Deciphering the Patterns of Genetic Admixture and Diversity in the Ecuadorian Creole Chicken
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sampling and DNA Extraction
2.3. Molecular Marker Analysis
2.3.1. Mitochondrial DNA D-Loop Analysis
2.3.2. Microsatellites Markers
2.4. Statistical and Genetic Analyses
2.4.1. Mitochondrial DNA D-Loop
2.4.2. Microsatellites Markers
3. Results
3.1. Mitochondrial DNA Phylogeny
3.2. Microsatellites Markers
3.2.1. Marker Polymorphism and Diversity
3.2.2. Population Structure
4. Discussion
4.1. Mitochondrial DNA D-loop Analysis
4.2. Microsatellites Markers
4.2.1. Genetic Diversity
4.2.2. Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amores Cárdenas, C.E. Determinación de la viabilidad financiera de la producción avícola mediante la utilización de planteles de crianza de pollo de engorde en la región oriental del Ecuador; Pontificia Universidad Católica del Ecuador: Quito, Ecuador, 2016. [Google Scholar]
- Seligmann, L.J. The Chicken in Andean History and Myth - the Quechua Concept of Wallpa. Ethnohistory 1987, 34, 139–170. [Google Scholar] [CrossRef]
- Dancause, K.N.; Vilar, M.G.; Steffy, R.; Lum, J.K. Characterizing genetic diversity of contemporary pacific chickens using mitochondrial DNA analyses. PLoS ONE 2011, 6, e16843. [Google Scholar] [CrossRef] [PubMed]
- Gongora, J.; Rawlence, N.J.; Mobegi, V.A.; Jianlin, H.; Alcalde, J.A.; Matus, J.T.; Hanotte, O.; Moran, C.; Austin, J.J.; Ulm, S.; et al. Indo-European and Asian origins for Chilean and Pacific chickens revealed by mtDNA. Proc. Natl. Acad. Sci. USA 2008, 105, 10308–10313. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.W.; Peng, M.S.; Wu, G.S.; Ouyang, Y.N.; Yang, Z.Y.; Yu, N.; Liang, J.P.; Pianchou, G.; Beja-Pereira, A.; Mitra, B.; et al. Chicken domestication: An updated perspective based on mitochondrial genomes. Heredity 2013, 110, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Storey, A.A.; Ramirez, J.M.; Quiroz, D.; Burley, D.V.; Addison, D.J.; Walter, R.; Anderson, A.J.; Hunt, T.L.; Athens, J.S.; Huynen, L.; et al. Radiocarbon and DNA evidence for a pre-Columbian introduction of Polynesian chickens to Chile. Proc. Natl. Acad. Sci. USA 2007, 104, 10335–10339. [Google Scholar] [CrossRef] [PubMed]
- Alderson, G.L.H. Conservation of breeds and maintenance of biodiversity: Justification and methodology for the conservation of Animal Genetic Resources. Arch. Zootec. 2018, 67. [Google Scholar]
- Carvalho, N.; Canela, F.M.; Leite, P.H.S.; Ferreira, M.A.; Oliveira, V.R.; Santos, M.F.; Souza, N.O.S.; Buso, G.S.C. Analysis of genetic variability of commercial melon cultivars using SSR molecular markers. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef] [PubMed]
- Ceccobelli, S.; Di Lorenzo, P.; Lancioni, H.; Ibáñez, L.V.M.; Tejedor, M.T.; Castellini, C.; Landi, V.; Martínez, A.M.; Bermejo, J.V.D.; Pla, J.L.V. Genetic diversity and phylogeographic structure of sixteen Mediterranean chicken breeds assessed with microsatellites and mitochondrial DNA. Livest. Sci. 2015, 175, 27–36. [Google Scholar] [CrossRef]
- Delgado, J.V.; Martinez, A.M.; Acosta, A.; Alvarez, L.A.; Armstrong, E.; Camacho, E.; Canon, J.; Cortes, O.; Dunner, S.; Landi, V.; et al. Genetic characterization of Latin-American Creole cattle using microsatellite markers. Anim. Genet. 2012, 43, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Ferrando, A.; Manunza, A.; Jordana, J.; Capote, J.; Pons, A.; Pais, J.; Delgado, T.; Atoche, P.; Cabrera, B.; Martinez, A.; et al. A mitochondrial analysis reveals distinct founder effect signatures in Canarian and Balearic goats. Anim. Genet. 2015, 46, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Pons, A.L.; Landi, V.; Martinez, A.; Delgado, J.V. The biodiversity and genetic structure of Balearic sheep breeds. J. Anim. Breed. Genet. = Zeitschrift fur Tierzuchtung und Zuchtungsbiologie 2015, 132, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.; Landi, V.; Martinez, A.; Gomez, M.; Camacho, M.E.; Alvarez, L.A.; Aguirre, L.; Delgado, J.V. Molecular Study of the Amazonian Macabea Cattle History. PLoS ONE 2016, 11, e0165398. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.S.; Varlaro, J.; Reynolds, R. A rapid chemiluminescent method for quantitation of human DNA. Nucleic Acids Res. 1992, 20, 5061–5065. [Google Scholar] [CrossRef] [PubMed]
- Nishibori, M.; Shimogiri, T.; Hayashi, T.; Yasue, H. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim. Genet. 2005, 36, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Ceccobelli, S.; Lorenzo, P.D.; Lancioni, H.; Castellini, C.; Ibáñez, L.V.M.; Sabbioni, A.; Sarti, F.M.; Weigend, S.; Lasagna, E. Phylogeny, genetic relationships and population structure of five Italian local chicken breeds. Ital. J. Anim. Sci. 2013, 12, e66. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Park, S.D.E. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection; University of Dublin: Dublin, Ireland, 2001. [Google Scholar]
- Langella, O. Population Genetic Software (Individuals or Populations Distances, Phylogenetic Trees). Available online: http://www.bioinformatics.org/download.php?fileid=430 (accessed on 12 May 2002).
- Kalinowski, S.T. hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. An exact test for population differentiation. Evolution 1995, 49, 1280–1283. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.W.; Thompson, E.A. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992, 48, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations; Laboratoire Génome, Populations, Interactions, CNRS UMR 5000; Université de Montpellier II: Montpellier, France, 2004; pp. 1996–2004. [Google Scholar]
- Reynolds, J.; Weir, B.S.; Cockerham, C.C. Estimation of the coancestry coefficient: Basis for a short-term genetic distance. Genetics 1983, 105, 767–779. [Google Scholar]
- Langella, P.; Le Loir, Y. Heterologous protein secretion in Lactococcus lactis: A novel antigen delivery system. Braz. J. Med. Biol. Res. 1999, 32. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2005, 23, 254–267. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- François, O.; Ancelet, S.; Guillot, G. Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics 2006, 174, 805–816. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Muchadeyi, F.C.; Eding, H.; Simianer, H.; Wollny, C.B.; Groeneveld, E.; Weigend, S. Mitochondrial DNA D-loop sequences suggest a Southeast Asian and Indian origin of Zimbabwean village chickens. Anim. Genet. 2008, 39, 615–622. [Google Scholar] [CrossRef]
- Liao, Y.; Mo, G.; Sun, J.; Wei, F.; Liao, D.J. Genetic diversity of Guangxi chicken breeds assessed with microsatellites and the mitochondrial DNA D-loop region. Mol. Biol. Rep. 2016, 43, 415–425. [Google Scholar] [CrossRef]
- Liu, Y.P.; Wu, G.S.; Yao, Y.G.; Miao, Y.W.; Luikart, G.; Baig, M.; Beja-Pereira, A.; Ding, Z.L.; Palanichamy, M.G.; Zhang, Y.P. Multiple maternal origins of chickens: Out of the Asian jungles. Mol. Phylogenet. Evol. 2006, 38, 12–19. [Google Scholar] [CrossRef]
- Komiyama, T.; Ikeo, K.; Gojobori, T. The evolutionary origin of long-crowing chicken: Its evolutionary relationship with fighting cocks disclosed by the mtDNA sequence analysis. Gene 2004, 333, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Luzuriaga-Neira, A.; Villacis-Rivas, G.; Cueva-Castillo, F.; Escudero-Sanchez, G.; Ulloa-Nunez, A.; Rubilar-Quezada, M.; Monteiro, R.; Miller, M.R.; Beja-Pereira, A. On the origins and genetic diversity of South American chickens: One step closer. Anim. Genet. 2017, 48, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Teinlek, P.; Siripattarapravat, K.; Tirawattanawanich, C. Genetic diversity analysis of Thai indigenous chickens based on complete sequences of mitochondrial DNA D-loop region. Asian-Australas J. Anim. Sci. 2018, 31, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.; Sharma, A.; Lee, S.; Cho, C.Y.; Kim, J.H.; Choi, S.B.; Kim, H.; Seong, H.H.; Yeon, S.H.; Kim, D.H.; et al. Genetic diversity and relationships of korean chicken breeds based on 30 microsatellite markers. Asian-Australas J. Anim. Sci. 2014, 27, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.-X.; Zhang, G.-X.; Wang, J.-Y.; Li, Y.; Zhang, L.-J.; Wei, Y.; Wang, H.-H.; Zhang, L.; Hou, Q.-R. Genetic Diversity of a Chinese Native Chicken Breed, Bian Chicken, Based on Twenty-nine Microsatellite Markers. Asian-Australas J. Anim. Sci. 2010, 23, 154–161. [Google Scholar] [CrossRef]
- Fathi, M.; El-Zarei, M.; Al-Homidan, I.; Abou-Emera, O. Genetic diversity of Saudi native chicken breeds segregating for naked neck and frizzle genes using microsatellite markers. Asian-Australas J. Anim. Sci. 2018, 31, 1871–1880. [Google Scholar] [CrossRef]
- Chen, G.; Bao, W.; Shu, J.; Ji, C.; Wang, M.; Eding, H.; Muchadeyi, F.; Weigend, S. Assessment of Population Structure and Genetic Diversity of 15 Chinese Indigenous Chicken Breeds Using Microsatellite Markers. Asian-Australas J. Anim. Sci. 2008, 21, 331–339. [Google Scholar] [CrossRef]
- Petit, R.J.; El Mousadik, A.; Pons, O. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 1998, 12, 844–855. [Google Scholar] [CrossRef]
- Ollivier, L.; Foulley, J.-L. Aggregate diversity: New approach combining within-and between-breed genetic diversity. Livest. Prod. Sci. 2005, 95, 247–254. [Google Scholar] [CrossRef]
- Hill, W.G.; Rasbash, J. Models of long term artificial selection in finite population. Genet. Res. 1986, 48, 41–50. [Google Scholar] [CrossRef]
- Mukesh; Fernandes, M.; Han, J.L.; Sathyakumar, S. Genetics Driven Interventions for Ex Situ Conservation of Red Junglefowl (Gallus gallus murghi) Populations in India. Zoo Biol. 2013, 32, 476–483. [Google Scholar] [CrossRef] [PubMed]
- De Meeus, T. Revisiting FIS, FST, Wahlund Effects, and Null Alleles. J. Hered. 2018, 109, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, K.B.; Schurr, T.G.; Long, J.C.; Rosenberg, N.A.; Crawford, M.H.; Tarskaia, L.A.; Osipova, L.P.; Zhadanov, S.I.; Smith, D.G. A private allele ubiquitous in the Americas. Biol. Lett.-UK 2007, 3, 218–223. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bianchi, M.; Ceccobelli, S.; Landi, V.; Di Lorenzo, P.; Lasagna, E.; Ciocchetti, M.; Şahin, E.; Mugnai, C.; Panella, F.; Sarti, F.M. A microsatellites-based survey on the genetic structure of two Italian local chicken breeds. Ital. J. Anim. Sci. 2011, 10, e39. [Google Scholar] [CrossRef][Green Version]
- Emara, M.G.; Kim, H.; Zhu, J.; Lapierre, R.R.; Lakshmanan, N.; Lillehojt, H.S. Genetic diversity at the major histocompatibility complex (B) and microsatellite loci in three commercial broiler pure lines. Poult. Sci. 2002, 81, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Toalombo Vargas, P.A.; Gavilánez, A.A.V.; Lara, J.C.B.; Carrillo, E.R.O. Caracterización del mercado de huevo comercial (gallina lohmann brown) versus el huevo criollo (gallina de campo) en la Provincia de Tungurahua. Comer. y Neg. 2018, 50–60. [Google Scholar]
- Pomboza-Tamaquiza, P.; Guerrero-López, R.; Guevara-Freire, D.; Rivera, V. Granjas avícolas y autosuficiencia de maíz y soya: Caso Tungurahua-Ecuador. Estud. Soc. (Hermosillo, Son.) 2018, 28, 0. [Google Scholar] [CrossRef][Green Version]
- Avilés Esquivel, D. Caracterización genética del cuy doméstico de América del Sur mediante marcadores moleculares; Universidad de Cordoba: Cordoba, Spain, 2016. [Google Scholar]
- Francesch, A.; Cartaña, M. Coeficientes de endogamia y diferenciación poblacional en cuatro variedades de gallina Penedesenca después de 25 años de reproducción en población cerrada. In Proceedings of the Congreso Cientifico de Avicoltura, Lleida, Spain, 2–4 October 2013. [Google Scholar]
- Kong, H.S.; Oh, J.D.; Lee, J.H.; Jo, K.J.; Sang, B.D.; Choi, C.H.; Kim, S.D.; Lee, S.J.; Yeon, S.H.; Jeon, G.J.; et al. Genetic Variation and Relationships of Korean Native Chickens and Foreign Breeds Using 15 Microsatellite Markers. Asian-Australas J. Anim. Sci. 2006, 19, 1546–1550. [Google Scholar] [CrossRef]
- Arcos-Burgos, M.; Muenke, M. Genetics of population isolates. Clin. Genet. 2002, 61, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Finzi, A. Integrated backyard system. A contribution to the special programme for food security. In Boletín técnico de la FAO; University of Tuscia: Viterbo, Italia, 2000. [Google Scholar]
- Revidatti, F.; Rafart, J.F.; Terraes, J.C.; Fernandez, R.J.; Sandoval, G.L.; Asiain, M.V.; Sindik, M.M. Rendimiento reproductivo en cruzamientos entre razas tradicionales de aves productoras de huevo y carne. InVet 2005, 7, 19–23. [Google Scholar]
- Ferreira, E.; Souto, L.; Soares, A.; Fonseca, C. Genetic structure of the wild boar (Sus scrofa L.) population in Portugal. Wildl. Biol. Pract. 2006, 2, 17–25. [Google Scholar]
- Wheeldon, T.; White, B.N. Genetic analysis of historic western Great Lakes region wolf samples reveals early Canis lupus/lycaon hybridization. Biol. Lett.-UK 2008, 5, 101–104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kanginakudru, S.; Metta, M.; Jakati, R.D.; Nagaraju, J. Genetic evidence from Indian red jungle fowl corroborates multiple domestication of modern day chicken. BMC Evol. Biol. 2008, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, H.; Bhatia, K.; Wang, K.; Wang, Z. BioPig: A Hadoop-based analytic toolkit for large-scale sequence data. Bioinformatics 2013, 29, 3014–3019. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Delgado Bermejo, J.V.; Martínez, A.; Camacho Vallejo, M.E.; Vega Pla, J.L. Capítulo 6: Conservación de Razas de especies Domésticas. In Genética de Animales Domésticos; Giovambattista, G., Peral-García, P., Eds.; Inte-Médica: Ciudad Autónoma de Buenos Aires, Argentina, 2010; pp. 105–121. [Google Scholar]
- Rodero Serrano, E.; Rodero Franganillo, A.; Delgado-Bermejo, J.V. Primitive andalusian livestock and their implications in the discovery of America. Arch. Zootec. 1992, 41, 10. [Google Scholar]
- Rosenberg, N.A.; Pritchard, J.K.; Weber, J.L.; Cann, H.M.; Kidd, K.K.; Zhivotovsky, L.A.; Feldman, M.W. Genetic structure of human populations. Science 2002, 298, 2381–2385. [Google Scholar] [CrossRef] [PubMed]
- Waltmann, A.; Koepfli, C.; Tessier, N.; Karl, S.; Fola, A.; Darcy, A.W.; Wini, L.; Harrison, G.L.A.; Barnadas, C.; Jennison, C.; et al. Increasingly inbred and fragmented populations of Plasmodium vivax associated with the eastward decline in malaria transmission across the Southwest Pacific. PLoS Negl. Trop. Dis. 2018, 12, e0006146. [Google Scholar] [CrossRef] [PubMed]
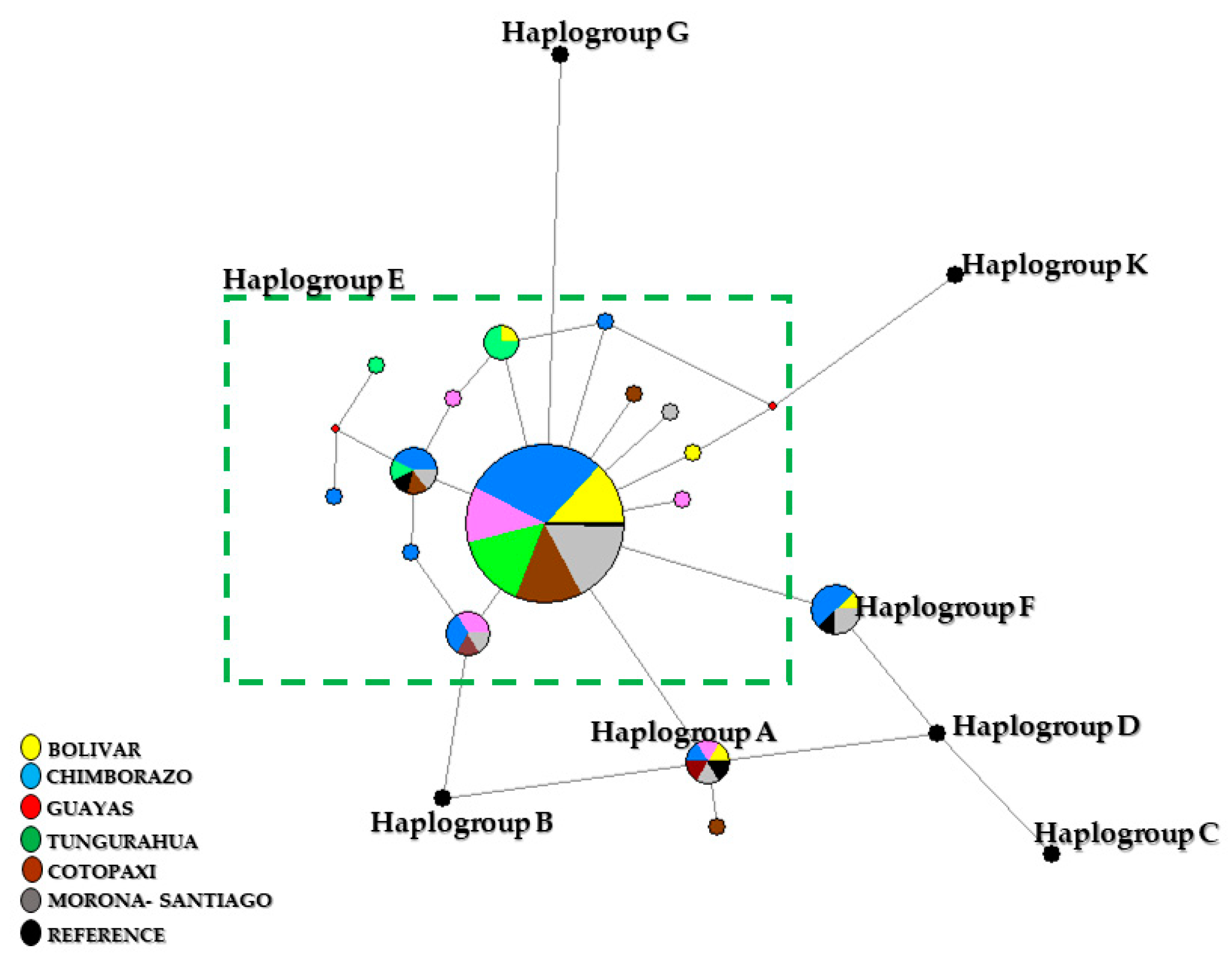
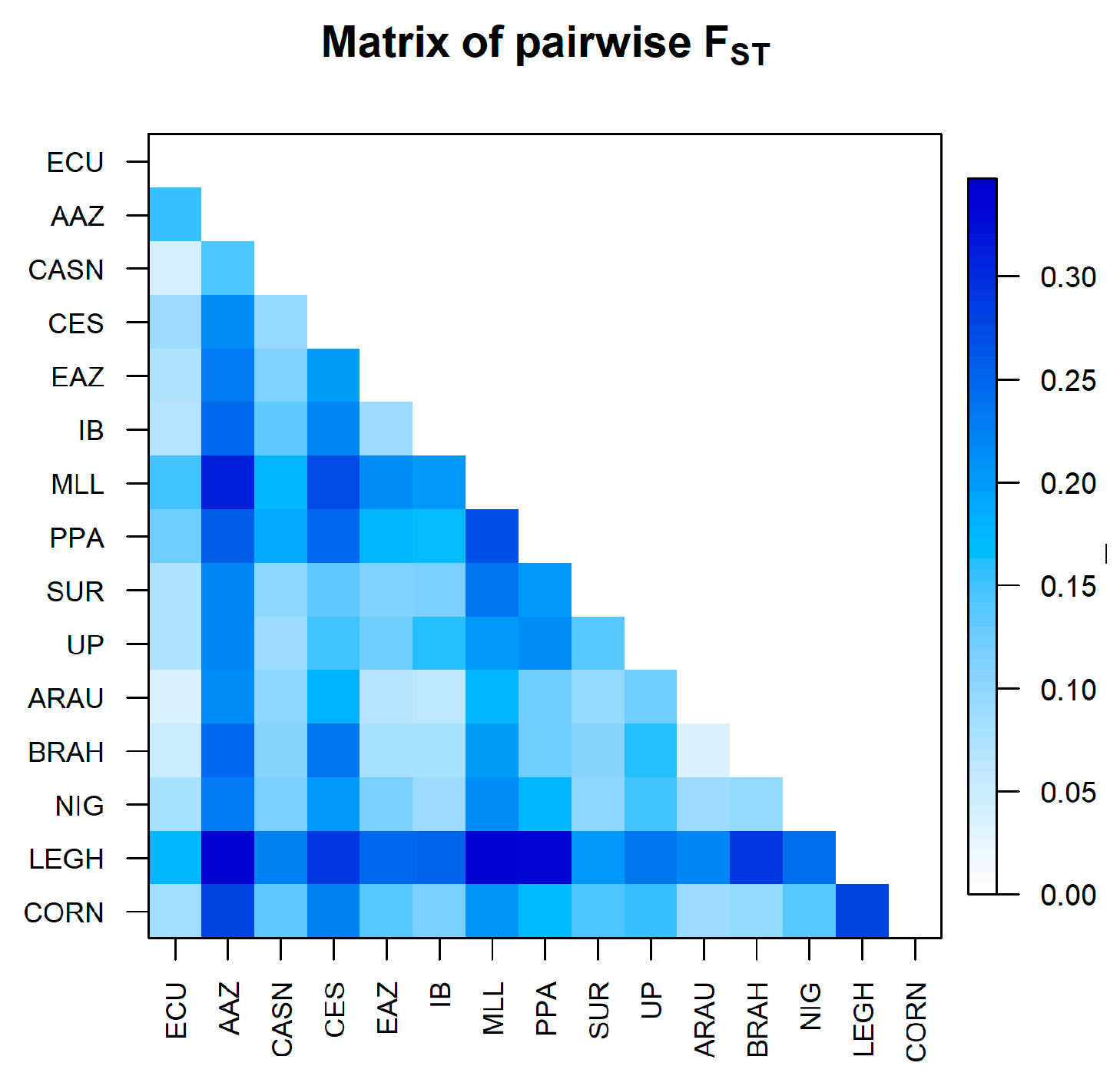
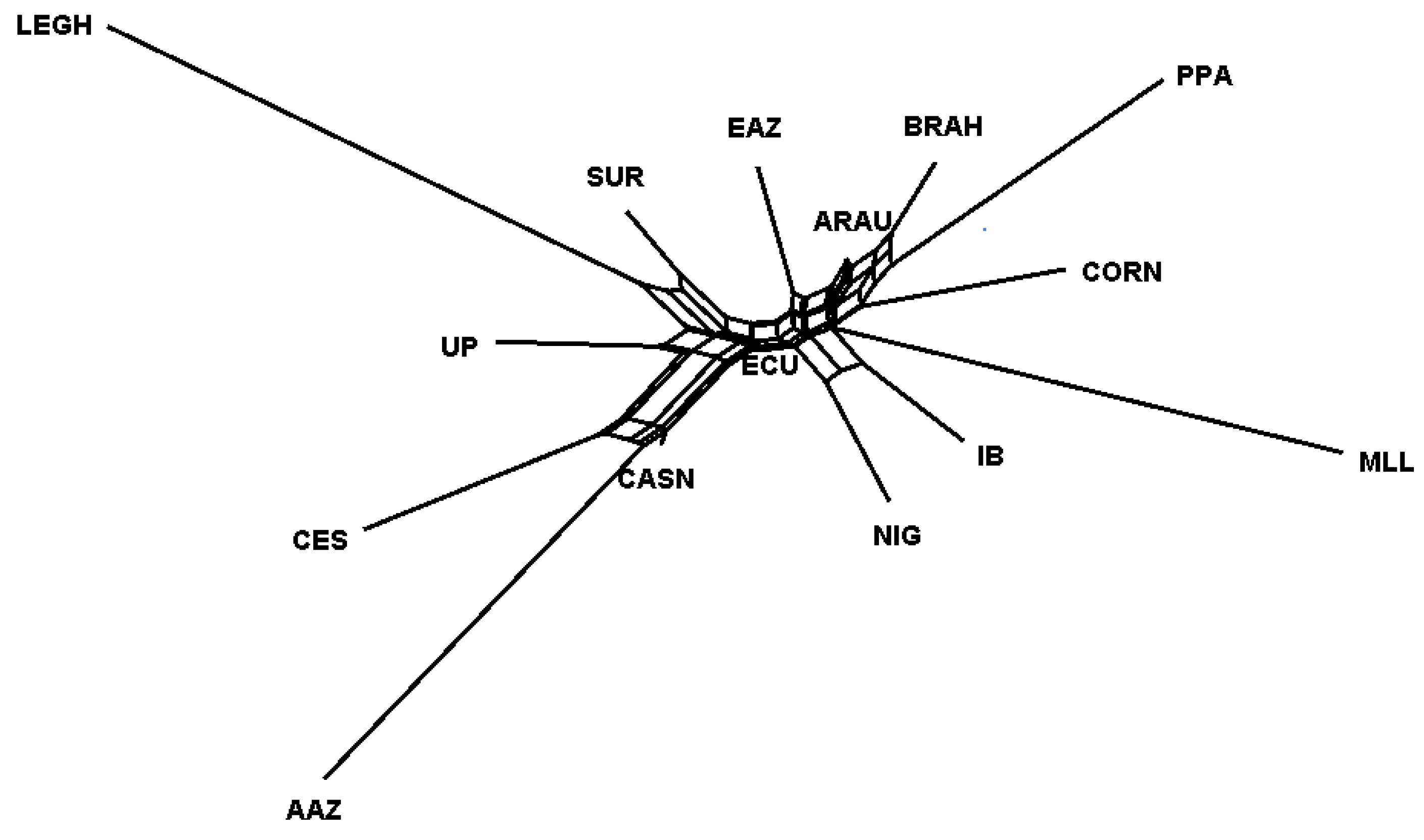


| Province | n | H | S | Hd | Π | D-Tajima |
|---|---|---|---|---|---|---|
| Bolívar | 31 | 6 | 5 | 0.301 | 0.00091 | −2.0081 * |
| Chimborazo | 70 | 9 | 84 | 0.358 | 0.00745 | −2.8870 *** |
| Guayas | 28 | 7 | 9 | 0.442 | 0.00235 | −2.01611 * |
| Tungurahua | 35 | 4 | 66 | 0.311 | 0.01121 | −2.76689 *** |
| Cotopaxi | 32 | 6 | 7 | 0.292 | 0.00140 | −2.07960* |
| Morona Santiago | 38 | 10 | 37 | 0.461 | 0.00577 | −2.70578 *** |
| All samples | 234 | 24 | 123 | 0.359 | 0.00542 | −2.85904 *** |
| Province | N | He | Ho | NA | AE | PAR | FIS |
|---|---|---|---|---|---|---|---|
| Bolivar | 35 | 0.6298 | 0.5500 | 5.80 | 3.58 | 0.25 | 0.128 * |
| Chimborazo | 72 | 0.6104 | 0.5341 | 6.20 | 3.46 | 0.23 | 0.126 * |
| Cotopaxi | 32 | 0.6039 | 0.5251 | 5.40 | 3.39 | 0.19 | 0.132 * |
| Guayas | 30 | 0.6531 | 0.5903 | 5.50 | 3.61 | 0.23 | 0.098 * |
| Morona-Santiago | 39 | 0.6426 | 0.5809 | 5.77 | 3.60 | 0.26 | 0.097 * |
| Tungurahua | 36 | 0.6284 | 0.5425 | 5.37 | 3.54 | 0.31 | 0.138 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toalombo Vargas, P.A.; León, J.M.; Fiallos Ortega, L.R.; Martinez, A.; Villafuerte Gavilanes, A.A.; Delgado, J.V.; Landi, V. Deciphering the Patterns of Genetic Admixture and Diversity in the Ecuadorian Creole Chicken. Animals 2019, 9, 670. https://doi.org/10.3390/ani9090670
Toalombo Vargas PA, León JM, Fiallos Ortega LR, Martinez A, Villafuerte Gavilanes AA, Delgado JV, Landi V. Deciphering the Patterns of Genetic Admixture and Diversity in the Ecuadorian Creole Chicken. Animals. 2019; 9(9):670. https://doi.org/10.3390/ani9090670
Chicago/Turabian StyleToalombo Vargas, Paula Alexandra, José Manuel León, Luis Rafael Fiallos Ortega, Amparo Martinez, Alex Arturo Villafuerte Gavilanes, Juan Vicente Delgado, and Vincenzo Landi. 2019. "Deciphering the Patterns of Genetic Admixture and Diversity in the Ecuadorian Creole Chicken" Animals 9, no. 9: 670. https://doi.org/10.3390/ani9090670
APA StyleToalombo Vargas, P. A., León, J. M., Fiallos Ortega, L. R., Martinez, A., Villafuerte Gavilanes, A. A., Delgado, J. V., & Landi, V. (2019). Deciphering the Patterns of Genetic Admixture and Diversity in the Ecuadorian Creole Chicken. Animals, 9(9), 670. https://doi.org/10.3390/ani9090670





