterial community dissimilarity (Bray–Curtis) for 10 sample sites (replicates combined) within three sampled locations; LR (A, high flow), DR (B, moderate flow), and LE (C, low flow). Only the LE location generated a significant positive relationship using a Mantel test (p = 0.007). The plotted line is the least-square best-fit linear regression line. The gray shading around the line represents a confidence interval of 95%.
Our directionality analysis of within-location bacterial community alpha diversity (Chao1) showed declining diversity with distance downstream at the LR location (p < 0.004, R2 = 0.62 (Chao1)) but not for either the DR or LE locations. The directionality analysis for beta diversity and distance downstream showed no significant linear relationship at any of the three locations; however, considerable variation was evident among sites within the three locations.
SIMPER analysis at the ASV level with the replicates combined showed an average similarity of 21.8 among sites for the Little River, 30.9 for the Detroit River, and 39.9 for the Lake Erie samples. The main ASV contributors to similarity within all three locations were from the families Comamonadaceae (class: Betaproteobacteria) and Flavobacteriaceae (class: Flavobacteriia).
4. Discussion
The composition of aquatic bacterial communities is determined by a variety of biotic and abiotic factors [4,37]. While published studies have reported diverse factors that contribute to aquatic bacterial community composition (e.g., [4,37], our study was designed to determine the contribution of the water-flow regime to variation in aquatic bacterial community composition and connectivity (i.e., similarity) in the Huron–Erie corridor region of the Great Lakes. Specifically, we assessed the connectivity and composition of aquatic bacterial community composition along transects in high-flow (Little River (LR)), moderate-flow (Detroit River (DR)), and low-flow (Lake Erie (LE)) aquatic habitats in Southern Ontario, Canada. The common core bacterial taxa found across locations and sites in our study (i.e., Betaproteobacteria, Flavobacteria, and Actinobacteria) are consistent with previous freshwater-ecosystem studies that reported similar conserved taxa [38,39]. However, those taxa are generally abundant in diverse aquatic ecosystems and are, thus, likely highly persistent [38], making them poor candidates for bacterial community connectivity analyses. The abundance of those taxa may have contributed to the lack of variation in bacterial community alpha diversity among sample sites or locations, despite our expectation that different flow regimes and other habitat characteristics would drive substantial differences in bacterial community diversity. There were, however, substantial differences in bacterial community composition among the three locations, and among sample sites within each location, likely reflecting habitat and hydrological differences among the sample sites (Table 1). We detected variation in bacterial community connectivity within our three sample locations, and the pattern of connectivity generally agreed with our predictions. The high-flow LR samples showed the highest connectivity (highest sample-site similarity), and the sample sites in the DR and LE exhibited similar levels of bacterial community connectivity. While we cannot conclude the observed differences were due to hydrology alone, the pattern of connectivity is consistent with a water-flow threshold effect where high flow rates (i.e., LR mean flow = 1.2 m/s) result in elevated connectivity while lower flows (DR = 0.67 m/s, LE = 0.51 m/s) result in similar connectivity. Our work serves to highlight the potential for hydrology to affect aquatic bacterial community connectivity and, hence, bacterial community composition.
We observed substantial variation in bacterial community composition but not diversity among the three sampled locations (LR, DR, and LE). While the pattern of location effects was consistent with our predictions based on hydrological differences, the three sampled locations likely also differed in other biotic and abiotic factors [5]. However, our reported patterns of community divergence are not likely due to spatial distance effects alone, as the three locations were separated by a maximum of 50 km and are connected as part of the Huron–Erie corridor [40]. Furthermore, other studies have generally shown that distance effects on bacterial community composition require greater spatial scales [4,37,41]. While we specifically selected the three locations to differ in water flow, we cannot conclusively determine the exact contribution of flow to the observed variation in the bacterial communities. The within-location patterns of bacterial community composition are suggestive of a hydrological effect, as the lower-flow locations (Detroit River and Lake Erie) showed the highest level of among-site divergence, which is consistent with the low connectivity at those low-flow locations.
Connectivity among ecological communities is the fundamental driver of the metacommunity concept in community ecology, where interactions among local communities determine the pattern of community composition [26]. Variation in microbial diversity and community composition in connected water systems is a well-known phenomenon. For example, researchers have reported a pattern of high diversity in headwater stream bacterial communities with decreasing diversity downstream [28,42], which is similar to the pattern we report for LR alpha diversity. This change in diversity downstream may be due to species sorting, where the community experiences a loss of terrestrial taxa and a gain in the prevalence of common aquatic taxa [14,43]. Read et al. [44] described a downstream shift in the River Thames Basin (UK) bacterial communities and speculated it was due to ecological succession effects. However, it is unlikely that the pattern of reduced downstream bacterial community diversity in LR is due to ecological succession, since the flow rate was high and the distance low.
Although all sites exhibited similar dominant taxa, we found significant variation in bacterial community composition between sites within a given location, which was an unexpected result, as our sampling transects had a maximum spatial extent of ~20km (Figure 1). The observed pattern of beta diversity variation was consistent with a water-flow rate effect on microbial connectivity as a high flow rate was associated with high microbial community connectivity (and thus low divergence), as demonstrated in the LR site samples. This agrees with the recent findings of Luo et al. [45], who reported that higher flow rates increased bacterial community coalescence and connectivity. This effect varied with season (with greater influence in summer than winter) and with habitat (affecting the water column and adjacent soil bacteria more than sediment and biofilm communities) [45]. Similarly, Stadler and del Giorgio [43] reported strong terrestrial–aquatic connectivity and the importance of the upstream bacterial communities in determining community composition and function; they also reported seasonal fluctuations in the extent and nature of the connectivity. It is possible that the temporal fluctuations reported in aquatic bacterial community composition may, in part, reflect changes in connectivity associated with hydrological seasonality. Finally, it is important to note that the patterns of variation in bacterial community composition reported here may reflect different sampling spatial scales—the Little River was sampled over ~5 km, while both the Detroit River and Lake Erie were sampled over ~20 km. While this transect length difference is substantial, the spatial scale of our sampling is relatively small, and hydrological flow is likely to be a larger factor in determining connectivity among sites.
Spatial surveys of aquatic bacterial community composition and diversity are common in the literature, and many have identified stark contrasts in occurrence patterns of some freshwater bacterial taxa [46,47,48]. Generally, those studies have concluded that environmental factors are dominant in determining the bacterial community composition of aquatic ecosystems [47,48] and that dispersal limitation and biogeography are less important [28,49]. However, robust biogeographic patterns, such as distance–decay of community similarity and taxa–area relationships, have been reported in aquatic microbes [50] and other microbial systems [51]. Bacterial community composition variation and distance–decay relationships are often a balancing act between dispersal rates and environmental selection, with the dominant force varying depending on the characteristics of the water body [52]. Nevertheless, in systems with high dispersal rates (i.e., high water flow) the composition of the bacterial community is expected to consist primarily of colonizers that have traveled from upstream communities [52]. In our study, the high flow rate (LR) and moderate flow rate (DR) locations consistently exhibited greater sample-site similarity, minimal beta diversity, and no evidence for distance–decay relationships relative to the relatively low-flow LE location. The role of hydrological flow in determining connectivity in aquatic microbial communities is potentially important, especially for stream rehabilitation applications, where bacterial community function is critical for stream health but could be affected by upstream conditions in complex ways.
The role of connectivity in the formation and maintenance of communities across all ecosystem scales is recognized as pivotal, especially for conservation. Given the fundamental role of microbial communities in aquatic ecosystem health, a more detailed picture of microbial connectivity in aquatic ecosystems needs to be developed. In this study, three aquatic ecosystems that are connected and geographically close were selected to determine the effects of hydrological flow on measures of bacterial community connectivity. We tested whether bacterial community composition and diversity would vary among and within the sampled locations despite the limited geographical scale and whether low-flow locations would exhibit lower connectivity relative to higher-flow locations. Additionally, our work highlights the potential for hydrologically driven variation in connectivity as a contributor to bacterial community composition variation and, hence, as an important factor to consider when interpreting community change in healthy and impacted aquatic ecosystems.
Author Contributions
J.S., C.J.V. and D.D.H. conceived and planned the experiments. J.S., S.W., J.W., D.M., E.B. and C.F. carried out fieldwork. J.S. carried out the wet laboratory sample preparations and experiments. All authors contributed to selecting the models and the computational framework for the data analysis. J.S., C.J.V. and D.D.H. contributed to the interpretation of the results. J.S. took the lead in performing the research and analyzing data. All the authors contributed to writing the manuscript. J.S., C.J.V. and D.D.H. provided critical feedback and helped shape the research, analysis, and manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by a Natural Science and Engineering Research Council of Canada grant (819047) to DDH and by the Department of Integrative Biology, University of Windsor.
Data Availability Statement
Sequences were deposited in the NCBI Short Read Archive (SRA) (BioProject ID: PRJNA983509).
Acknowledgments
We thank the students of the 2019 Evolutionary, Ecological, and Environmental Genetics special topics class for their assistance with sampling, sample processing, and analyses.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The Microbial Engines That Drive Earth’s Biogeochemical Cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A.; Cram, J.A.; Needham, D.M. Marine Microbial Community Dynamics and Their Ecological Interpretation. Nat. Rev. Microbiol. 2015, 13, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Kaunzinger, C.M.K.; Morin, P.J. Productivity Controls Food-Chain Properties in Microbial Communities. Nature 1998, 395, 495–497. [Google Scholar] [CrossRef]
- Sadeghi, J.; Chaganti, S.R.; Shahraki, A.H.; Heath, D.D. Microbial Community and Abiotic Effects on Aquatic Bacterial Communities in North Temperate Lakes. Sci. Total Environ. 2021, 781, 146771. [Google Scholar] [CrossRef] [PubMed]
- Shahraki, A.H.; Chaganti, S.R.; Heath, D. Spatio-Temporal Dynamics of Bacterial Communities in the Shoreline of Laurentian Great Lake Erie and Lake St. Clair’s Large Freshwater Ecosystems. BMC Microbiol. 2021, 21, 253. [Google Scholar] [CrossRef] [PubMed]
- Pearman, J.K.; Thomson-Laing, G.; Thompson, L.; Waters, S.; Vandergoes, M.J.; Howarth, J.D.; Duggan, I.C.; Hogg, I.D.; Wood, S.A. Human Access and Deterministic Processes Play a Major Role in Structuring Planktonic and Sedimentary Bacterial and Eukaryotic Communities in Lakes. PeerJ 2022, 10, e14378. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Meng, Z.; Liu, X.; Zhang, X. Microbial Assembly, Interaction, Functioning, Activity and Diversification: A Review Derived from Community Compositional Data. Mar. Life Sci. Technol. 2019, 1, 112–128. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, S.; Liu, X.; Yao, P.; Ge, T.; Zhang, X.H. Spatiotemporal Dynamics of the Archaeal Community in Coastal Sediments: Assembly Process and Co-Occurrence Relationship. ISME J. 2020, 14, 1463–1478. [Google Scholar] [CrossRef]
- Curren, E.; Leong, S.C.Y. Natural and Anthropogenic Dispersal of Cyanobacteria: A Review. Hydrobiologia 2020, 847, 2801–2822. [Google Scholar] [CrossRef]
- Jiao, S.; Liu, Z.; Lin, Y.; Yang, J.; Chen, W.; Wei, G. Bacterial Communities in Oil Contaminated Soils: Biogeography and Co-Occurrence Patterns. Soil Biol. Biochem. 2016, 98, 64–73. [Google Scholar] [CrossRef]
- Comte, J.; Culley, A.I.; Lovejoy, C.; Vincent, W.F. Microbial Connectivity and Sorting in a High Arctic Watershed. ISME J. 2018, 12, 2988–3000. [Google Scholar] [CrossRef] [PubMed]
- Argiroff, W.A.; Zak, D.R.; Lanser, C.M.; Wiley, M.J. Microbial Community Functional Potential and Composition Are Shaped by Hydrologic Connectivity in Riverine Floodplain Soils. Microb. Ecol. 2017, 73, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Paver, S.F.; Newton, R.J.; Coleman, M.L. Microbial Communities of the Laurentian Great Lakes Reflect Connectivity and Local Biogeochemistry. Environ. Microbiol. 2020, 22, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.S.; Lu, N.; Sangwan, N.; Li, A.; Dsouza, M.; Stumpf, A.J.; Sevilla, T.; Culotti, A.; Keefer, L.L.; Kelly, J.J.; et al. Microbial Diversity in an Intensively Managed Landscape Is Structured by Landscape Connectivity. FEMS Microbiol. Ecol. 2017, 93, fix120. [Google Scholar] [CrossRef] [PubMed]
- Laperriere, S.M.; Hilderbrand, R.H.; Keller, S.R.; Trott, R.; Santoro, A.E. Headwater Stream Microbial Diversity and Function across Agricultural and Urban Land Use Gradients. Appl. Environ. Microbiol. 2020, 86, e00018-20. [Google Scholar] [CrossRef]
- Varliero, G.; Bienhold, C.; Schmid, F.; Boetius, A.; Molari, M. Microbial Diversity and Connectivity in Deep-Sea Sediments of the South Atlantic Polar Front. Front. Microbiol. 2019, 10, 665. [Google Scholar] [CrossRef] [PubMed]
- Stanish, L.F.; Hull, N.M.; Robertson, C.E.; Harris, J.K.; Stevens, M.J.; Spear, J.R.; Pace, N.R. Factors Influencing Bacterial Diversity and Community Composition in Municipal Drinking Waters in the Ohio River Basin, USA. PLoS ONE 2016, 11, e0157966. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbruck, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I.; et al. Defining Seasonal Marine Microbial Community Dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef]
- Ghosh, S.; Leff, L.G. Impacts of Labile Organic Carbon Concentration on Organic and Inorganic Nitrogen Utilization by a Stream Biofilm Bacterial Community. Appl. Environ. Microbiol. 2013, 79, 7130–7141. [Google Scholar] [CrossRef]
- Kent, A.D.; Yannarell, A.C.; Rusak, J.A.; Triplett, E.W.; McMahon, K.D. Synchrony in Aquatic Microbial Community Dynamics. ISME J. 2007, 1, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, R.; Stocker, R. Microbes in Flow. Curr. Opin. Microbiol. 2015, 25, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Campen, R.; Kowalski, J.; Lyons, W.B.; Tulaczyk, S.; Dachwald, B.; Pettit, E.; Welch, K.A.; Mikucki, J.A. Microbial Diversity of an Antarctic Subglacial Community and High-Resolution Replicate Sampling Inform Hydrological Connectivity in a Polar Desert. Environ. Microbiol. 2019, 21, 2290–2306. [Google Scholar] [CrossRef]
- Lehtola, M.J.; Laxander, M.; Miettinen, I.T.; Hirvonen, A.; Vartiainen, T.; Martikainen, P.J. The Effects of Changing Water Flow Velocity on the Formation of Biofilms and Water Quality in Pilot Distribution System Consisting of Copper or Polyethylene Pipes. Water Res. 2006, 40, 2151–2160. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.E.; Capocchi, J.K.; Albright, M.B.N.; Hao, Z.; Brodie, E.L.; Martiny, J.B.H. Routes and Rates of Bacterial Dispersal Impact Surface Soil Microbiome Composition and Functioning. ISME J. 2022, 16, 2295–2304. [Google Scholar] [CrossRef] [PubMed]
- Crevecoeur, S.; Edge, T.A.; Watson, L.C.; Watson, S.B.; Greer, C.W.; Ciborowski, J.J.H.; Diep, N.; Dove, A.; Drouillard, K.G.; Frenken, T.; et al. Spatio-Temporal Connectivity of the Aquatic Microbiome Associated with Cyanobacterial Blooms Along a Great Lake Riverine-Lacustrine Continuum. Front. Microbiol. 2023, 14, 1073753. [Google Scholar] [CrossRef]
- Choudoir, M.J.; DeAngelis, K.M. A Framework for Integrating Microbial Dispersal Modes into Soil Ecosystem Ecology. iScience 2022, 25, 103887. [Google Scholar] [CrossRef] [PubMed]
- Crump, B.C.; Amaral-Zettler, L.A.; Kling, G.W. Microbial Diversity in Arctic Freshwaters Is Structured by Inoculation of Microbes from Soils. ISME J. 2012, 6, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Lapointe, N.W.; Corkum, L.D.; Mandrak, N.E. Macrohabitat Associations of Fishes in Shallow Waters of the Detroit River. J. Fish. Biol. 2010, 76, 446–466. [Google Scholar] [CrossRef]
- Shahraki, A.H.; Chaganti, S.R.; Heath, D. Assessing High-Throughput Environmental DNA Extraction Methods for Meta-Barcode Characterization of Aquatic Microbial Communities. J. Water Health 2019, 17, 37–49. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using Qiime 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. Dada2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An Improved Greengenes Taxonomy with Explicit Ranks for Ecological and Evolutionary Analyses of Bacteria and Archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16s Rrna Gene Database and Workbench Compatible with Arb. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.R.; Ray, N.G.; Paul, J.S.; Richard, M.W. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation; Primer-E Ltd.: Plymouth, UK, 2014; p. 260. [Google Scholar]
- Langenheder, S.; Lindstrom, E.S. Factors Influencing Aquatic and Terrestrial Bacterial Community Assembly. Environ. Microbiol. Rep. 2019, 11, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Dann, L.M.; Smith, R.J.; Jeffries, T.C.; McKerral, J.C.; Fairweather, P.G.; Oliver, R.L.; Mitchell, J.G. Persistence, Loss and Appearance of Bacteria Upstream and Downstream of a River System. Mar. Freshw. Res. 2016, 68, 851–862. [Google Scholar] [CrossRef][Green Version]
- Doherty, M.; Yager, P.L.; Moran, M.A.; Coles, V.J.; Fortunato, C.S.; Krusche, A.V.; Medeiros, P.M.; Payet, J.P.; Richey, J.E.; Satinsky, B.M.; et al. Bacterial Biogeography across the Amazon River-Ocean Continuum. Front. Microbiol. 2017, 8, 882. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Schwab, D.J.; Lang, G.A. Real-Time Hydraulic and Hydrodynamic Model of the St. Clair River, Lake St. Clair, Detroit River System. J. Hydraul. Eng. 2010, 136, 507–518. [Google Scholar] [CrossRef]
- Shahraki, A.H.; Chaganti, S.R.; Heath, D.D. Diel Dynamics of Freshwater Bacterial Communities at Beaches in Lake Erie and Lake St. Clair, Windsor, Ontario. Microb. Ecol. 2021, 81, 1–13. [Google Scholar] [CrossRef]
- Savio, D.; Sinclair, L.; Ijaz, U.Z.; Parajka, J.; Reischer, G.H.; Stadler, P.; Blaschke, A.P.; Bloschl, G.; Mach, R.L.; Kirschner, A.K.; et al. Bacterial Diversity Along a 2600 km River Continuum. Environ. Microbiol. 2015, 17, 4994–5007. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M.; Del Giorgio, P.A. Terrestrial Connectivity, Upstream Aquatic History and Seasonality Shape Bacterial Community Assembly within a Large Boreal Aquatic Network. ISME J. 2022, 16, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Read, D.S.; Gweon, H.S.; Bowes, M.J.; Newbold, L.K.; Field, D.; Bailey, M.J.; Griffiths, R.I. Catchment-Scale Biogeography of Riverine Bacterioplankton. ISME J. 2015, 9, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Xiang, X.; Yang, Y.; Huang, G.; Fu, K.; Che, R.; Chen, L. Seasonal Effects of River Flow on Microbial Community Coalescence and Diversity in a Riverine Network. FEMS Microbiol. Ecol. 2020, 96, fiaa132. [Google Scholar] [CrossRef] [PubMed]
- Simek, K.; Kasalicky, V.; Jezbera, J.; Hornak, K.; Nedoma, J.; Hahn, M.W.; Bass, D.; Jost, S.; Boenigk, J. Differential Freshwater Flagellate Community Response to Bacterial Food Quality with a Focus on Limnohabitans Bacteria. ISME J. 2013, 7, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Yannarell, A.C.; Triplett, E.W. Geographic and Environmental Sources of Variation in Lake Bacterial Community Composition. Appl. Environ. Microbiol. 2005, 71, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Lindström, E.S.; Agterveld, M.P.K.-V.; Zwart, G. Distribution of Typical Freshwater Bacterial Groups Is Associated with Ph, Temperature, and Lake Water Retention Time. Appl. Environ. Microbiol. 2005, 71, 8201–8206. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; McMahon, K.D. Species-Sorting May Explain an Apparent Minimal Effect of Immigration on Freshwater Bacterial Community Dynamics. Environ. Microbiol. 2009, 11, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Milke, F.; Wagner-Doebler, I.; Wienhausen, G.; Simon, M. Selection, Drift and Community Interactions Shape Microbial Biogeographic Patterns in the Pacific Ocean. ISME J. 2022, 16, 2653–2665. [Google Scholar] [CrossRef]
- Green, J.L.; Holmes, A.J.; Westoby, M.; Oliver, I.; Briscoe, D.; Dangerfield, M.; Gillings, M.; Beattie, A.J. Spatial Scaling of Microbial Eukaryote Diversity. Nature 2004, 432, 747–750. [Google Scholar] [CrossRef]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B. Beyond Biogeographic Patterns: Processes Shaping the Microbial Landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).