MicroRNA-152 Promotes Slow-Twitch Myofiber Formation via Targeting Uncoupling Protein-3 Gene
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Porcine Myoblasts Isolation and Culture
2.3. Cell Transfection
2.4. Real-Time PCR
2.5. Western Blot Analysis
2.6. Luciferase Reporter Assay
2.7. Metachromatic ATPase Staining
2.8. Metabolic-Related Enzymes Activities Analysis
2.9. Statistical Analysis
3. Results and Discussion
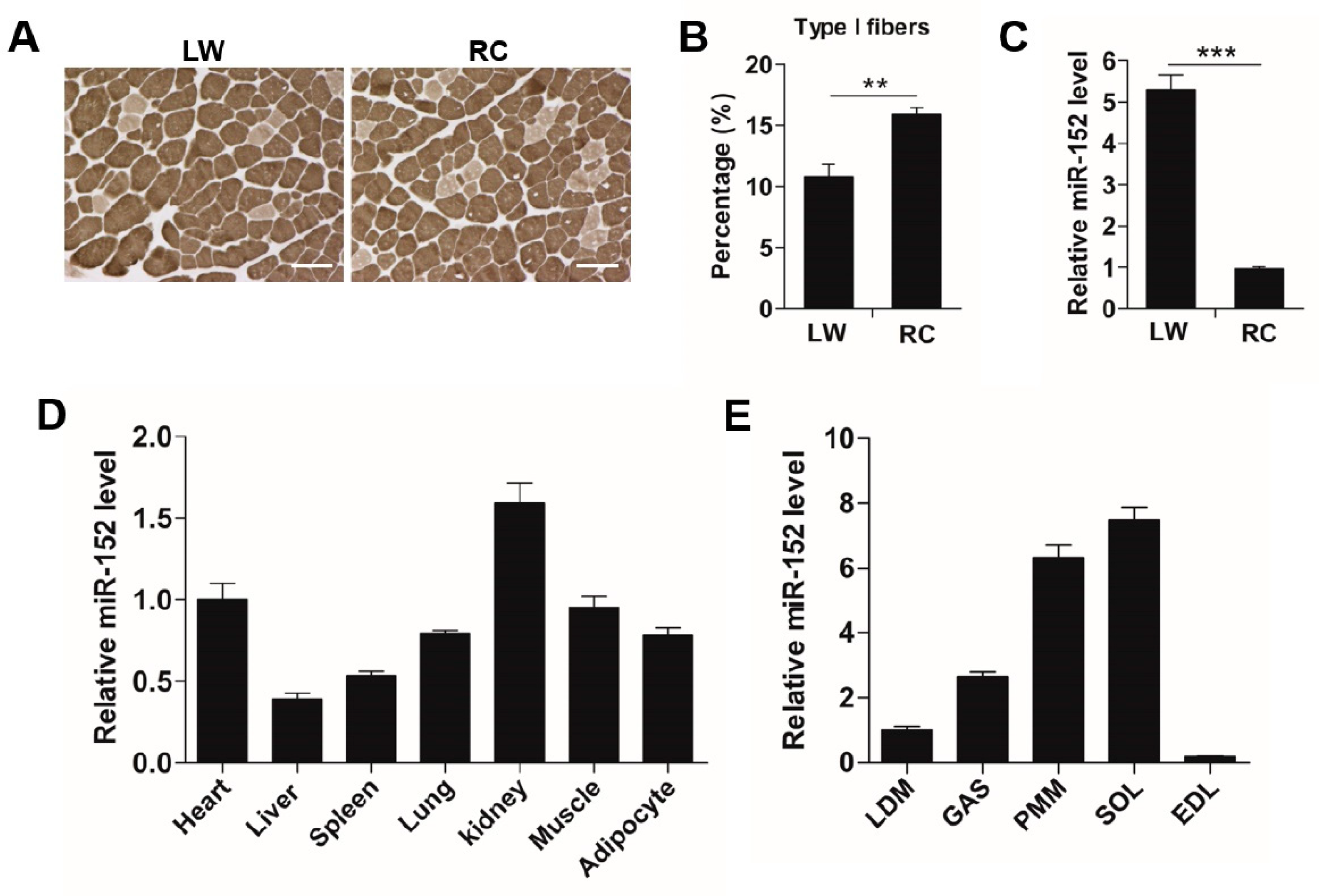
3.1. MiR-152 is More Abundant in Slow-Twitch Myofiber-Enriched Muscles
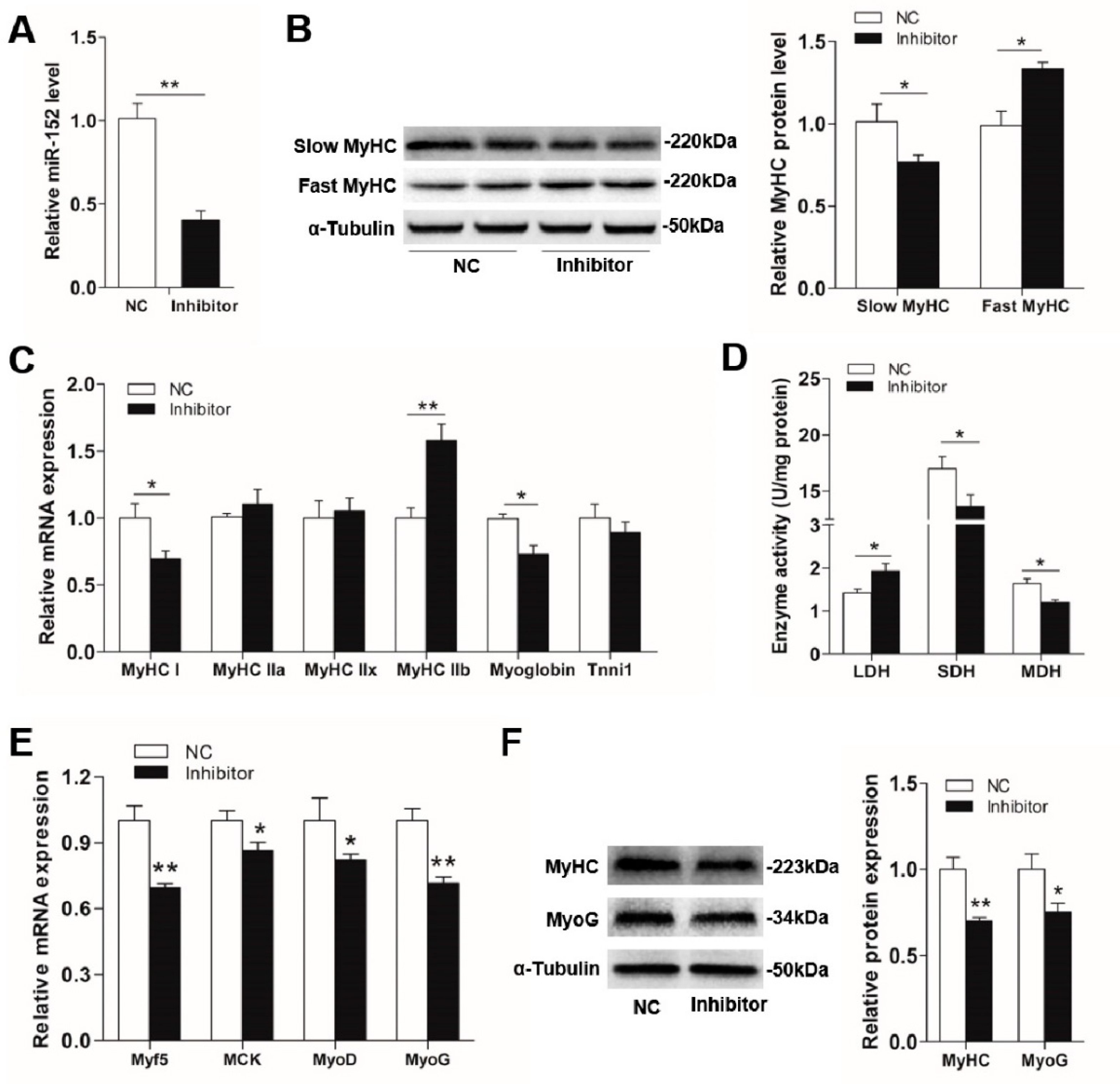
3.2. MiR-152 Promotes Slow-Switch Myofiber Formation and Skeletal Myogenesis
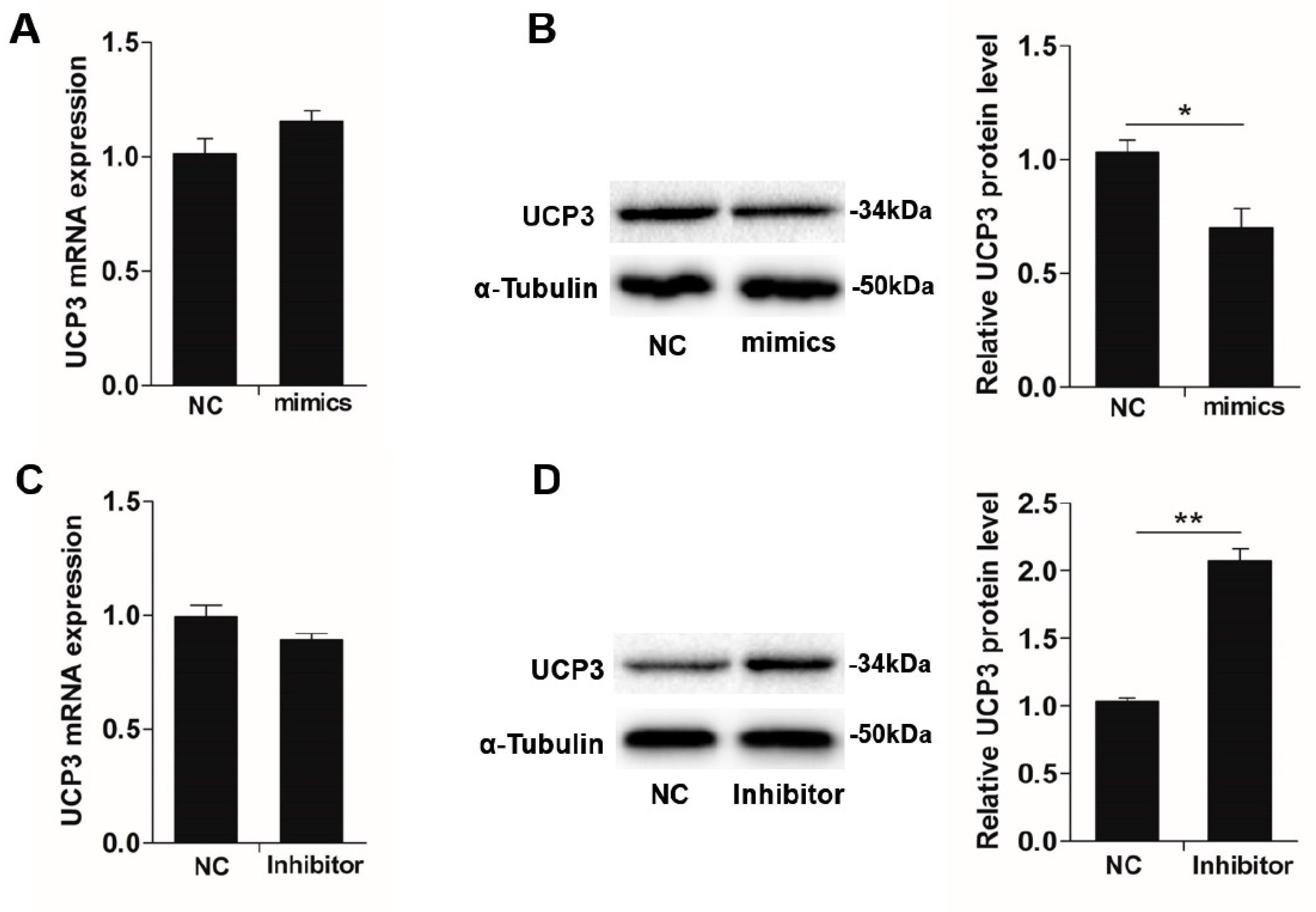
3.3. UCP3 is a Direct Target of MiR-152
3.4. UCP3 Mediates MiR-152 Action in Slow-Twitch Myofiber Formation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, C.; Luo, J.; Zheng, P.; Yu, B.; Huang, Z.; Mao, X.; He, J.; Yu, J.; Chen, J.; Chen, D. Differential expression of lipid metabolism-related genes and myosin heavy chain isoform genes in pig muscle tissue leading to different meat quality. Animal 2015, 9, 1073–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Z.; Wang, L.; Xu, Z.; Huang, J.; Wang, Y. Developmental changes of carcass composition, meat quality and organs in the Jinhua pig and Landrace. Animal 2009, 3, 468–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Lei, H.; Zhang, S.; Li, X.; Li, M.; Jiang, X.; Zhu, K.; Zhu, L. The comparison of energy metabolism and meat quality among three pig breeds. Anim. Sci. J. 2014, 85, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.; Choi, Y.; Lee, S.; Shin, H.; Choe, J.; Kim, J.; Hong, K.C.; Kim, B.-C. Comparing the histochemical characteristics and meat quality traits of different pig breeds. Meat Sci. 2008, 80, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Wu, T.; Reng, Y.; Wang, Y. Breed difference and regulation of the porcine adipose triglyceride lipase and hormone sensitive lipase by TNFα. Anim. Genet. 2009, 40, 863–870. [Google Scholar] [CrossRef]
- Lefaucheur, L.; Milan, D.; Ecolan, P.; Le Callennec, C. Myosin heavy chain composition of different skeletal muscles in Large White and Meishan pigs. J. Anim. Sci. 2004, 82, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, J.; Zhu, R.; Guo, J.; Wu, Y. Effect of myosin heavy chain composition of muscles on meat quality in Laiwu pigs and Duroc. Sci. China Life Sci. 2008, 51, 127–132. [Google Scholar] [CrossRef]
- Pette, D.; Staron, R.S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 2000, 50, 500–509. [Google Scholar] [CrossRef]
- Zierath, J.R.; Hawley, J.A. Skeletal muscle fiber type: Influence on contractile and metabolic properties. PLoS Biol. 2004, 2, e348. [Google Scholar] [CrossRef]
- Bowker, B.C.; Swartz, D.R.; Grant, A.L.; Gerrard, D.E. Myosin heavy chain isoform composition influences the susceptibility of actin-activated S1 ATPase and myofibrillar ATPase to pH inactivation. Meat Sci. 2005, 71, 342–350. [Google Scholar] [CrossRef]
- Choe, J.H.; Choi, Y.M.; Lee, S.H.; Shin, H.G.; Ryu, Y.C.; Hong, K.C.; Kim, B.C. The relation between glycogen, lactate content and muscle fiber type composition, and their influence on postmortem glycolytic rate and pork quality. Meat Sci. 2008, 80, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.T.; Kim, G.D.; Hwang, Y.H.; Ryu, Y.C. Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Sci. 2013, 95, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-D.; Jeong, J.-Y.; Jung, E.-Y.; Yang, H.-S.; Lim, H.-T.; Joo, S.-T. The influence of fiber size distribution of type IIB on carcass traits and meat quality in pigs. Meat Sci. 2013, 94, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.H.; Kim, B.C. Association of blood glucose, blood lactate, serum cortisol levels, muscle metabolites, muscle fiber type composition, and pork quality traits. Meat Sci. 2014, 97, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.C.; Kim, B.C. The relationship between muscle fiber characteristics, postmortem metabolic rate, and meat quality of pig longissimus dorsi muscle. Meat Sci. 2005, 71, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Rooij, E.V.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J.; Olson, N.E. A Family of microRNAs Encoded by Myosin Genes Governs Myosin Expression and Muscle Performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, Z.; John, R.; Hazen, B.C.; Ling, L.; Leone, T.C.; Vega, R.B.; Hui, X.; Conley, K.E.; Johan, A.; Smith, S.R. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J. Clin. Investig. 2013, 123, 2564–2575. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Bezprozvannaya, S.; Shelton, J.M.; Frisard, M.I.; Hulver, M.W.; McMillan, R.P.; Wu, Y.; Voelker, K.A.; Grange, R.W.; Richardson, J.A. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J. Clin. Investig. 2011, 121, 3258. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, X.; Li, Y.; Zhao, L.; Lu, M.; Yao, X.; Xia, H.; Wang, Y.-C.; Liu, M.-F.; Jiang, J. Thyroid hormone regulates muscle fiber type conversion via miR-133a1. J. Cell Biol. 2014, 207, 753–766. [Google Scholar] [CrossRef]
- Nielsen, M.; Hansen, J.; Hedegaard, J.; Nielsen, R.; Panitz, F.; Bendixen, C.; Thomsen, B. MicroRNA identity and abundance in porcine skeletal muscles determined by deep sequencing. Anim. Genet. 2010, 41, 159–168. [Google Scholar] [CrossRef]
- He, D.; Zou, T.; Gai, X.; Ma, J.; Li, M.; Huang, Z.; Chen, D. MicroRNA expression profiles differ between primary myofiber of lean and obese pig breeds. PLoS ONE 2017, 12, e0181897. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, Q.-S.; Li, W.-B.; Wei, J.; Chang, W.-K.; Chen, Z.; Qiao, H.-Y.; Jia, Y.-W.; Tian, J.-H.; Liang, B.-S. miRNA targeted signaling pathway in the early stage of denervated fast and slow muscle atrophy. Neural Regen. Res. 2016, 11, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Gan, M.; Du, J.; Shen, L.; Yang, D.; Jiang, A.; Li, Q.; Jiang, Y.; Tang, G.; Li, M.; Wang, J. miR-152 regulates the proliferation and differentiation of C2C12 myoblasts by targeting E2F3. In Vitro Cell. Dev. Biol. Anim. 2018, 54, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ying, Z.-Z.; Tang, Z.-L.; Long, L.-Q.; Li, K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 2012, 287, 21093–21101. [Google Scholar] [CrossRef] [PubMed]
- Boss, O.; Samec, S.; Paoloni-Giacobino, A.; Rossier, C.; Dulloo, A.; Seydoux, J.; Muzzin, P.; Giacobino, J.-P. Uncoupling protein-3: A new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997, 408, 39–42. [Google Scholar] [CrossRef]
- Boss, O.; Hagen, T.; Lowell, B.B. Uncoupling proteins 2 and 3: Potential regulators of mitochondrial energy metabolism. Diabetes 2000, 49, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Schrauwen, P.; Saris, W.H.; Hesselink, M.K. An alternative function for human uncoupling protein 3: Protection of mitochondria against accumulation of nonesterified fatty acids inside the mitochondrial matrix. FASEB J. 2001, 15, 2497–2502. [Google Scholar] [CrossRef]
- Russell, A.; Wadley, G.; Hesselink, M.; Schaart, G.; Lo, S.; Leger, B.; Garnham, A.; Kornips, E.; Cameron-Smith, D.; Giacobino, J.-P. UCP3 protein expression is lower in type I, IIa and IIx muscle fiber types of endurance-trained compared to untrained subjects. Pflug. Arch. Eur. J. Phys. 2003, 445, 563–569. [Google Scholar] [CrossRef]
- Hesselink, M.; Keizer, H.; Borghouts, L.; Schaart, G.; Kornips, C.; Slieker, L.; Sloop, K.; Saris, W.; Schrauwen, P. Protein expression of UCP3 differs between human type 1, type 2a, and type 2b fibers. FASEB J. 2001, 15, 1071–1073. [Google Scholar] [CrossRef]
- MacLellan, J.D.; Gerrits, M.F.; Gowing, A.; Smith, P.J.; Wheeler, M.B.; Harper, M.-E. Physiological increases in uncoupling protein 3 augment fatty acid oxidation and decrease reactive oxygen species production without uncoupling respiration in muscle cells. Diabetes 2005, 54, 2343–2350. [Google Scholar] [CrossRef]
- Toime, L.J.; Brand, M.D. Uncoupling protein-3 lowers reactive oxygen species production in isolated mitochondria. Free Radic. Biol. Med. 2010, 49, 606–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, R.; Tresini, M. Oxidative stress and gene regulation. Free Radic. Biol. Med. 2000, 28, 463–499. [Google Scholar] [CrossRef]
- Droge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Hawley, J.A.; Zierath, J.R. Integration of metabolic and mitogenic signal transduction in skeletal muscle. Exerc. Sport Sci. Rev. 2004, 32, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Z.; Yang, K.; Shu, G.; Wang, S.; Gao, P.; Zhu, X.; Xi, Q.Y.; Zhang, Y.; Jiang, Q. EGCG reduces slow-twitch muscle fiber formation and mitochondrial biosynthesis in C2C12 cells by repressing AMPK activity and PGC-1α expression. J. Agric. Food Chem. 2016, 64, 6517–6523. [Google Scholar] [CrossRef] [PubMed]
- Canepari, M.; Pellegrino, M.; d’Antona, G.; Bottinelli, R. Skeletal muscle fibre diversity and the underlying mechanisms. Acta Physiol. 2010, 199, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Flueck, M. Tuning of mitochondrial pathways by muscle work: From triggers to sensors and expression signatures. Appl. Physiol. Nutr. Metab. 2009, 34, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Yan, H.; Cao, S.; Liu, J.; Zhang, H. Effect of feed intake level on the determination of apparent and standardized total tract digestibility of phosphorus for growing pigs. Anim. Feed Sci. Tech. 2018, 246, 137–143. [Google Scholar] [CrossRef]
- Yang, T.; Chen, X.; Huang, Z.; Wen, W.; Xu, M.; Chen, D.; Yu, B.; He, J.; Luo, J.; Yu, J. MicroRNA-27a promotes porcine myoblast proliferation by downregulating myostatin expression. Animal 2014, 8, 1867–1872. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Li, Y.; Yan, H.; Zhang, H. L-tryptophan enhances intestinal integrity in diquat-challenged piglets associated with improvement of redox status and mitochondrial function. Animals 2019, 9, 266. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, B.; Yu, J.; Zheng, P.; Huang, Z.; Luo, Y.; Luo, J.; Mao, X.; Yan, H.; He, J.; et al. Butyrate promotes slow-twitch myofiber formation and mitochondrial biogenesis in finishing pigs via inducing specific microRNAs and PGC-1α expression. J. Anim. Sci. 2019, 97, 3180–3192. [Google Scholar] [CrossRef]
- Yan, H.; Cao, S.; Li, Y.; Zhang, H.; Liu, J. Reduced meal frequency alleviates high-fat diet-induced lipid accumulation and inflammation in adipose tissue of pigs under the circumstance of fixed feed allowance. Eur. J. Nutr. 2019, in press. [Google Scholar] [CrossRef]
- Hintz, C.S.; Coyle, E.F.; Kaiser, K.K.; Chi, M.M.; Lowry, O.H. Comparison of muscle fiber typing by quantitative enzyme assays and by myosin ATPase staining. J. Histochem. Cytochem. 1984, 32, 655–660. [Google Scholar] [CrossRef]
- Liu, J.B.; Yan, H.L.; Zhang, Y.; Hu, Y.D.; Zhang, H.F. Effects of dietary energy and protein content and lipid source on growth performance and carcass traits in Pekin ducks. Poultry. Sci. 2019, in press. [Google Scholar] [CrossRef]
- Jia, A.; Feng, J.; Zhang, M.; Chang, Y.; Li, Z.; Hu, C.; Zhen, L.; Zhang, S.; Peng, Q. Effects of immunological challenge induced by lipopolysaccharide on skeletal muscle fiber type conversion of piglets. J. Anim. Sci. 2015, 93, 5194–5203. [Google Scholar] [CrossRef]
- Peter, J.B.; Barnard, R.J.; Edgerton, V.R.; Gillespie, C.A.; Stempel, K.E. Metabolic profiles of three fiber types of skeletal muscle in guinea pigs and rabbits. Biochemistry 1972, 11, 2627–2633. [Google Scholar] [CrossRef]
- Chen, X.; Guo, Y.; Jia, G.; Liu, G.; Zhao, H.; Huang, Z. Arginine promotes skeletal muscle fiber type transformation from fast-twitch to slow-twitch via Sirt1/AMPK pathway. J. Nutr. Biochem. 2018, 61, 155–162. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber tpes in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, B.; He, J.; Chen, D. From nutrient to microRNA: A novel insight into cell signaling involved in skeletal muscle development and disease. Int. J. Biol. Sci. 2016, 12, 1247–1261. [Google Scholar] [CrossRef]
- Putman, C.T.; Dixon, W.T.; Pearcey, J.A.; MacLean, I.M.; Jendral, M.J.; Kiricsi, M.; Murdoch, G.K.; Pette, D. Chronic low-frequency stimulation upregulates uncoupling protein-3 in transforming rat fast-twitch skeletal muscle. Am. J. Physiol. 2004, 287, R1419–R1426. [Google Scholar] [CrossRef]
- Joubert, R.; Collin, A.; Berri, C.; Lefaucheur, L.; Vincent, A.; Fillaut, M.; Ecolan, P.; Mur, L.; Godet, E.; Crochet, S. Expression of Uncoupling Proteins and Mitochondrial Activity Are Dependent on Muscular Fibre Type In Rabbits and Chickens, 2nd ed.; EAPP Publication: Vichy, France, 2007. [Google Scholar]
- Russell, A.P.; Somm, E.; Praz, M.; Crettenand, A.; Hartley, O.; Melotti, A.; Giacobino, J.P.; Muzzin, P.; Gobelet, C.; Dériaz, O. UCP3 protein regulation in human skeletal muscle fibre types I, IIa and IIx is dependent on exercise intensity. J. Physiol. 2003, 550, 855–861. [Google Scholar] [CrossRef]




| Gene | Primer Sequence (5′ to 3′) | Accession No. |
|---|---|---|
| MyHC I | F: GTTTGCCAACTATGCTGGGG | NM_213855.1 |
| R: TGTGCAGAGCTGACACAGTC | ||
| MyHC IIa | F: CTCTGAGTTCAGCAGCCATGA | NM214136.1 |
| R: GATGTCTTGGCATCAAAGGGC | ||
| MyHC IIx | F: TTGACTGGGCTGCCATCAAT | NM_001104951.2 |
| R: GCCTCAATGCGCTCCTTTTC | ||
| MyHC IIb | F: GAGGTACATCTAGTGCCCTGC | NM_001123141.1 |
| R: GCAGCCTCCCCAAAAATAGC | ||
| Myoglobin | F: GGAAGGTGGAGGCTGATGTC | NM_214236.1 |
| R: CCGTGCTTCTTCAGGTCCTC | ||
| Tnni1 | F: AGGAACACGAGGAGCGAGAG | NM_213912.3 |
| R: CACCTTGGCGTGAAGTTCC | ||
| Myf5 | F: GAGTTCGGGGACGAGTTTGA | NM_001278775.1 |
| R: TTTCCTCTTGCACGCTTTGC | ||
| MCK | F: CACCCCTTCATGTGGAACGA | NM_001129949.1 |
| R: TCGAACTTGGGATGCTTGCT | ||
| MyoD | F: TGCCCAAGGTGGAAATCCTG | NM_001002824.1 |
| R: GCTGTAATAGGTGCCGTCGT | ||
| MyoG | F: GAAAACTACCTGCCCGTCCA | NM_001012406.1 |
| R: CCACAGACACGGACTTCCTC | ||
| β-Actin | F: TGTACACACCTCATGCCAGC | EU655628.1 |
| R: GAGCCGCGTGTGTGTAACTA | ||
| miR-152 | RT: GTCGTATCCAGTGCAGGGTCCGAG GTATTCGCACTGGATACGACCCAAGT | |
| F: CGCGACAGTGCATGACAGA | ||
| R: AGTGCAGGGTCCGAGGTATT | ||
| U6 | F: CTCGCTTCGGCAGCACA | |
| R: AACGCTTCACGAATTTGCGT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Yan, H.; Zhou, P.; Zhang, Z.; Liu, J.; Zhang, H. MicroRNA-152 Promotes Slow-Twitch Myofiber Formation via Targeting Uncoupling Protein-3 Gene. Animals 2019, 9, 669. https://doi.org/10.3390/ani9090669
Zhang Y, Yan H, Zhou P, Zhang Z, Liu J, Zhang H. MicroRNA-152 Promotes Slow-Twitch Myofiber Formation via Targeting Uncoupling Protein-3 Gene. Animals. 2019; 9(9):669. https://doi.org/10.3390/ani9090669
Chicago/Turabian StyleZhang, Yong, Honglin Yan, Pan Zhou, Zhenzhen Zhang, Jingbo Liu, and Hongfu Zhang. 2019. "MicroRNA-152 Promotes Slow-Twitch Myofiber Formation via Targeting Uncoupling Protein-3 Gene" Animals 9, no. 9: 669. https://doi.org/10.3390/ani9090669





