Low Abundant Bacteria Reflect Soil Specificity—Analysis of Bacterial Communities from Archaeological Investigation of Pre-Industrial Saline Ash Deposits of Bad Dürrenberg (Germany)
Abstract
1. Introduction
2. Experimental
2.1. Soil Samples
2.2. DNA Extraction, Sequencing and Data Processing
3. Results and Discussions
3.1. Composition of Soil Bacterial Communities on the Level of Phyla and Selected Classes, Orders and Families
3.2. Comparison of Soil Bacterial Communities on the Level of Operation Taxonomical Units (OTUs)
3.2.1. General Correlation of OTU Abundances
3.2.2. Dominant OTUs
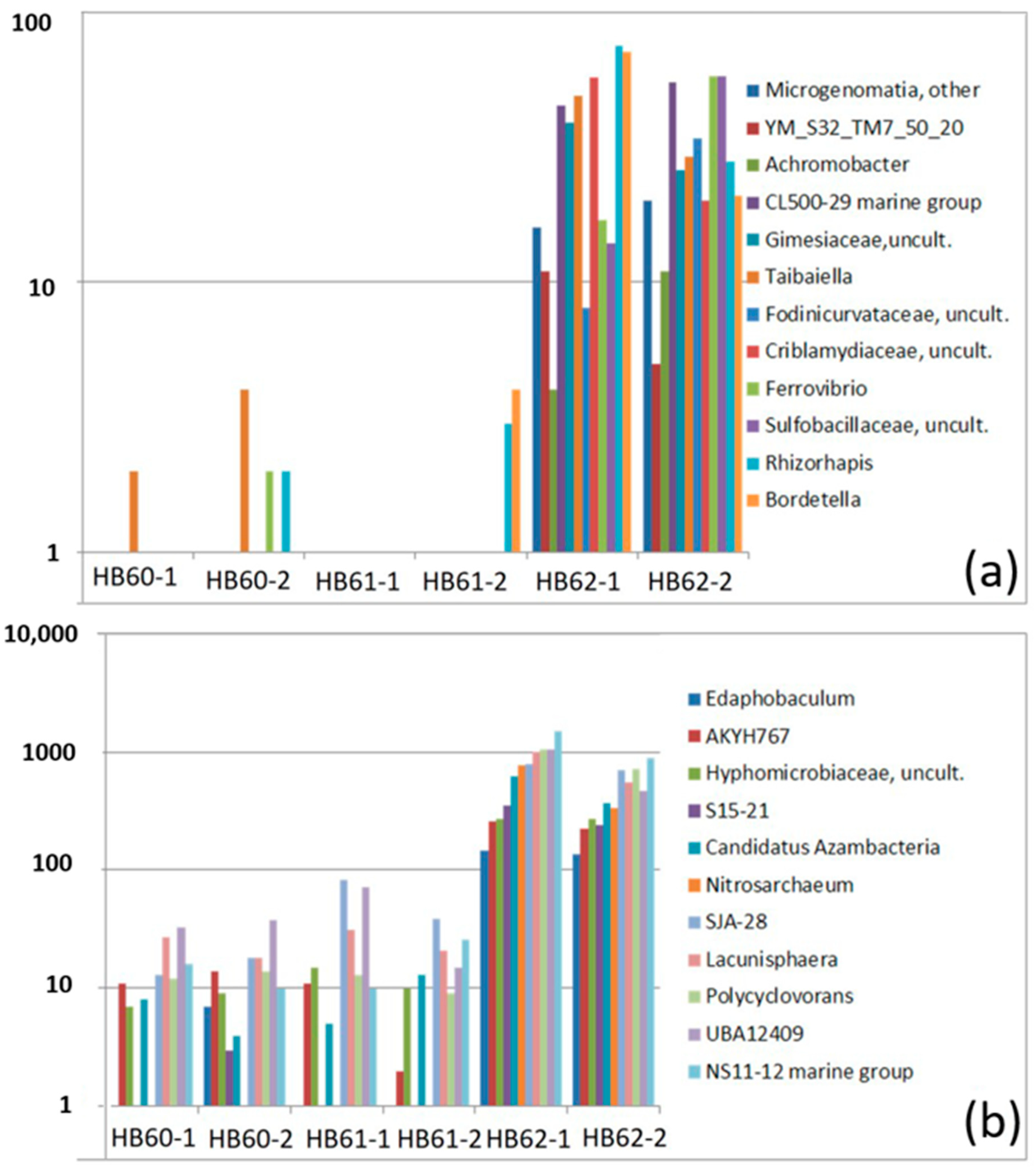
3.2.3. Sample-Specific Types
3.3. General Sample Specificity of Low Abundant OTUs Shown by Comparative Principle Component Analyses (PCA) and Comparison with Soil Bacterial Communities from an Archaeological Excavation Site near Bennstedt (Germany)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmid, M.W.; Van Moorsel, S.J.; Hahkl, T.; De LUca, E.; De Deyn, G.B.; Wagg, C.; Niklaus, P.A.; Schmid, B. Effects of plant community history, soil legacy and plant diversity on soil microbial communities. J. Ecol. 2021, 109, 3007–3023. [Google Scholar] [CrossRef]
- Bartelt-Ryser, J.; Joshi, J.; Schmid, B.; Brandl, H.; Balser, T. Soil feedbacks of plant diversity on soil microbial communities and subsequent plant growth. Perspect. Plant Ecol. Evol. Syst. 2005, 7, 27–49. [Google Scholar] [CrossRef]
- Schippers, A.; Jozsa, P.; Sand, W. Sulfur chemistry in bacterial leaching of pyrite. Appl. Environ. Microbiol. 1996, 62, 3424–3431. [Google Scholar] [CrossRef]
- Haferburg, G.; Kothe, E. Metallomics: Lessons for metalliferous soil remediation. Appl. Microbiol. Biotechnol. 2010, 87, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Mesa, V.; Gallego, J.L.R.; Gonzáles-Gil, R.; Lauga, B.; Sánchez, J.; Méndez-Garcia, C.; Peláez, A.I. Bacterial, archaeal, and eukaryotic diversity across distinct microhabits in an acid mine drainage. Front. Microbiol. 2018, 8, 1. [Google Scholar]
- Mourinha, C.; Palma, P.; Alexandre, C.; Cruz, N.; Rodrigues, S.M.; Alvarenga, P. Potentially toxic elements contamination of soils affected by mining activities in the Portugese sector of the Iberian pyrite belt and optional remediation actions, a review. Environments 2022, 9, 11. [Google Scholar] [CrossRef]
- Chernysheva, E.; Korobov, D.; Borisov, A. Thermophilic microorganisms in arable land around medieval archaeological sites in Northern Caucasus, Russia: Novel evidence of past manuring practices. Geoarchaeol. Int. J. 2017, 32, 494–501. [Google Scholar] [CrossRef]
- Margesin, R.; Siles, J.A.; Cajthaml, T.; Ohlinger, B.; Kistler, E. Microbiology meets archaeology: Soil microbial communities reveal different human activities at archaic Monte Iato (sixth century BC). Microb. Ecol. 2017, 73, 925–938. [Google Scholar] [CrossRef]
- Wegner, C.E.; Liesack, W. Unexpected dominance of elusive acidobacteria in early industrial soft coal slags. Front. Microbiol. 2017, 8, 1023. [Google Scholar] [CrossRef]
- Köhler, J.M.; Kalensee, F.; Günther, P.M.; Schüler, T.; Cao, J. The local ecological memory of soil: Majority and minority components of bacterial communities in prehistoric urns from Schöps (Germany). Int. J. Environ. Res. 2018, 12, 575–684. [Google Scholar] [CrossRef]
- Singer, D.; Herndon, E.; Zemanek, L.; Kortney, C.; Sander, T.; Senko, J.; Perdrial, N. Biogeochemical controls on the potential for long-term contaminated leaching from soils developing on historical coal mine spoil. Soil Syst. 2021, 5, 3. [Google Scholar] [CrossRef]
- Ehrhardt, L.; Günther, P.M.; Böhme, M.; Köhler, J.M.; Cao, J. Three Soil Bacterial Communities from an Archaeological Excavation Site of an Ancient Coal Mine near Bennstedt (Germany) Characterized by 16S r-RNA Sequencing. Environments 2022, 9, 115. [Google Scholar] [CrossRef]
- Olías, M.; Nieto, J.M. Background Conditions and Mining Pollution throughout History in the Río Tinto (SW Spain). Environments 2015, 2, 295–316. [Google Scholar] [CrossRef]
- Liu, J.; Hua, Z.-S.; Chen, L.-X.; Kuang, J.-L.; Li, S.-J.; Shu, W.-S.; Huang, L.-N. Correlating Microbial Diversity Patterns with Geochemistry in an Extreme and Heterogeneous Environment of Mine Tailings. Appl. Environ. Microbiol. 2014, 80, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Bickel, S.; Or, D. The chosen few—Variations in common and rare soil bacteria across biomes. ISME J. 2021, 15, 3315–3325. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Dini-Andreote, F.; Salles, J.F. Community assembly processes of the microbial rare biosphere. Trends Microbiol. 2018, 26, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Jones, S.E.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N.; Gilbert, J.A. Conditionally rare taxa disproportionately contribute to temporal changes in microbialdiversity. MBio 2014, 5, e01371-14. [Google Scholar] [CrossRef] [PubMed]
- Pascoal, F.; Costa, R.; Magallhanes, C. The microbial rare biosphere: Current concepts, methods and ecological principles. FEMS Microbiol. Ecol. 2021, 97, fiaa227. [Google Scholar]
- Kurm, V.; VanderPutten, W.; De Boer, W.; Naus-Wiezer, S.; Hol, W.H.G. Low abundant soil bacteria can be metabollically versatile and fast growing. Ecology 2017, 98, 555–564. [Google Scholar] [CrossRef]
- Matthias, W. Die Salzproduktion-ein bedeutender Faktor in der Wirtschaft der frühbronzezeitlichen Bevölkerung an der mittleren Saale. Jahresschr. f. Mitteldtsch. Vorgesch. 1976, 60, 373–394. (In German) [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schwee, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequenc-ing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.-W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glockner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acid Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Shinohara, A.; Fukui, M. Sulfurifustis variabilis gen. nov., sp. nov., a sulfur oxidizer isolated from a lake, and proposal of Acidiferrobacteraceae fam. nov. and Acidiferrobacterales ord. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 3709–3713. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cánovas, M.J.; Quesada, E.; Martínez-Checa, F.; Del Moral, A.; Béjar, V. Salipiger mucescens gen. nov., sp. nov., a moderately halophilic, exopolysaccharide-producing bacterium isolated from hypersaline soil, belonging to the alpha-Proteobacteria. Int. J. Syst. Evol. Microbiol. 2004, 54, 1735–1740. [Google Scholar] [CrossRef]
- Tang, S.-K.; Zhi, X.-Y.; Wang, Y.; Shi, R.; Lou, K.; Xu, L.H.; Li, W.-J. Haloactinopolyspora alba gen. nov., sp. nov., a halophilic filamentous actinomycete isolated from a salt lake, with proposal of Jiangellaceae fam. nov. and Jiangellineae subord. nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Katayama, T.; Yamagishi, T.; Hanada, S.; Tamaki, H.; Kamagata, Y.; Oshima, K.; Hattori, M.; Marumo, K.; Nedachi, M.; et al. Methyloceanibacter caenitepidi gen. nov., sp. nov., a facultatively methylotrophic bacterium isolated from marine sediments near a hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2014, 64, 462–468. [Google Scholar] [CrossRef]
- Mu, Y.; Zhou, L.; Zeng, X.-C.; Liu, L.; Pan, Y.; Chen, X.; Wang, J.; Li, S.; Li, W.-J.; Wang, Y. Arsenicitalea aurantiaca gen. nov., sp. nov., a new member of the family Hyphomicrobiaceae, isolated from high-arsenic sediment. Int. J. Syst. Evol. Microbiol. 2016, 66, 5478–5484. [Google Scholar] [CrossRef]
- Tiago, I.; Pires, C.; Mendes, V.; Morais, P.V.; Da Costa, M.; Veríssimo, A. Microcella putealis gen. nov., sp. nov., a gram-positive alkaliphilic bacterium iso-lated from a nonsaline alkaline groundwater. Syst. Appl. Microbiol. 2005, 28, 479–487. [Google Scholar]
- Roh, S.W.; Kim, K.-H.; Nam, Y.D.; Chang, H.-W.; Kim, M.S.; Shin, K.-S.; Yoon, J.-H.; Oh, H.-M.; Bae, J.W. Aliihoeflea aestuarii gen. nov., sp. nov., a novel bacterium isolated from tidal flat sediment. J. Microbiol. 2008, 46, 594–598. [Google Scholar] [CrossRef]
- Xu, X.-W.; Huo, Y.-Y.; Wang, C.-S.; Oren, A.; Cui, H.-L.; Vedler, E.; Wu, M. Pelagibacterium halotolerans gen. nov., sp. nov. and Pelagibacterium luteolum sp. nov., novel members of the family Hyphomicrobiaceae. Int. J. Syst. Evol. Microbiol. 2011, 61, 1817–1822. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Van Pelt, S.; Tourova, T.P.; Evtushenko, L.I. Nitriliruptor alkaliphilus gen. nov., sp. nov., a deep-lineage haloalkaliphilic actinobacterium from soda lakes capable of growth on aliphatic nitriles, and proposal of Nitriliruptoraceae fam. nov. and Nitriliruptorales ord. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-G.; Chen, J.Y.; Wang, H.-F.; Xiao, M.; Yang, L.-L.; Guo, J.-W.; Zhou, E.M.; Zhang, Y.-M.; Li, W.-J. Egicoccus halophilus gen. nov., sp. nov., a halophilic, alkalitolerant actinobacterium and proposal of Egicoccaceae fam. nov. and Egicoccales ord. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.-Y.; Islam, M.A.; Gwak, J.-H.; Kim, J.-G.; Rhee, S.-K. Nitrosarchaeum koreense gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon member of the phylum Thaumarchaeota isolated from agricultural soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, A.Y.; Chernousova, E.Y.; Dubinina, G.A. Ferrovibrio denitrificans gen. nov., sp. nov., a novel neutrophilic facultative anaerobic Fe(II)-oxidizing bacterium. FEMS Microbiol Lett. 2012, 335, 19–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gutierrez, T.; David, H.; Green, D.H.; Nichols, P.D.; Whitman, W.B.; Semple, K.T.; Aitken, M.D. Polycyclovorans algicola gen. nov., sp. nov., an aromatic-hydrocarbon-degrading marine bacterium found associated with laboratory cultures of marine phytoplankton. Appl. Environ. Microbiol. 2013, 79, 205–214. [Google Scholar] [CrossRef]
- Köhler, J.M.; Ehrhardt, L.; Ca, J.; Möller, F.; Schüler, T.; Günther, P.M. Beta-Diversity Enhancement by Archaeological Structures: Bacterial Communities of an Historical Tannery Area of the City of Jena (Germany) Reflect the Ancient Human Impact. Ecologies 2023, 4, 325–343. [Google Scholar] [CrossRef]
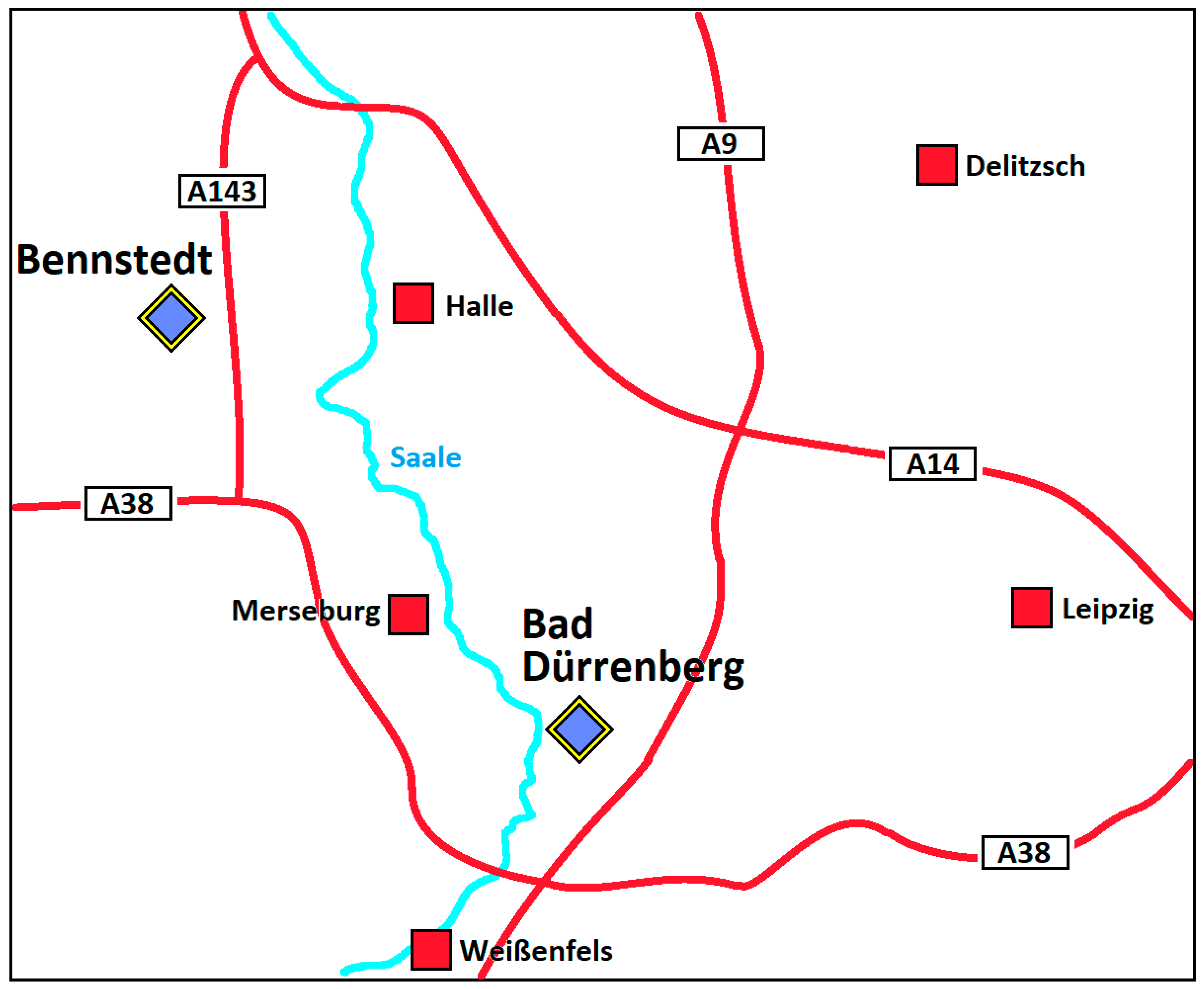










| Sample-No | Origin | pH-Value | El. Cond. (µS/cm) |
|---|---|---|---|
| HB60-1 | Bad Dürrenberg, top soil | 7.7 | 1068 |
| HB60-2 | Bad Dürrenberg, top soil | 7.7 | 1068 |
| HB61-1 | Bad Dürrenberg, short–term deposit | 8.2 | 2100 |
| HB61-2 | Bad Dürrenberg, short-term deposit | 8.2 | 2100 |
| HB62-1 | Bad Dürrenberg, long-term deposit | 8.1 | 2337 |
| HB62-2 | Bad Dürrenberg, long-term deposit | 8.1 | 2337 |
| HB57-1 | Bennstedt, coal seam | 4.2 | 115 |
| HB57-2 | Bennstedt, coal seam | 4.2 | 115 |
| HB58-1 | Bennstedt, bright sediment | 4.1 | 43 |
| HB58-2 | Bennstedt, bright sediment | 4.1 | 43 |
| HB59-1 | Bennstedt, top soil | 4.05 | 45 |
| HB59-2 | Bennstedt, top soil | 4.05 | 45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Köhler, J.M.; Ehrhardt, L.; Günther, P.M.; Böhme, M.; Cao, J. Low Abundant Bacteria Reflect Soil Specificity—Analysis of Bacterial Communities from Archaeological Investigation of Pre-Industrial Saline Ash Deposits of Bad Dürrenberg (Germany). Environments 2024, 11, 42. https://doi.org/10.3390/environments11030042
Köhler JM, Ehrhardt L, Günther PM, Böhme M, Cao J. Low Abundant Bacteria Reflect Soil Specificity—Analysis of Bacterial Communities from Archaeological Investigation of Pre-Industrial Saline Ash Deposits of Bad Dürrenberg (Germany). Environments. 2024; 11(3):42. https://doi.org/10.3390/environments11030042
Chicago/Turabian StyleKöhler, Johann Michael, Linda Ehrhardt, Peter Mike Günther, Manfred Böhme, and Jialan Cao. 2024. "Low Abundant Bacteria Reflect Soil Specificity—Analysis of Bacterial Communities from Archaeological Investigation of Pre-Industrial Saline Ash Deposits of Bad Dürrenberg (Germany)" Environments 11, no. 3: 42. https://doi.org/10.3390/environments11030042
APA StyleKöhler, J. M., Ehrhardt, L., Günther, P. M., Böhme, M., & Cao, J. (2024). Low Abundant Bacteria Reflect Soil Specificity—Analysis of Bacterial Communities from Archaeological Investigation of Pre-Industrial Saline Ash Deposits of Bad Dürrenberg (Germany). Environments, 11(3), 42. https://doi.org/10.3390/environments11030042







