Integrated Genomic Analysis Reveals the Synergistic Role of PNPLA3 and ABCC8 Variants in Diabetic MASLD in Pakistan
Abstract
1. Introduction
2. Methodology
2.1. Patient Selection
2.2. Study Phases
2.2.1. Whole Exome Sequencing (WES) Discovery Cohort
2.2.2. Sanger Sequencing (Validation Cohort)
3. Results
3.1. Baseline Characteristics and Analysis of the Discovery Cohort
3.2. Baseline Characteristics of the Validation Cohort
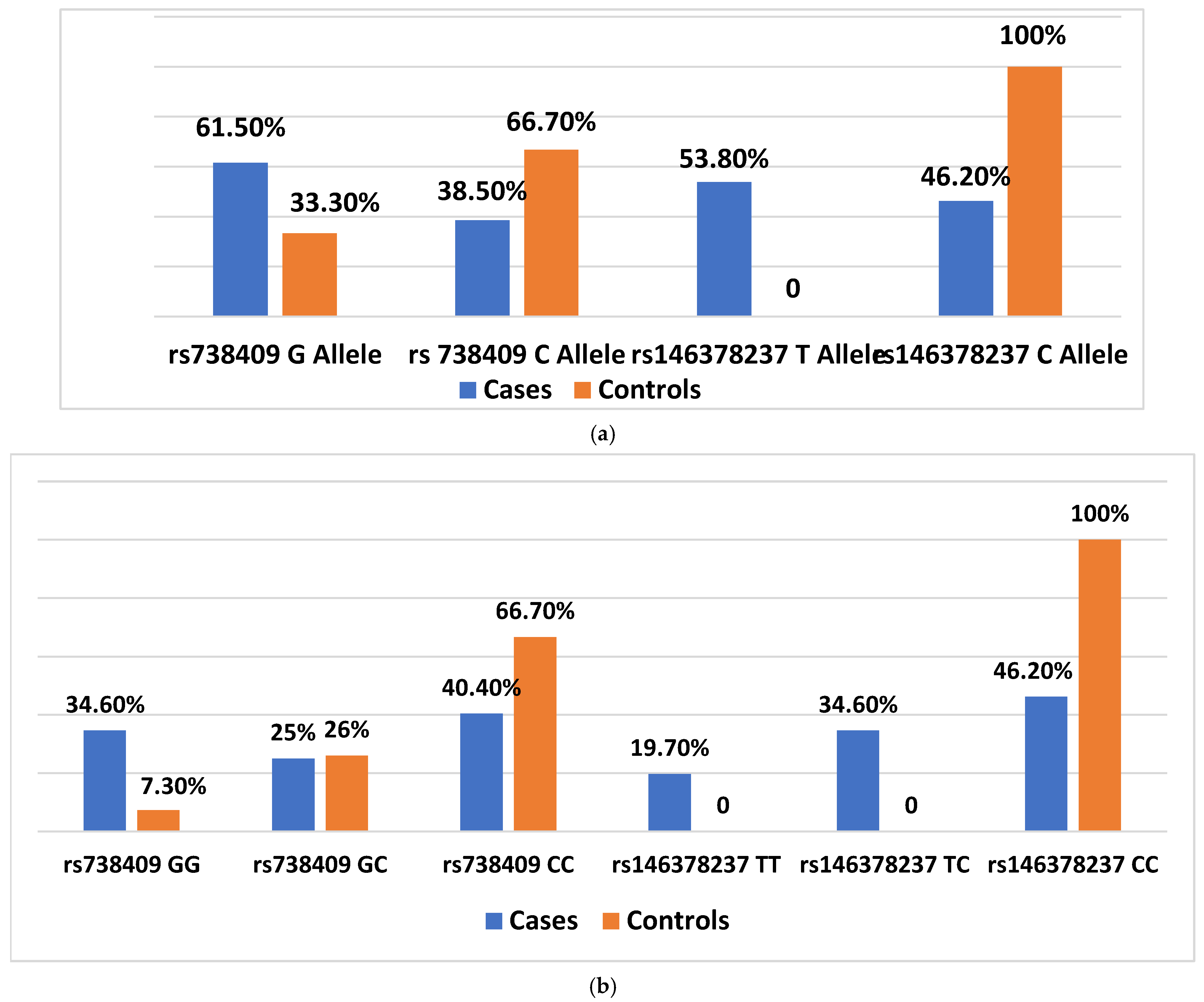
3.3. Association of rs738409 and rs146378237 Polymorphisms with Disease Susceptibility
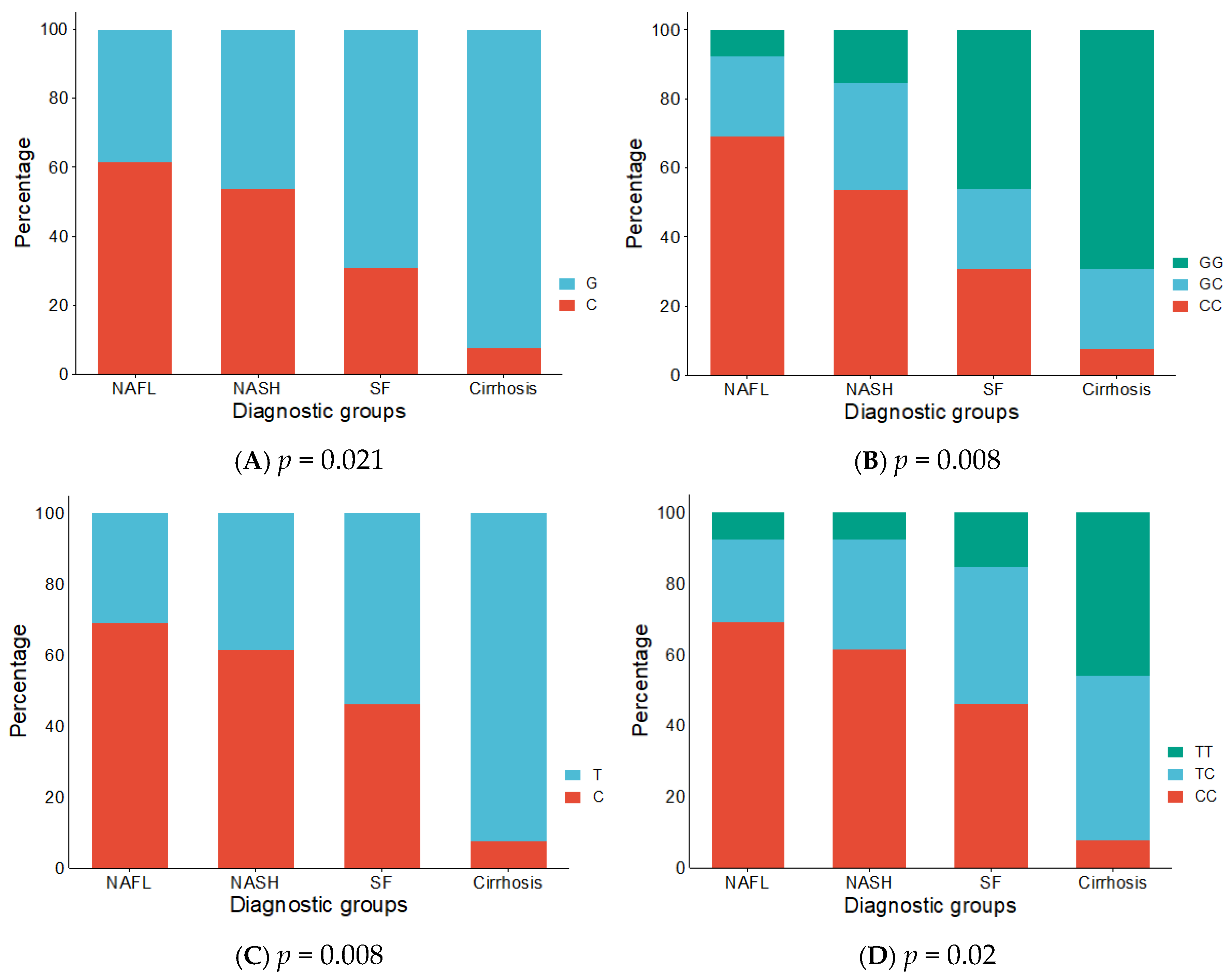
3.4. Distribution of Alleles and Genotypes Across Disease Subtypes
3.5. Analysis of rs738409 and rs146378237 Variants in Relation to Demographics and Comorbidities
3.6. Potential Synergistic Effect of rs738409 and rs146378237 in MASLD Cases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NAFLD | Non-alcoholic fatty liver disease |
| NAFL | Non-alcoholic fatty liver |
| SF | Steatofibrosis |
| NASH | Non-alcoholic steatohepatitis |
| MASLD | Metaboloic dysfunction associated steatotic liver disease |
| BMI | body mass index |
| ALT | alanine aminotransferase |
| AST | Aspartate transaminase |
| ALP | Alkaline phosphatase |
| GGT | gamma-glutamyl transpeptidase |
| FBS | fasting blood sugar |
| TG | Triglyceride |
| HDL | High density lipoprotein |
| LDL | Low density lipoprotein |
References
- Stefan, N.; Cusi, K. A global view of the interplay between non-alcoholic fatty liver disease and diabetes. Lancet Diabetes Endocrinol. 2022, 10, 284–296. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 2023, 78, 1966–1986. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335. [Google Scholar] [CrossRef]
- Fan, J.-G.; Kim, S.-U.; Wong, V.W.-S. New trends on obesity and NAFLD in Asia. J. Hepatol. 2017, 67, 862–873. [Google Scholar] [CrossRef]
- Battistella, S.; D’Arcangelo, F.; Grasso, M.; Zanetto, A.; Gambato, M.; Germani, G.; Senzolo, M.; Russo, F.P.; Burra, P. Liver transplantation for non-alcoholic fatty liver disease: Indications and post-transplant management. Clin. Mol. Hepatol. 2023, 29, S286. [Google Scholar] [CrossRef]
- Kang, J.-S. Non-Alcoholic Fatty Liver Disease-New Insight and Glance Into Disease Pathogenesis; IntechOpen: London, UK, 2023. [Google Scholar]
- Salari, N.; Darvishi, N.; Mansouri, K.; Ghasemi, H.; Hosseinian-Far, M.; Darvishi, F.; Mohammadi, M. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease: A systematic review and meta-analysis. BMC Endocr. Disord. 2021, 21, 125. [Google Scholar] [CrossRef]
- Trépo, E.; Romeo, S.; Zucman-Rossi, J.; Nahon, P. PNPLA3 gene in liver diseases. J. Hepatol. 2016, 65, 399–412. [Google Scholar] [CrossRef]
- Chen, V.L.; Oliveri, A.; Miller, M.J.; Wijarnpreecha, K.; Du, X.; Chen, Y.; Cushing, K.C.; Lok, A.S.; Speliotes, E.K. PNPLA3 genotype and diabetes identify patients with nonalcoholic fatty liver disease at high risk of incident cirrhosis. Gastroenterology 2023, 164, 966–977.e917. [Google Scholar] [CrossRef]
- Butnariu, L.I.; Bizim, D.A.; Păduraru, G.; Păduraru, L.; Moisă, Ș.M.; Popa, S.; Gimiga, N.; Ghiga, G.; Bădescu, M.C.; Lupu, A. Congenital hyperinsulinism caused by mutations in ABCC8 gene associated with early-onset neonatal hypoglycemia: Genetic heterogeneity correlated with phenotypic variability. Int. J. Mol. Sci. 2024, 25, 5533. [Google Scholar] [CrossRef]
- Srivastava, A.; Gailer, R.; Tanwar, S.; Trembling, P.; Parkes, J.; Rodger, A.; Suri, D.; Thorburn, D.; Sennett, K.; Morgan, S. Prospective evaluation of a primary care referral pathway for patients with non-alcoholic fatty liver disease. J. Hepatol. 2019, 71, 371–378. [Google Scholar] [CrossRef]
- Karlas, T.; Petroff, D.; Sasso, M.; Fan, J.G.; Mi, Y.Q.; de Lédinghen, V.; Kumar, M.; Lupsor-Platon, M.; Han, K.H.; Cardoso, A.C.; et al. Individual Patient Data Meta-Analysis of Controlled Attenuation Parameter (CAP) Technology for Assessing Steatosis. J. Hepatol. 2017, 66, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines on Non-Invasive Tests for Evaluation of Liver Disease Severity and Prognosis—2021 Update. J. Hepatol. 2021, 75, 659–689. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Kozlitina, J.; Sookoian, S. Global epidemiological impact of PNPLA3 I148M on liver disease. Liver Int. 2025, 45, e16123. [Google Scholar] [CrossRef] [PubMed]
- 1000 Genomes Project Consortium, A global reference for human genetic variation. Nature 2015, 526, 68–74.
- WPcalc. Hardy-Weinberg Equilibrium Calculator. Available online: https://wpcalc.com/en/equilibrium-hardy-weinberg/ (accessed on 20 June 2025).
- SRplot. Available online: https://www.bioinformatics.com.cn/en (accessed on 13 May 2025).
- BioRender-Science Illustration Tool. 2025. Available online: https://biorender.com (accessed on 22 June 2025).
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Hassan, F.; Farman, M.; Khan, K.A.; Awais, M.; Akhtar, S. Prevalence of nonalcoholic fatty liver disease in Pakistan: A systematic review and meta-analysis. Sci. Rep. 2024, 14, 19573. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef]
- Narayanasamy, K.; Karthick, R.; Panneerselvam, P.; Mohan, N.; Ramachandran, A.; Prakash, R.; Rajaram, M. Association of metabolic syndrome and patatin-like phospholipase 3–rs738409 gene variant in non-alcoholic fatty liver disease among a Chennai-based south Indian population. J. Gene Med. 2020, 22, e3160. [Google Scholar] [CrossRef]
- Kanth, V.V.R.; Sasikala, M.; Rao, P.N.; Avanthi, U.S.; Rao, K.R.; Reddy, D.N. Pooled genetic analysis in ultrasound measured non-alcoholic fatty liver disease in Indian subjects: A pilot study. World J. Hepatol. 2014, 6, 435. [Google Scholar] [CrossRef]
- Mattaparthi, N.S.; Velaga, L.; Achanta, C.R.; Chadram, B.; Porupureddy, M.S.; Panthagada, H. Association of PNPLA3 Polymorphisms in Patients with Metabolic Dysfunction Associated Steatotic Liver Disease in North Coastal Andhra Pradesh: A Case-Control Study. J. Chem. Health Risk 2024, 14, 1800–1809. [Google Scholar]
- Das, M.; Biswas, A.; Goswami, S.; Deb, R.; Das, S.; Ray, D. Association of the rs738409 polymorphism in PNPLA3 with development and severity of non-alcoholic fatty liver disease in ethnic Bengali population of West Bengal. medRxiv 2024. [Google Scholar] [CrossRef]
- Zain, S.M.; Mohamed, R.; Mahadeva, S.; Cheah, P.L.; Rampal, S.; Basu, R.C.; Mohamed, Z. A multi-ethnic study of a PNPLA3 gene variant and its association with disease severity in non-alcoholic fatty liver disease. Hum. Genet. 2012, 131, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Elmansoury, N.; Megahed, A.A.; Kamal, A.; El-Nikhely, N.; Labane, M.; Abdelmageed, M.; Daly, A.K.; Wahid, A. Relevance of PNPLA3, TM6SF2, HSD17B13, and GCKR Variants to MASLD Severity in an Egyptian Population. Genes 2024, 15, 455. [Google Scholar] [CrossRef]
- Altaf, B.; Jawed, S.; Ghazali, W.S.W.; Aziz, A.A.A.; Salam, R.M.T.; Rasheed, A.; Mohamed, M. Patatin-like phospholipase domain-containing 3 (PNPLA3) variants rs 738408 and rs 738409 single nucleotide polymorphism as predictor of metabolic associated fatty liver disease and its progression. Pak. J. Med. Sci. 2025, 41, 1322–1330. [Google Scholar] [CrossRef]
- NCBI. db SNP:rs146378237. Available online: https://www.ncbi.nlm.nih.gov/snp/rs146378237 (accessed on 30 May 2025).
- Chen, Y.-Y.; Chen, C.-S.; Huang, J.-F.; Su, W.-H.; Li, C.-Y.; Chen, W.-S.; Lin, E.-S.; Chuang, W.-L.; Yu, M.-L.; Wang, S.-C. The obesity-related mutation gene on nonalcoholic fatty liver disease. Hum. Genet. 2025, 144, 1–14. [Google Scholar] [CrossRef]
- Rosso, C.; Caviglia, G.P.; Birolo, G.; Armandi, A.; Pennisi, G.; Pelusi, S.; Younes, R.; Liguori, A.; Perez-Diaz-del-Campo, N.; Nicolosi, A. Impact of PNPLA3 rs738409 polymorphism on the development of liver-related events in patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2023, 21, 3314–3321.e3313. [Google Scholar] [CrossRef]
- Shabbir, A.; Abbas, Z.; Khatoon, A.; Mirza, T. Role of Alanine Transaminase and Transient Elastography in Categorising Nonalcoholic Fatty Liver Disease Subgroups. J. Coll. Physicians Surg. Pak. 2024, 34, 22–26. [Google Scholar] [CrossRef]
- Xia, M.; Varmazyad, M.; Pla-Palacín, I.; Gavlock, D.C.; DeBiasio, R.; LaRocca, G.; Reese, C.; Florentino, R.M.; Faccioli, L.A.; Brown, J.A. Comparison of wild-type and high-risk PNPLA3 variants in a human biomimetic liver microphysiology system for metabolic dysfunction-associated steatotic liver disease precision therapy. Front. Cell Dev. Biol. 2024, 12, 1423936. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Darlay, R.; Cockell, S.; Meroni, M.; Govaere, O.; Tiniakos, D.; Burt, A.D.; Bedossa, P.; Palmer, J.; Liu, Y.-L. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J. Hepatol. 2020, 73, 505–515. [Google Scholar] [CrossRef]
- Bale, G.; Mitnala, S.; Padaki, N.R.; Sharma, M.; Kulkarni, A.V.; Pawar, S.C.; Reddy, N.; Vishnubhotla, R. I148M variant of PNPLA3-gene is not associated with metabolic syndrome in patients with NAFLD in the Indian ethnicity. Hum. Gene 2022, 33, 201073. [Google Scholar] [CrossRef]
- Tran, N.Q.; Truong, S.D.; Ma, P.T.; Hoang, C.K.; Le, B.H.; Dinh, T.T.N.; Van Tran, L.; Tran, T.V.; Le, L.H.G.; Le, K.T. Association of KCNJ11 and ABCC8 single-nucleotide polymorphisms with type 2 diabetes mellitus in a Kinh Vietnamese population. Medicine 2022, 101, e31653. [Google Scholar] [CrossRef] [PubMed]
- National Centre for Biotechnology Information (NCBI). ABCC8 ATP Binding Cassette Subfamily C Member 8 [Homo sapiens (Human)]. Gene ID: 6833. Available online: https://www.ncbi.nlm.nih.gov/gene/6833 (accessed on 23 June 2025).
- Balamurugan, K.; Kavitha, B.; Yang, Z.; Mohan, V.; Radha, V.; Shyng, S.L. Functional characterization of activating mutations in the sulfonylurea receptor 1 (ABCC8) causing neonatal diabetes mellitus in Asian Indian children. Pediatr. Diabetes 2019, 20, 397–407. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Qi, X.; Chao, Y.; Chen, Q.; Cheng, P.; Yu, X.; Kuai, M.; Wu, J.; Li, W.; Zhang, Q. IRS1/PI3K/AKT pathway signal involved in the regulation of glycolipid metabolic abnormalities by Mulberry (Morus alba L.) leaf extracts in 3T3-L1 adipocytes. Chin. Med. 2020, 15, 1. [Google Scholar] [CrossRef]
- Marcondes-de-Castro, I.A.; Reis-Barbosa, P.H.; Marinho, T.S.; Aguila, M.B.; Mandarim-de-Lacerda, C.A. AMPK/mTOR pathway significance in healthy liver and non-alcoholic fatty liver disease and its progression. J. Gastroenterol. Hepatol. 2023, 38, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Reyna-Neyra, A.; Philippe, L.; Thoreen, C.C. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 2017, 19, 1083–1090. [Google Scholar] [CrossRef]
- Szwed, A.; Kim, E.; Jacinto, E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 2021, 101, 1371–1426. [Google Scholar] [CrossRef]

| Gene | SNP ID | Primer Sequence | Length (bp) | Tm | GC% |
|---|---|---|---|---|---|
| PNPLA3 | rs738409 | ||||
| Forward Primer | GCATTTTCAAGTTTGTTGCCCTG | 23 | 60 | 43 | |
| Reverse Primer | CTGAAAGGCAGTGAGGCATGG | 21 | 61 | 57 | |
| ABCC8 | rs146378237 | ||||
| Forward Primer | GCATGCAGCTTTCTGGCTTTC | 21 | 61 | 52 | |
| Reverse Primer | TGAGGGGTGTCTCTGTGCTTC | 21 | 61 | 57 |
| Characteristics | Total (n = 52) Median [IQR]/Mean ± SD |
|---|---|
| Age | 49.46 ± 13.64 |
| Weight | 80 [21] |
| Height | 165.23 ± 9.95 |
| BMI | 28.65 [9.2] |
| ALT | 27.5 [37] |
| AST | 34.5 [21] |
| ALP | 78 [55.38] |
| GGT | 35.5 [49] |
| Total Bilirubin | 0.6 [0.48] |
| Direct Bilirubin | 0.2 [0.17] |
| Albumin | 3.89 ± 0.79 |
| Platelet count | 253 [133] |
| HbA1c | 6.03 ± 1.02 |
| FBS | 96 [26] |
| Serum Insulin | 14.31 [7.38] |
| HOMA-IR | 3.50 [2.22] |
| Total Cholesterol | 158.48 ± 37.35 |
| Triglycerides | 140 [70] |
| HDL | 42 [13] |
| LDL | 108.15 ± 34.77 |
| Steatosis score | 296.38 ± 28.94 |
| Fibrosis score | 9.05 [8.1] |
| Variables | Variants | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs738409 Allele | rs738409 Genotype | rs146378237 Allele | rs146378237 Genotype | |||||||
| C (Ref.) | G (Alt.) | CC (Ref.) | GC (Alt.) | GG (Alt.) | C (Ref.) | T (Alt.) | CC (Ref.) | TC (Alt.) | TT (Alt.) | |
| HTN n (%) | ||||||||||
| Present | 13 (40.6) | 19 (59.4) | 13 (40.6) | 7 (21.9) | 12 (37.5) | 13 (40.6) | 19 (59.4) | 13 (40.6) | 13 (40.6) | 6 (18.8) |
| Absent | 7 (35) | 13 (65) | 8 (40) | 6 (30) | 6 (30) | 11 (55) | 9 (45) | 11 (55) | 5 (25) | 4 (20) |
| p = 0.685 a 0.056 b | p = 0.769 a 0.101 b | p = 0.312 a 0.140 b | p = 0.489 a 0.166 b | |||||||
| DM n (%) | ||||||||||
| Present | 12 (42.9) | 16 (57.1) | 12 (42.9) | 4 (14.3) | 12 (42.9) | 9 (32.1) | 19 (67.9) | 9 (32.1) | 9 (32.1) | 10 (35.7) |
| Absent | 8 (33.3) | 16 (66.7) | 9 (37.5) | 9 (37.5) | 6 (25) | 15 (62.5) | 9 (37.5) | 15 (62.5) | 9 (37.5) | 0 |
| p = 0.482 a 0.098 b | p = 0.131 a 0.280 b | p = 0.029 a,* 0.304 b | p = 0.004 a,* 0.465 b | |||||||
| Dyslipidemia n (%) | ||||||||||
| Present | 10 (32.3) | 21 (67.7) | 11 (35.5) | 8 (25.8) | 12 (38.7) | 14 (45.2) | 17 (54.8) | 14 (45.2) | 11 (35.5) | 6 (19.4) |
| Absent | 10 (47.6) | 11 (52.4) | 10 (47.6) | 5 (23.8) | 6 (28.6) | 10 (47.6) | 11 (52.4) | 10 (47.6) | 7 (33.3) | 4 (19) |
| p = 0.264 a 0.155 b | p = 0.654 a 0.128 b | p = 0.862 a 0.024 b | p = 0.983 a 0.025 b | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shabbir, A.; Khatoon, A.; Abbas, Z.; Srivastava, S.; Mirza, T. Integrated Genomic Analysis Reveals the Synergistic Role of PNPLA3 and ABCC8 Variants in Diabetic MASLD in Pakistan. Med. Sci. 2025, 13, 178. https://doi.org/10.3390/medsci13030178
Shabbir A, Khatoon A, Abbas Z, Srivastava S, Mirza T. Integrated Genomic Analysis Reveals the Synergistic Role of PNPLA3 and ABCC8 Variants in Diabetic MASLD in Pakistan. Medical Sciences. 2025; 13(3):178. https://doi.org/10.3390/medsci13030178
Chicago/Turabian StyleShabbir, Asma, Ambrina Khatoon, Zaigham Abbas, Sucheta Srivastava, and Talat Mirza. 2025. "Integrated Genomic Analysis Reveals the Synergistic Role of PNPLA3 and ABCC8 Variants in Diabetic MASLD in Pakistan" Medical Sciences 13, no. 3: 178. https://doi.org/10.3390/medsci13030178
APA StyleShabbir, A., Khatoon, A., Abbas, Z., Srivastava, S., & Mirza, T. (2025). Integrated Genomic Analysis Reveals the Synergistic Role of PNPLA3 and ABCC8 Variants in Diabetic MASLD in Pakistan. Medical Sciences, 13(3), 178. https://doi.org/10.3390/medsci13030178








