Four Novel SNPs of MYO1A Gene Associated with Heat-Tolerance in Chinese Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Samples and Data Collection
2.2. SNP Detecting and Genotyping
2.3. Statistical Analysis
3. Results
3.1. SNPs Screening and Genotyping
3.2. Haplotype Analysis and Linkage Disequilibrium
3.3. Association Analysis of SNPs and Haplotype Combination
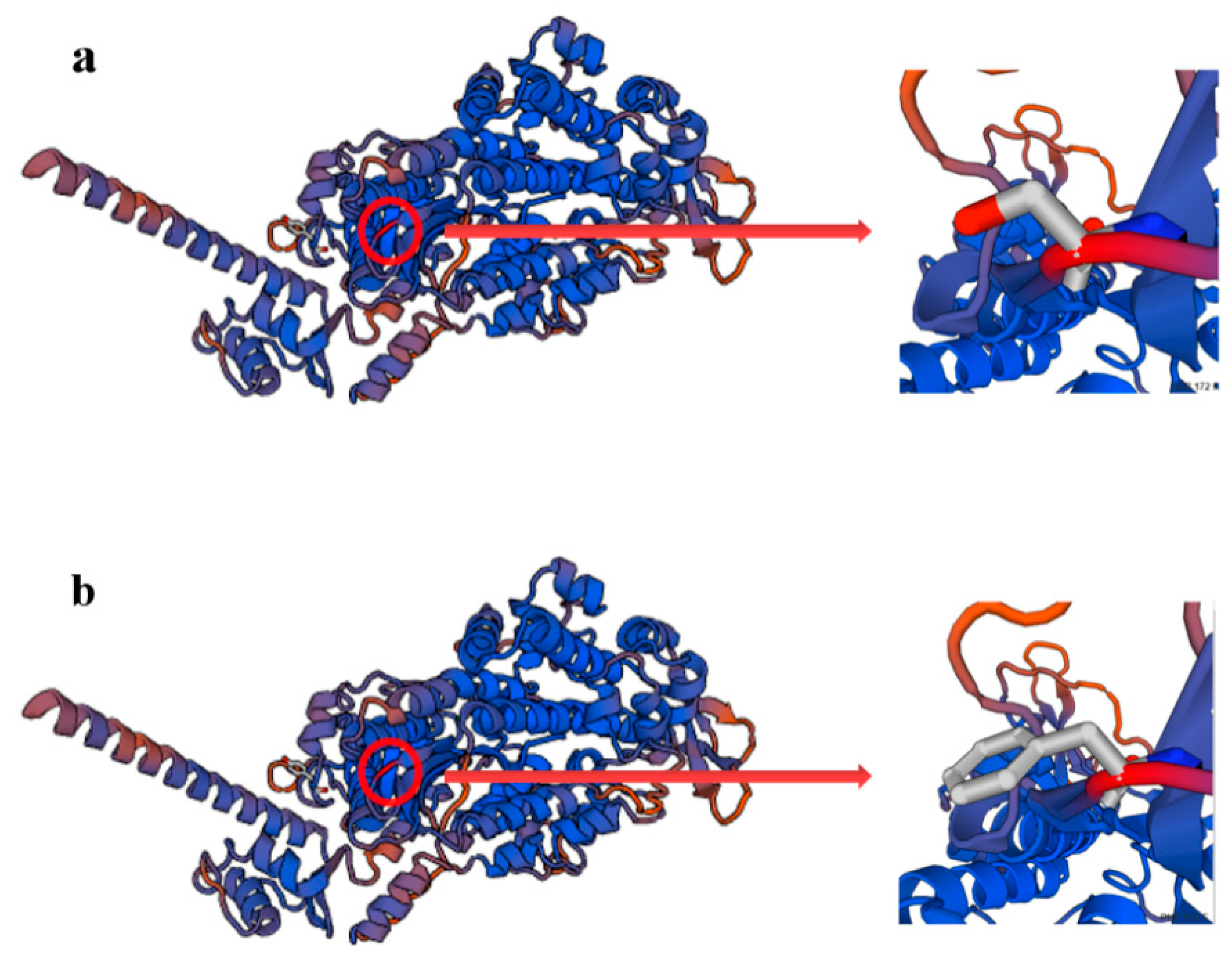
3.4. Mutation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wheelock, J.B.; Rhoads, R.P.; VanBaale, M.J.; Sanders, S.R.; Baumgard, L.H. Effects of heat stress on energetic metabolism in lactating Holstein cows. J. Dairy Sci. 2010, 93, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Garner, J.B.; Douglas, M.L.; Williams, S.O.; Wales, W.J.; Marett, L.C.; Nguyen, T.T.T.; Reich, C.M.; Hayes, B.J. Genomic Selection Improves Heat Tolerance in Dairy Cattle. Sci. Rep. 2016, 6, 34114. [Google Scholar] [CrossRef] [PubMed]
- Hansen, P.J. Effects of coat colour on physiological responses to solar radiation in Holsteins. Vet. Rec. 1990, 127, 333–334. [Google Scholar] [PubMed]
- Hansen, P.J. Physiological and cellular adaptations of zebu cattle to thermal stress. Anim. Reprod. Sci. 2004, 82, 349–360. [Google Scholar] [CrossRef]
- Hartman, M.A.; Spudich, J.A. The myosin superfamily at a glance. J. Cell Sci. 2012, 125, 1627–1632. [Google Scholar] [CrossRef]
- Heissler, S.M.; Sellers, J.R. Various Themes of Myosin Regulation. J. Mol. Biol. 2016, 428, 1927–1946. [Google Scholar] [CrossRef]
- Attanapola, S.L.; Alexander, C.J.; Mulvihill, D.P. Ste20-kinase-dependent TEDS-site phosphorylation modulates the dynamic localisation and endocytic function of the fission yeast class I myosin, Myo1. J. Cell Sci. 2009, 122, 3856–3861. [Google Scholar] [CrossRef]
- Hammer, J.A.; Sellers, J.R. Walking to work: Roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 2012, 13, 13–26. [Google Scholar] [CrossRef]
- Edea, Z.; Dadi, H.; Dessie, T.; Uzzaman, M.R.; Rothschild, M.F.; Kim, E.S.; Sonstegard, T.S.; Kim, K.S. Genome-wide scan reveals divergent selection among taurine and zebu cattle populations from different regions. Anim. Genet. 2018, 49, 550–563. [Google Scholar] [CrossRef]
- Chen, N.; Fu, W.; Zhao, J.; Shen, J.; Chen, Q.; Zheng, Z.; Chen, H.; Sonstegard, T.S.; Lei, C.; Jiang, Y. The Bovine Genome Variation Database (BGVD): Integrated Web-database for Bovine Sequencing Variations and Selective Signatures. BioRxiv 2019, 802223. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, X.; Zhang, Y.; Zhao, Y.; Zhang, J.; Jia, Y.; Zhu, B.; Xu, L.; Zhang, L.; Gao, H.; et al. Genome-wide assessment of genetic diversity and population structure insights into admixture and introgression in Chinese indigenous cattle. BMC Genet. 2018, 19, 114. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Gautier, M.; Ding, X.; Zhang, H.; Wang, Y.; Wang, X.; Faruque, M.O.; Li, J.; Ye, S.; Gou, X.; et al. Species composition and environmental adaptation of indigenous Chinese cattle. Sci. Rep. 2017, 7, 16196. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.J.; Liu, Y.P.; Liu, Y.X.; Li, X.W.; Yao, Y.G. Genetic diversity and origin of Chinese cattle revealed by mtDNA D-loop sequence variation. Mol. Phylogenet. Evol. 2006, 38, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.Z.; Chen, H.; Zhang, H.C.; Cai, X.; Liu, R.Y.; Luo, L.Y.; Wang, C.F.; Zhang, W.; Ge, Q.L.; Zhang, R.F.; et al. Origin and phylogeographical structure of Chinese cattle. Anim. Genet. 2010, 37, 579–582. [Google Scholar] [CrossRef]
- Li, R.; Zhang, X.M.; Campana, M.G.; Huang, J.P.; Chang, Z.H.; Qi, X.B.; Shi, H.; Su, B.; Zhang, R.F.; Lan, X.Y.; et al. Paternal origins of Chinese cattle. Anim. Genet. 2013, 44, 446–449. [Google Scholar] [CrossRef]
- Denman, A.M. Molecular Cloning: A Laboratory Manual. Immunology 1983, 49, 411. [Google Scholar]
- Yong, Y.O.N.G.; Lin, H.E. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005, 15, 97. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Mcdowell, R.E.; Hooven, N.W.; Camoens, J.K. Effect of Climate on Performance of Holsteins in First Lactation. J. Dairy Sci. 1976, 59, 965–971. [Google Scholar] [CrossRef]
- Sun, Y.; Sun, Y.; Sun, Z.; Li, P.; Li, C. Mapping monthly distribution of daily light integrals across China. Hunan For. Sci. Technol. 2015, 42, 43–47. [Google Scholar]
- Dikmen, S.; Hansen, P.J. Is the temperature-humidity index the best indicator of heat stress in lactating dairy cows in a subtropical environment? J. Dairy Sci. 2009, 92, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Ardlie, K.G.; Leonid, K.; Mark, S. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 2002, 3, 299–309. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, C.J.; Swain, D.L.; Kadarmideen, H.N. Evolutionary process of Bos taurus cattle in favourable versus unfavourable environments and its implications for genetic selection. Evol. Appl. 2010, 3, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Mazzolini, R.; Dopeso, H.; Mateo-Lozano, S.; Chang, W.; Rodrigues, P.; Bazzocco, S.; Alazzouzi, H.; Landolfi, S.; Hernández-Losa, J.; Andretta, E.; et al. Brush border myosin Ia has tumor suppressor activity in the intestine. Proc. Natl. Acad. Sci. USA 2012, 109, 1530–1535. [Google Scholar] [CrossRef]
- Donaudy, F.; Ferrara, A.; Esposito, L.; Hertzano, R.; Ben-David, O.; Bell, R.E.; Melchionda, S.; Zelante, L.; Avraham, K.B.; Gasparini, P. Multiple Mutations of MYO1A, a Cochlear-Expressed Gene, in Sensorineural Hearing Loss. Am. J. Hum. Genet. 2003, 72, 1571–1577. [Google Scholar] [CrossRef]
- Bennett, D.C.; MLynn, L. The color loci of mice—A genetic century. Pigment Cell Res. 2010, 16, 333–344. [Google Scholar] [CrossRef]
- O’Sullivan, T.N.; Wu, X.S.; Rachel, R.A.; Huang, J.D.; Swing, D.A.; Matesic, L.E.; Hammer, J.A.; Copeland, N.G.; Jenkins, N.A. Dsu functions in a MYO5A-independent pathway to suppress the coat color of dilute mice. Proc. Natl. Acad. Sci. USA 2004, 101, 16831–16836. [Google Scholar] [CrossRef]
- Wu, X.S.; Rao, K.; Zhang, H.; Wang, F.; Sellers, J.R.; Matesic, L.E.; Copeland, N.G.; Jenkins, N.A.; Hammer, J.A., III. Identification of an organelle receptor for myosin-Va. Nat. Cell Biol. 2002, 4, 271–278. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, X.; Guan, L.; Xu, S.; Yuan, Y.; Lv, D.; He, X.; Zhan, J.; Kong, Y.; Guo, J.; et al. Unconventional myosin VIIA promotes melanoma progression. J. Cell Sci. 2018, 131, 209924. [Google Scholar] [CrossRef]
- Gutiérrez-Gil, B.; Wiener, P.; Williams, J.L. Genetic effects on coat colour in cattle: Dilution of eumelanin and phaeomelanin pigments in an F2-Backcross Charolais x Holstein population. BMC Genet. 2007, 8, 56. [Google Scholar] [CrossRef]
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef] [PubMed]
- Wallberg, A.; Schöning, C.; Webster, M.T. Two extended haplotype blocks are associated with adaptation to high altitude habitats in East African honey bees. PLoS Genet. 2017, 13, e1006792. [Google Scholar] [CrossRef] [PubMed]
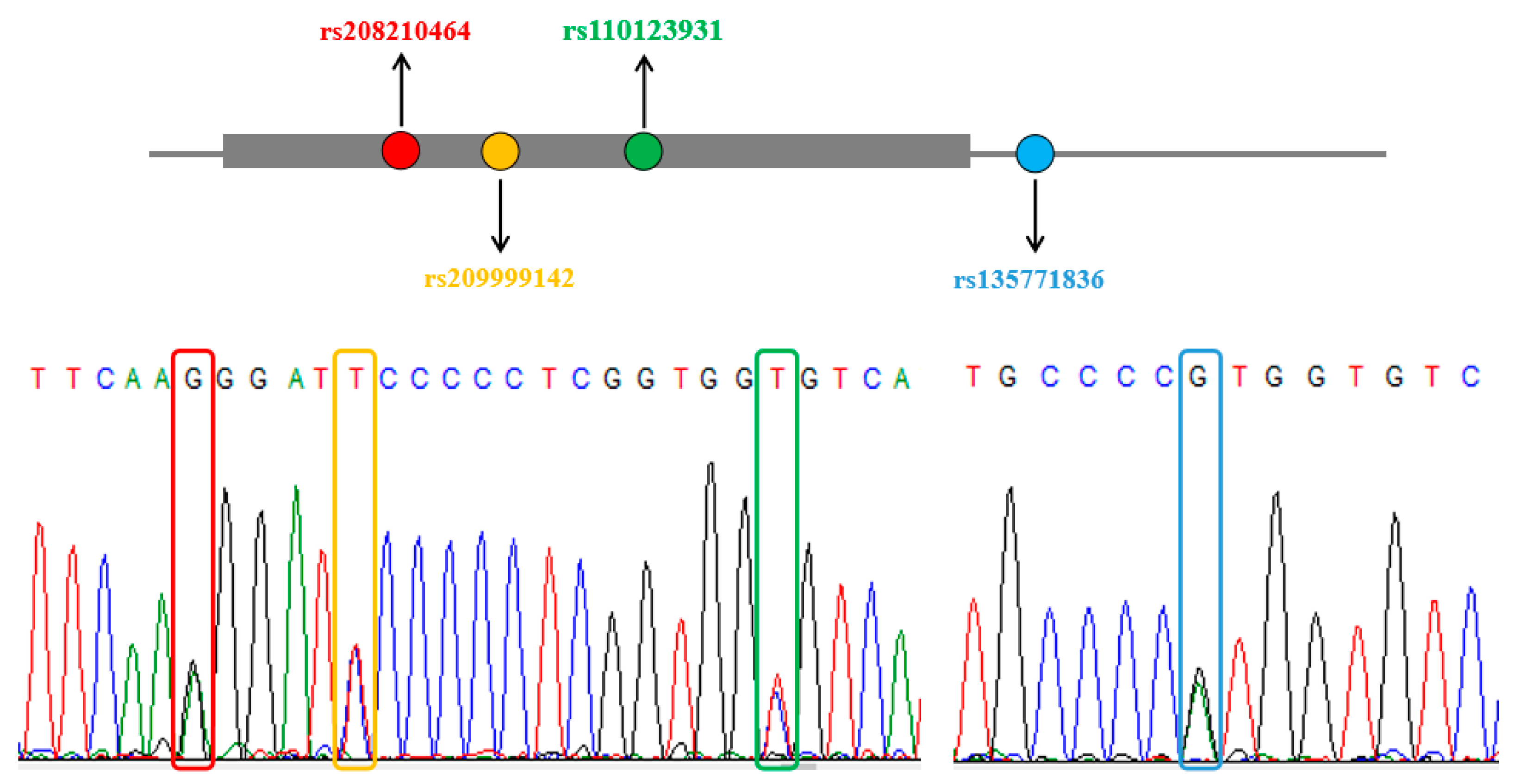
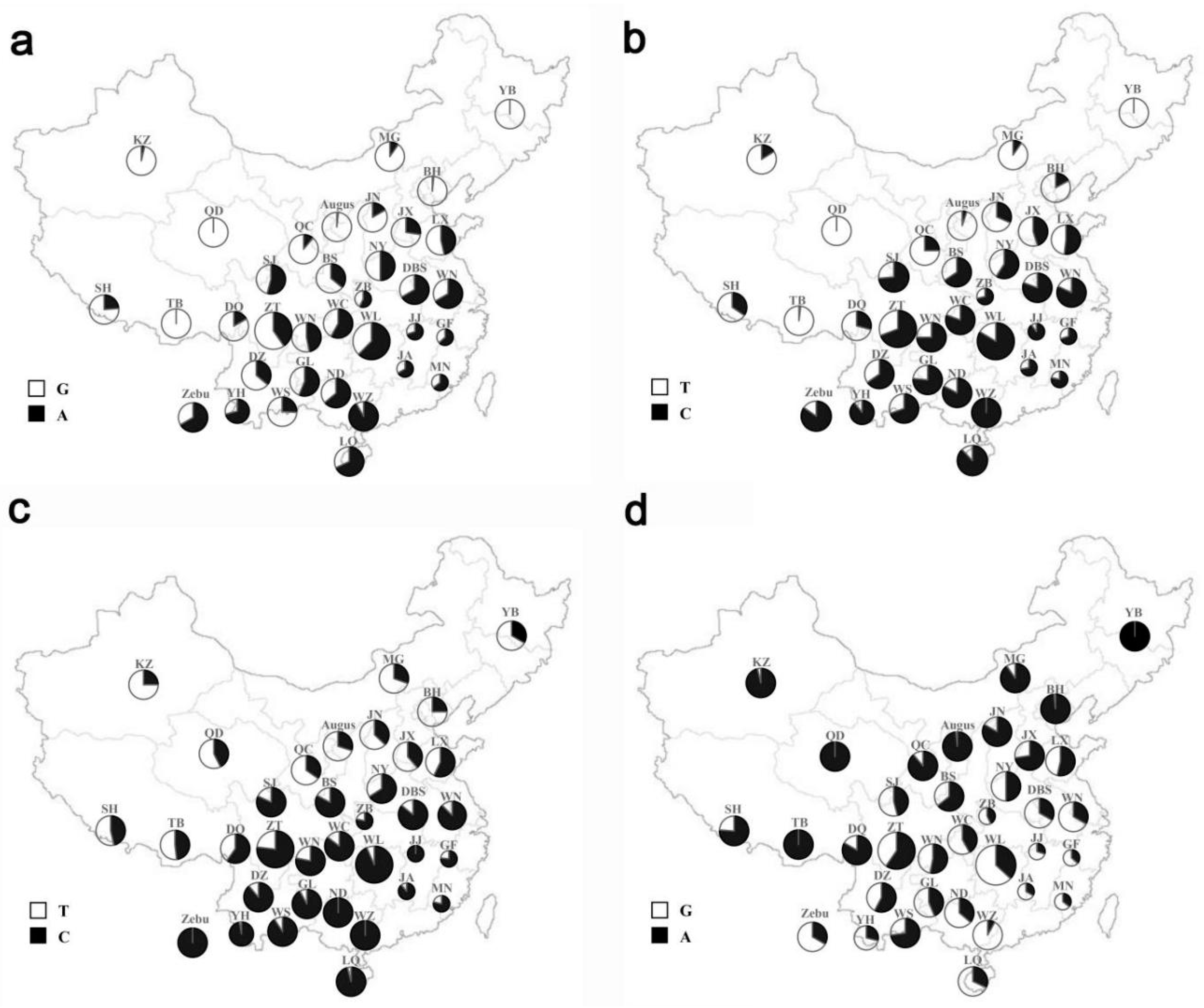
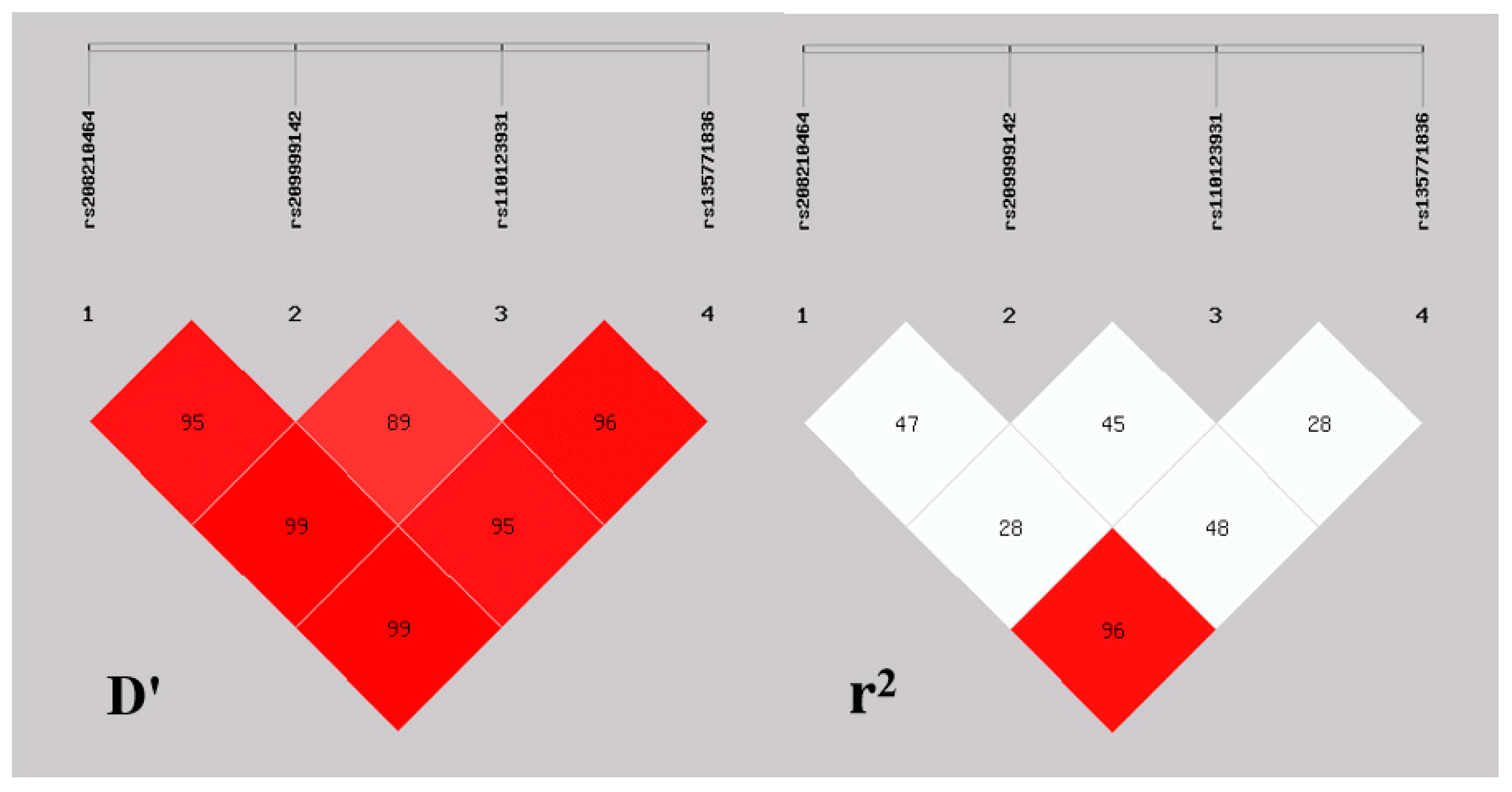
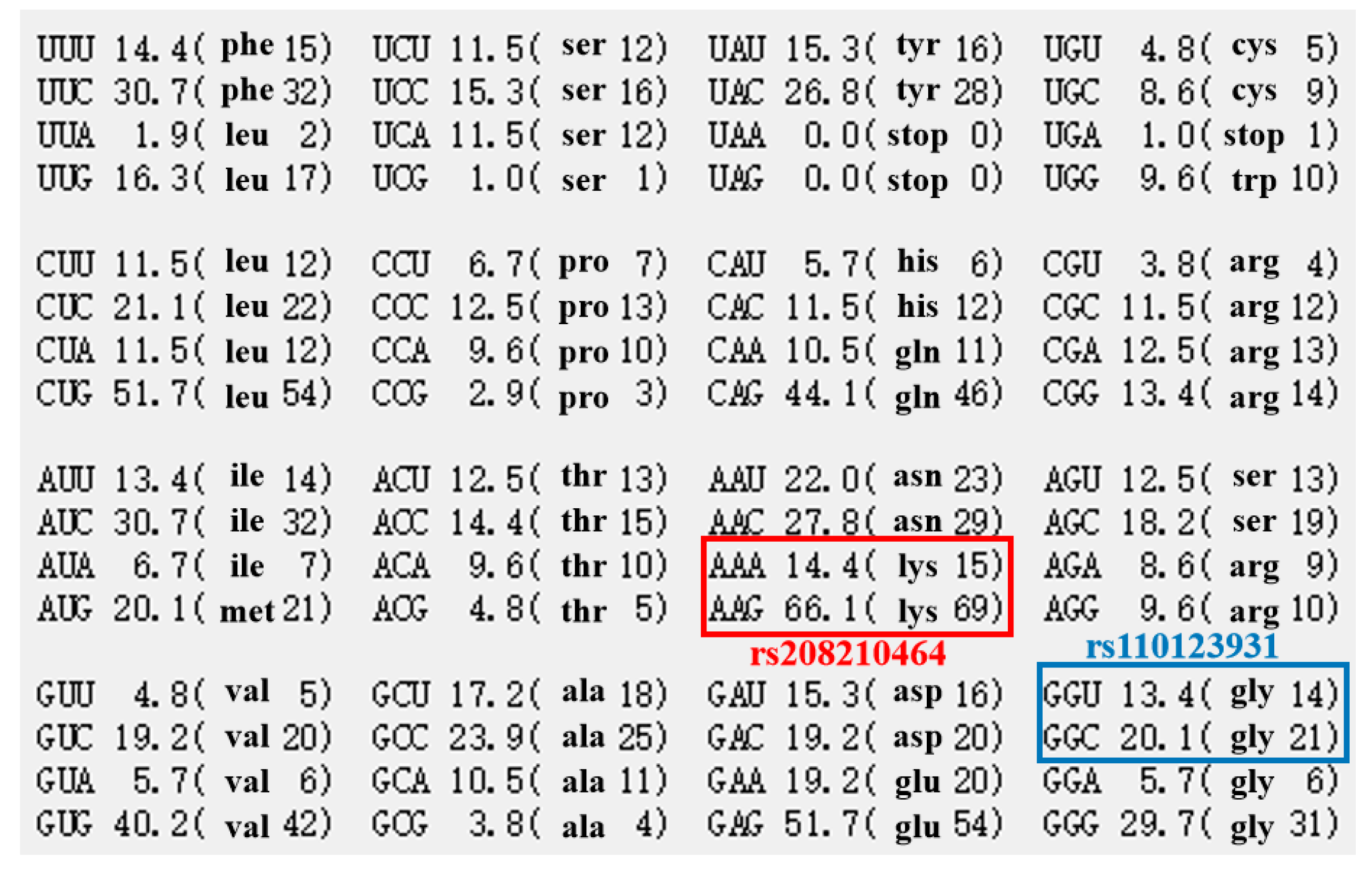
| D′/r2 | rs209999142 | rs110123931 | rs135771836 |
|---|---|---|---|
| rs208210464 | 0.96/0.48 | 0.99/0.29 | 0.99/0.96 |
| rs209999142 | - | 0.90/0.46 | 0.95/0.49 |
| rs110123931 | - | - | 0.97/0.28 |
| Haplotype | rs208210464 | rs209999142 | rs110123931 | rs135771836 | Frequency |
|---|---|---|---|---|---|
| Hap1 | A | C | C | G | 0.39 |
| Hap2 | G | T | T | A | 0.29 |
| Hap3 | G | C | C | A | 0.15 |
| Hap4 | G | T | C | A | 0.14 |
| Loci | Genotype (n) | T (°C) (LSM ± SE) | H (%) (LSM ± SE) | THI (LSM ± SE) | SR (%) (LSM ± SE) |
|---|---|---|---|---|---|
| rs208210464 (g.56383560 G > A) | AA (222) | 17.05 ± 0.30 A | 76.33 ± 0.49 A | 61.06 ± 0.41 A | 42.79 ± 0.60 c |
| AG (359) | 13.93 ± 0.26 B | 72.43 ± 0.46 B | 57.20 ± 0.38 B | 44.90 ± 0.54 B | |
| GG (425) | 10.39 ± 0.25 C | 65.08 ± 0.58 C | 52.42 ± 0.35 C | 53.14 ± 0.53 a | |
| rs209999142 (g.56383565 T > C) | CC (413) | 16.19 ± 0.23 A | 75.69 ± 0.38 A | 60.06 ± 0.33 A | 42.89 ± 0.46 C |
| CT (306) | 13.20 ± 0.27 B | 70.80 ± 0.48 B | 56.26 ± 0.40 B | 47.00 ± 0.59 B | |
| TT (285) | 8.59 ± 0.26 C | 61.55 ± 0.71 C | 49.95 ± 0.36 C | 56.22 ± 0.61 A | |
| rs110123931 (g.56383578 T > C) | CC (553) | 15.62 ± 0.21 A | 74.87 ± 0.36 A | 59.36 ± 0.30 A | 43.81 ± 0.42 C |
| CT (288) | 10.67 ± 0.28 B | 65.53 ± 0.68 b | 52.85 ± 0.40 B | 50.89 ± 0.63 B | |
| TT (165) | 9.02 ± 0.32 C | 62.60 ± 0.73 c | 50.46 ± 0.24 C | 56.46 ± 0.77 A | |
| rs135771836 (g.56383635 A > G) | AA (417) | 10.22 ± 0.25 C | 64.81 ± 0.52 C | 52.17 ± 0.34 C | 53.18 ± 0.54 A |
| AG (362) | 13.95 ± 0.26 B | 72.47 ± 0.45 B | 57.24 ± 0.37 B | 44.90 ± 0.54 b | |
| GG (227) | 17.13 ± 0.30 A | 76.41 ± 0.48 A | 61.25 ± 0.41 A | 43.04 ± 0.60 b |
| Combined Haplotypes | T (°C) (LSM ± SE) | H (%) (LSM ± SE) | THI (LSM ± SE) | SR (%) (LSM ± SE) |
|---|---|---|---|---|
| Hap 1/1 (0.22) | 16.98 ± 0.31 a | 76.29 ± 0.50 a | 60.9 ± 0.42 a | 42.83 ± 0.61 D |
| Hap 1/3 (0.16) | 15.23 ± 0.40 b | 74.96 ± 0.65 a | 58.8 ± 0.56 a | 42.67 ± 0.80 D |
| Hap 2/2 (0.15) | 8.38 ± 0.31 D | 61.69 ± 0.78 C | 49.5 ± 0.43 C | 57.60 ± 0.79 a |
| Hap 1/2 (0.13) | 12.00 ± 0.42 C | 68.88 ± 0.74 B | 54.5 ± 0.61 B | 48.12 ± 0.89 c |
| Hap 2/4 (0.10) | 7.88 ± 0.41 D | 59.33 ± 1.44 C | 49.1 ± 0.56 C | 55.81 ± 1.10 a |
| Hap 1/4 (0.07) | 14.66 ± 0.58 b | 73.87 ± 1.05 a | 58.3 ± 0.84 a | 42.88 ± 1.32 D |
| Hap 2/4 (0.05) | 12.25 ± 0.65 C | 68.72 ± 1.03 B | 54.9 ± 0.95 B | 48.56 ± 1.37 c |
| Hap 4/4 (0.04) | 10.75 ± 1.03 C | 66.28 ± 2.24 B | 53.0 ± 1.46 B | 53.31 ± 1.80 b |
| Hap 3/3 (0.04) | 15.37 ± 0.93 a | 75.14 ± 1.47 a | 59.1 ± 1.35 a | 42.79 ± 1.72 D |
| Hap 3/4 (0.03) | 14.82 ± 0.79 b | 74.81 ± 1.22 a | 58.3 ± 1.14 a | 46.33 ± 1.73 c |
| Type | Seq Identity | QMEAN | C-Beta | All Atom | Solvation | Torsion |
|---|---|---|---|---|---|---|
| Wild type | 66.53% | 2.98 | 1.71 | 0.56 | 0.66 | 2.89 |
| Mutant type | 66.53% | 2.99 | 1.71 | 0.55 | 0.63 | 2.89 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, P.; Cai, C.; Qu, K.; Chen, N.; Jia, Y.; Hanif, Q.; Liu, J.; Zhang, J.; Chen, H.; Huang, B.; et al. Four Novel SNPs of MYO1A Gene Associated with Heat-Tolerance in Chinese Cattle. Animals 2019, 9, 964. https://doi.org/10.3390/ani9110964
Jia P, Cai C, Qu K, Chen N, Jia Y, Hanif Q, Liu J, Zhang J, Chen H, Huang B, et al. Four Novel SNPs of MYO1A Gene Associated with Heat-Tolerance in Chinese Cattle. Animals. 2019; 9(11):964. https://doi.org/10.3390/ani9110964
Chicago/Turabian StyleJia, Peng, Cuicui Cai, Kaixing Qu, Ningbo Chen, Yutang Jia, Quratulain Hanif, Jianyong Liu, Jicai Zhang, Hong Chen, Bizhi Huang, and et al. 2019. "Four Novel SNPs of MYO1A Gene Associated with Heat-Tolerance in Chinese Cattle" Animals 9, no. 11: 964. https://doi.org/10.3390/ani9110964
APA StyleJia, P., Cai, C., Qu, K., Chen, N., Jia, Y., Hanif, Q., Liu, J., Zhang, J., Chen, H., Huang, B., & Lei, C. (2019). Four Novel SNPs of MYO1A Gene Associated with Heat-Tolerance in Chinese Cattle. Animals, 9(11), 964. https://doi.org/10.3390/ani9110964





