Simple Summary
There is increased interest in determining the effect of the biological clock system on reproduction, but how this biological system affects mammalian fertility and the regulation by clock genes on key genes of reproduction is poorly understood. This study examined the function of Leptin on reproduction through interaction with the Leptin receptor (Lepr) and the regulation of the key clock gene brain and muscle ARNT-like 1 (Bmal1) on Lepr. The results suggested that estrogen (E2) synthesis is regulated by Bmal1 through the Leptin–Lepr pathway as part of the regulatory mechanism of the circadian system on the fertility of female mammals.
Abstract
Chronobiology affects female fertility in mammals. Lepr is required for leptin regulation of female reproduction. The presence of E-box elements in the Lepr promoter that are recognized and bound by clock genes to initiate gene transcription suggested that circadian systems might regulate fertility through Lepr. However, it is unclear whether Bmal1, a key oscillator controlling other clock genes, is involved in leptin regulation in hormone synthesis through Lepr. In this study, serum estradiol (E2) concentration and the expressions of Bmal1, Lepr, Cyp19a1, and Cyp11a1 genes were found to display well-synchronized circadian rhythms. Knockdown of Bmal1 significantly reduced expression levels of Lepr, Fshr, and Cyp19a1 genes; protein production of Bmal1, Lepr, and Cyp19a1; and the E2 concentration in granulosa cells. Knockdown of Lepr reduced the expression levels of Cyp19a1 and Cyp11a1 genes and Cyp19a1 protein, and also reduced E2 concentration. Addition of leptin affected the expression of Cyp19a1, Cyp11a1, and Fshr genes. Bmal1 deficiency counteracted leptin-stimulated upregulation of the genes encoding E2 synthesis in granulosa cells. These results demonstrated that Bmal1 participates in the process by which leptin acts on Lepr to regulate E2 synthesis.
1. Introduction
Female fertility depends on a functional circadian system, which has an intimate association with the reproduction processes [1]. At the molecular level, the circadian clock is composed of a network of transcription-translation feedback loops. Heterodimers of Clock (circadian locomotor output cycles kaput) and Bmal1 bind to E-box elements on the genome to activate the transcription of a number of clock control genes [2]. These clock control genes are expressed and coordinate with rhythmic physiological functions in cells and tissues [3]. Bmal1 is the key gene of this oscillator, and its deficiency results in severe clock disruption, which has been associated with decreased fertility [4,5,6,7]. The extent of illumination as well as exposure to light at night can cause circadian clock dysfunction, which increases rates of menstrual disruption, odds ratio, and subfertility in women [8]. However, the details of the regulation of steroidogenesis by the circadian clock have not been determined.
Leptin, encoded by the obese gene [9], is a metabolic hormone [10]. Leptin is predominantly produced by adipose tissues, with some amount produced by other tissues, such as the stomach, muscle, ovary, and placenta [11,12]. Via its specific receptors, leptin acts to regulate food intake and energy metabolism [13]. In addition, leptin is essential for fertility due to its ability to affect the functions of the hypothalamus, pituitary, and reproductive organs [10]. Leptin mutants (ob/ob) mice are infertile, but the administration of exogenous leptin raises FSH and LH concentrations, increases gonadal weight, and restores fertility [14,15]. Lepr is encoded by the diabetes gene [16] and belongs to the class-1 cytokine receptor superfamily [17]. Transcription and translation of Lepr occur in different tissues, including hypothalamus, pituitary, ovary, and adrenal glands [18]. The activation of specific membrane-associated receptors is required for leptin function [13,19]. Crucially, human chorionic gonadotropin (HCG) stimulates Lepr expression in granulosa cells [18] and Lepr null mice are infertile [20]. In some mammal granulosa cells, leptin can promote cell proliferation, impair cell apoptosis, and stimulate steroidogenesis through the MAPK and PKA pathways in a dose-dependent manner [21]. In oocytes, leptin enhances nuclear and cytoplasmic maturation [9]. Mice treated with leptin antagonists exhibit significantly reduced ovulation rates [18].
Leptin works through Lepr in the ovary [22]. Measurements of mRNA and protein of Leptin and ovarian cells suggest that leptin directly regulates ovarian cell functions. Although the Lepr promoter includes potential binding sites for Bmal1/Clock heterodimers, whether and how Bmal1 participates in the leptin-regulated synthesis of E2 by affecting the expression of Lepr has been determined. Therefore, in this study, we focused on the role of Bmal1 in the process of leptin-regulated E2 synthesis.
2. Materials and Methods
2.1. Animals, Welfare Assurance, and Management
The use of animals and the experimental protocol were approved by the Northwest Agriculture and Forestry University Animal Research Ethics Committee (Yangling, Shaanxi, China).
Three-week old, Kunming background female mice were purchased from the Laboratory Animal Center of the Fourth Military Medical University (Xi’an, China). Rodents were housed at 2–5 per cage, fed ad libitum a standard laboratory chow diet, and had free access to fresh water. The animal room was maintained at a constant temperature of 25 ± 1 °C and humidity at 55% ± 5% with 12 h light/12 h dark cycles (light, ZT0–12; dark, ZT12–24; ZT = zeitgeber time).
2.2. Granulosa Cell Culture and Treatment
Granulosa cells were harvested as described previously [23] with some minor changes. Briefly, mice (21–23 d of age) were injected subcutaneously with 1 mg of diethylstilbestrol (DES; Sigma-Aldrich, Inc., St. Louis, MO, USA) for 3 d. Granulosa cells were harvested from ovary follicles on day 4 at ZT1. Ovaries were incubated with DMEM/F12 containing 6 mM EGTA for 20 min and then incubated in DMEM/F12 medium containing 0.5 M sucrose at 37 °C for 15 min before granulosa cells were collected. After washing with PBS, follicles were punctured with a 27-gauge needle in DMEM/F12 medium and then passed through a 70 and then 200-mesh sieve. Granulosa cells were washed by centrifugation at 1000 rpm for 10 min and seeded in culture dishes. The cells were cultured in DMEM/F12 medium supplemented with 10% FBS (Gibco Laboratories, Gaithersburg, MD, USA) at 37 °C in 5% CO2 for 24 h and then the medium was substituted with serum-free medium containing 0.3% Albumin Bovine V (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) for an additional 12 h. For leptin treatment, cells were cultured in fresh media containing 0, 0.1, 0.5, 1, 5, and 10 ng/mL of recombinant mouse leptin (R&D Systems, Inc., Minneapolis, MN, USA) for 24 h.
2.3. Transfection with siRNA
The siRNA technique was used to assess the effects of Bmal1 and Lepr on the oscillation-related genes in granulosa cells to determine Bmal1 and Lepr function for E2 synthesis. The siRNA sequences of Bmal1, Lepr, and a scrambled negative control (NC) were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences of siRNA used in this study are as follows:
Bmal1: 5′-CCUCCACAAUCAGUGACUUTT-3′ (sense) and 5′-AAGUCACUGAUUGUGGAGGTT-3′ (antisense),
Lepr: 5′-GCUGGAGUCCCAAACAAUATT-3′ (sense) and 5′-UAUUGUUUGGGACUCCAGCTT-3′ (antisense), and
NC: 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense).
The transfection protocol was performed according to the manufacturer’s guidelines. When the cell density reached 50%–60%, the cell culture medium was replaced with new medium without serum. The siRNA and NC were diluted into Opti-MEM (Gibco Laboratories) and compounded with X-tremeGENE siRNA transfection reagent (Roche Applied Science, Indianapolis, IN, USA) and delivered into the cells. After exposure for 24 h, the medium was replaced by DMEM/F12 medium containing 0.3% bovine albumin fraction V (Beijing Solarbio Science and Technology Co., Ltd.) and incubated for 24 h.
2.4. RNA Extraction and Quantitative Real-Time PCR Measurement
Total RNA samples were isolated from ovaries and cultured cells using RNAiso Plus (TaKaRa Bio Inc., Shiga, Japan) according to the manufacturer’s protocol [24]. After extraction, 500 ng of total RNA was used to produce cDNA with a Prime Scrip RT Kit (TaKaRa Bio Inc.). Specific primers were designed using Primer 5.0 software and GAPDH used as a housekeeping gene for comparison. Real-time PCR was carried out with IQ5 system using SYBR Green in a mixture system including 5 μL of SYBR (Vazyme Biotech Co., Ltd., Nanjing, China), 0.5 μL of forward primers, 0.5 μL of reverse primers, 1 μL of template cDNA, and 3 μL of ddH2O. Data were calculated with the 2-∆∆t method. Primer sequences are listed in Table 1 and Table 2.

Table 1.
Primers of mice genes.

Table 2.
qRT-PCR primers of rats genes.
2.5. Western Blotting
Cellular proteins were extracted from ovaries and cultured granulosa cells with RIPA (Applygen Technologies Inc., Beijing, China) supplemented with 1 mM PMSF serine protease inhibitor. The protein content was measured using a BCA Protein Assay Kit according to the manufacturer’s guidelines (CWBIO, Beijing, China). Western blotting was conducted as described in a previous report [24]. A total of 20 micrograms of protein was electrophoresed on a 12% SDS-polyacrylamide gel and transferred to PVDF membranes. The levels of the Bmal1 and other proteins involved in Lepr function and E2 synthesis were measured using specific antibodies. Antibodies against Bmal1 (1/500; sc-365645, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), Lepr (1/500; ab5593, Abcam, Inc., Cambridge, MA, USA), Fshr (1/1000; ab75200, Abcam, Inc.), Cyp19a1 (1/1000; ab124776, Abcam, Inc.), and Cyp11a1 (1/500; #14217, CST, Inc., Danvers, MA, USA) were used to detect the levels of these proteins in granulosa cells.
2.6. Estradiol Assay
The E2 concentrations in the mouse serum samples and cell culture medium were detected using an ELISA assay (Nanjing Jiancheng Bio-Engineering Institute Co., Ltd., Jiancheng, China) in accordance with the manufacturer’s protocol.
2.7. Statistical Analysis
Statistical analysis was performed using Graphpad Prim6 [25]. Student’s t-test and one-way ANOVA were used to analyze paired groups and more than two groups. Data are presented as mean ± SE. Significances were indicated as *, p < 0.05 and **, p < 0.01. Relative gene expression was analyzed by the 2-∆∆ ct method.
3. Results
3.1. Leptin Function in Steroidogenesis-Related Genes
To investigate the action of leptin on the expression of steroidogenesis genes, varied leptin doses (0, 0.1, 0.5, 1, 5, and 10 ng/mL) were added into cell culture media for 24 h and the expression levels of the E2 synthesis-related genes of Cyp19a1, Cyp11a1, and Fshr were determined. The observed changes of gene expression with changes in the leptin concentration series were different for different genes (Figure 1). For the Cyp19a1 gene, the maximum expression appeared at 0.5-ng/mL leptin (p < 0.05), and the minimum expression was at 10-ng/mL leptin (p < 0.05, Figure 1A). The expression of Cyp11a1 was higher (p < 0.05, Figure 1B) at 0.1, 0.5, and 1-ng/mL leptin concentrations compared to the levels at the other leptin concentrations. Fshr expression was similar at 0, 0.1, 0.5, and 1 ng/mL leptin but was decreased at 5 and 10 ng/mL leptin (p < 0.05, Figure 1C).
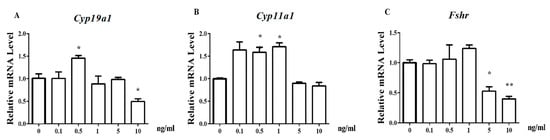
Figure 1.
Leptin regulates the expression of E2 synthesis-related genes of Cyp19a1, Cyp11a1, and Fshr in a dose-dependent manner in granulosa cells after exposure to leptin (0, 0.1, 0.5, 1, 5, and 10 ng/mL) for 24 h. (A–C): relative mRNA expression of Cyp19a1, Cyp11a1, and Fshr, respectively. Statistics are showed as mean ± SEM (n = 3 per group). Significances are expressed as follows: * p < 0.05, ** p < 0.01.
3.2. Lepr is Necessary for E2 Synthesis
To determine if the observed biological activity of leptin on steroidogenesis required interaction with Lepr, the expression levels of Lepr, Cyp19a1, Cyp11a1, and Fshr were measured before or after the silencing of Lepr by siRNA in the presence or absence of leptin. Lepr siRNA treatment suppressed the leptin-stimulated upregulation of Cyp19a1, Cyp11a1, and Fshr (p < 0.05, Figure 2), as evident by comparison to cells without Lepr siRNA treatment in the presence of leptin. The results indicated that Lepr siRNA treatment weakened the leptin contribution to regulation of E2 synthesis-related genes.
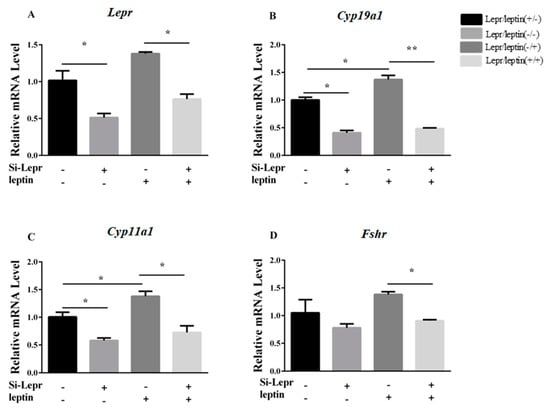
Figure 2.
The interference of Lepr impaired the action of leptin on synthesis genes. After treatment with Lepr siRNA for 24 h, granulosa cells were cultured with or without leptin (0.5 ng/mL) for 24 h. The transcription levels of Lepr, Cyp19a1, and Cyp11a1 were determined by qRT-PCR. (A–D): relative mRNA expression of Lepr, Cyp19a1, Cyp11a1, and Fshr. Statistics are presented as mean values ± SEM (n = 3 per group). Significance values are expressed as follows: * p < 0.05, ** p < 0.01 (si-Lepr: Lepr siRNA treatment group, NC: negative control group).
Granulosa cells were treated with Lepr siRNA to further explore the effects of Lepr on E2 synthesis. With the disturbance of Lepr and Cyp19a1 were decreased at both the RNA and the protein level. Cyp11a1 was inhibited by Lepr siRNA at the RNA level but not at the protein level. Since the RNA level of Fshr did not change significantly, its protein concentrations were not detected (Figure 3A–C). The E2 concentration in the medium was reduced significantly by si-Lepr (p < 0.01, Figure 3D).
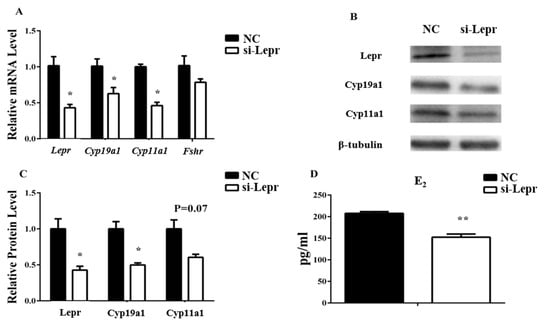
Figure 3.
Lepr siRNA treatment reduces E2 production in granulosa cells. The transcription levels of ovarian genes were ascertained by qRT-PCR. The protein levels were analyzed by western blot. The E2 concentrations in the cell medium were measured by ELISA. (A): relative mRNA expression of Lepr, Cyp19a1, and Cyp11a1. (B,C): relative protein expression of Lepr, Cyp19a1, and Cyp11a1. (D): E2 concentration in cell medium. Results are presented as mean values ± SEM (n = 3 per group). The significance of the results is expressed as follows: * p < 0.05, ** p < 0.01 (si-Lepr: Lepr siRNA treatment group, NC: negative control group).
3.3. E2 Concentration in Serum and Lepr Expression in Ovaries Presented Evident Rhythm
Bioinformatics analysis identified eleven E-box like elements in the 1500 bp upstream of the Lepr promoter (Figure 4). Therefore, we next asked if Lepr might be regulated by the clock system. To determine whether Lepr transcription and E2 production were affected by the circadian system, the E2 concentrations and mRNA concentrations of ovarian genes (Bmal1, Lepr, Fshr, Cyp19a1, and Cyp11a1) were tested.
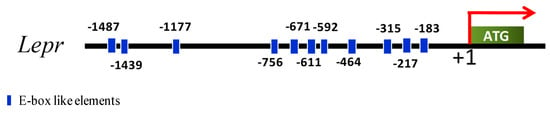
Figure 4.
Schematic representation of the porcine Lepr gene promoter. Eleven E-box like elements are located upstream of the promoter.
Eight-week-old female mice living in 12 h light/12 h dark cycles were euthanized at ZT 0, 4, 8, 12, 16, 20, and 24 (ZT 0 is 8 am; n = 6 per group). The rhythmicity change of Bmal1 indicated effective function of the circadian system in ovaries of Kunming background female mice (Figure 5A). The Lepr and E2 mRNA levels showed robust rhythms and both peaked during the dark phase at ZT16 (Figure 5B,F), which suggested that Lepr was sensitive to light rhythm and might act in the ovary at ZT16.
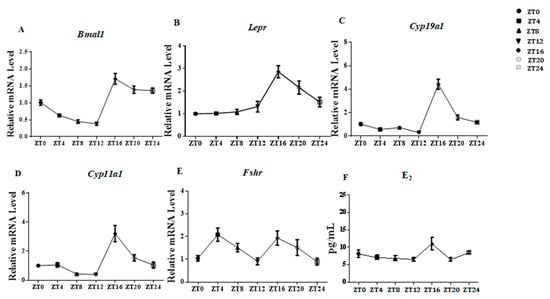
Figure 5.
The rhythm of E2 concentration in serum and related gene expression in ovaries. Samples were from eight-week-old female mice subjected to light/dark cycles. The transcription levels of ovarian genes were ascertained by qRT-PCR at ZT0, ZT4, ZT8, ZT12, ZT16, ZT20, and ZT24. The E2 concentrations in blood sera were measured by ELISA at ZT0, ZT4, ZT8, ZT12, ZT16, ZT20, and ZT24. (A–E): the relative mRNA expression levels of ovarian genes. (F): E2 concentration in blood serum. Statistics are showed as mean values ± SEM (n = 6; ZT: zeitgeber time).
The Bmal1 siRNA was used to interfere with Bmal1 function in granulosa cells to confirm whether the E2 concentration was affected by Bmal1. The results showed decreased Lepr and Cyp19a1 RNA and protein levels in the presence of the Bmal1 siRNA. Fshr was inhibited by Bmal1 siRNA at the RNA level but not at the protein level. Since Cyp11a1 did not change significantly with the Bmal1 siRNA, its protein concentration was not detected. The results showed that transcription of the Clock gene was not affected by Bmal1 interference, which was consistent with the previous report (Figure 6A–C). Consistently, the E2 level was significantly suppressed by the Bmal1 siRNA (Figure 6D).
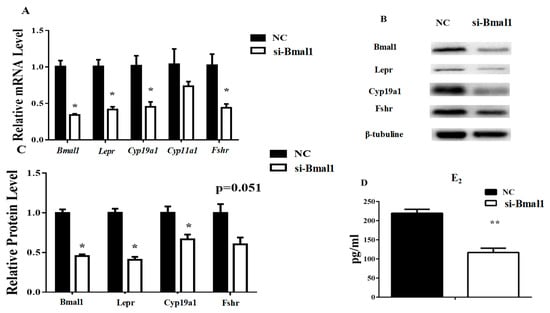
Figure 6.
Silencing of Bmal1 impairs the expression of Lepr and the secretion of E2 in granulosa cells. The transcription levels of ovarian genes were measured by qRT-PCR and the protein levels were analyzed by western blot. The E2 concentration in cell medium was measured by ELISA. (A): relative mRNA expression of clock and ovarian genes. (B,C): relative protein expression of Bmal1, Lepr, Fshr, and Cyp19a1. (D): E2 concentration in cell medium. Statistics are showed as mean ± SEM (n = 3 per group). Significances are expressed as follows: * p < 0.05, ** p < 0.01 (si-Bmal1: Bmal1 siRNA treatment group, NC: negative control group).
3.4. Bmal1 siRNA Repressed Leptin-Induced E2 Synthesis-Related Genes
We next asked if leptin is required for the observed effect of silencing Bmal1. To determine if Bmal1 is involved in the hormone synthesis process that is promoted by leptin, granulosa cells were cultured with Bmal1 siRNA for 24 h and then treated with 0.5-ng/mL leptin for 24 h. The results showed that Bmal1 siRNA prevented the leptin-promoted upregulation of Lepr, Cyp19a1, Cyp11a1, and Fshr (p < 0.05, Figure 7A–E).
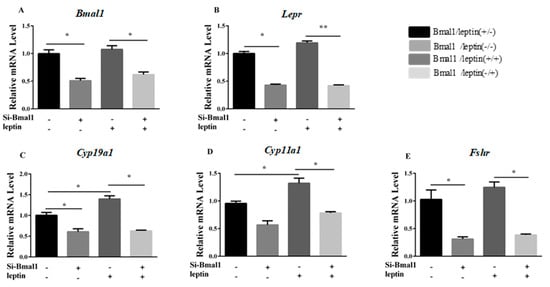
Figure 7.
Silencing of Bmal1 limited the action of leptin on synthesis genes. After treatment with Bmal1 siRNA for 24 h, granulosa cells were cultured with or without leptin (0.5 ng/mL) for 24 h. (A–E): relative mRNA expression of Bmal1, Lepr, Cyp19a1, Cyp11a1, and Fshr were determined by qRT-PCR. Statistics are shown as mean ± SEM (n = 3 per group). Significance values are expressed as follows: * p < 0.05, ** p < 0.01 (si-Bmal1: Bmal1 siRNA treatment group, NC: negative control group).
4. Discussion
E2 concentration and E2 synthesis-related gene expression were altered by adding leptin to cell media, indicating that leptin affected E2 synthesis. The down regulation of E2 synthesis-related gene expression caused by Lepr siRNA indicated the involvement of Lepr in E2 synthesis regulation in granulosa cells. This observation was consistent with the effect of leptin on granulosa cells, as previously reported. For example, in cultured rabbit granulosa cells, addition of 10 ng/mL leptin promoted proliferation, reduced apoptosis, and increased E2 generation via excitation of the MAPK and PKA signaling pathways [16]. The present results indicated that Fshr down regulation with 10 ng/mL leptin was consistent with the previous report that FSH stimulation of steroidogenes is inhibited by high doses of exogenous leptin [26].
The expression patterns of Bmal1, Lepr, Fshr, Cyp19a1, and Cyp11a1 genes in ovarian cells were found to exhibit circadian rhythms within the 12 h light/12 h dark cycle, as did the serum E2 concentration. All patterns were well-synchronized, and peaked at ZT16 (dark period). Thus, the findings suggested the involvement of Lepr in E2 synthesis and that this physiological process might be regulated by the circadian system.
Expression of ovarian genes and E2 concentrations in cultured granulosa cells were reduced with Bmal1 deficiency, which suggested that Bmal1 was essential to maintain the functions of genes for ovarian steroidogenesis. These results were consistent with a previous report that specific knockout of Bmal1 in ovarian steroidogenesis cells resulted in abnormalities of mice estrus cycles [27]. Wang proposed that Bmal1 deficiency increased cell apoptosis and inhibited steroidogenesis in porcine granulosa cells [24]. Lepr transcription in ovarian cells was regulated by Bmal1 and displayed rhythmicity because Bmal1 deficiency impaired Lepr expression and counteracted leptin-promoted expression of the E2 synthesis genes in granulosa cells. Both Lepr and E2 synthesis-related genes were repressed when Bmal1 was inhibited by siRNA even in the presence of leptin, but this repression did not occur when Bmal1 was normally expressed. Thus, the results indicated that Bmal1 is necessary for leptin action in the process of E2 synthesis.
5. Conclusions
In conclusion, the results indicate that Lepr was involved in the stimulation of E2 synthesis in granulosa cells by leptin, and this process might be regulated by the biological clock gene Bmal1 (Figure 8). There were eleven circadian binding sites in the promoter of lepr, but whether Bmal1 directly regulates Lepr and the details of the regulatory mechanism must be further verified.
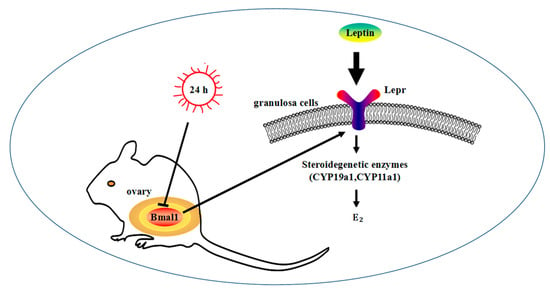
Figure 8.
Lepr was regulated by Bmal1 in the leptin-regulated process of E2 synthesis. Lepr and E2 synthesis-related genes are regulated by Bmal1.
Author Contributions
Conceptualization, G.C. and S.S.; methodology, G.M.; software, A.X.; validation, G.C., G.M.; formal analysis, G.C.; investigation, G.M.; resources, S.S.; data curation, G.C.; writing—original draft preparation, G.C. and G.M.; writing—review and editing, G.C.; visualization, J.S. and Y.Z.; supervision, G.Y.; project administration, G.C. and S.S.; funding acquisition, G.C. and S.S.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31802047), the National Key Technology R&D Program of China (Grant No. 2015BAD03B01-10), and Agricultural Science and Technology Innovation Transformation Project in Shaanxi Province of China (Grant/Award No. NYKJ-2016-08).
Conflicts of Interest
All authors declare no conflict of interest.
References
- Sen, A.; Sellix, M.T. The Circadian Timing System and Environmental Circadian Disruption: From Follicles to Fertility. Endocrinology 2016, 157, 3366–3373. [Google Scholar] [CrossRef] [PubMed]
- Amaral, F.G.; Castrucci, A.M.; Cipolla-Neto, J.; Poletini, M.O.; Mendez, N.; Richter, H.G.; Sellix, M.T. Environmental control of biological rhythms: Effects on development, fertility and metabolism. J. Neuroendocrinol. 2014, 26, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, T.; Hermann-Luibl, C.; Helfrich-Forster, C. Circadian light-input pathways in Drosophila. Commun. Integr. Biol. 2016, 9, e1102805. [Google Scholar] [CrossRef] [PubMed]
- Chu, A.; Zhu, L.; Blum, I.D.; Mai, O.; Leliavski, A.; Fahrenkrug, J.; Oster, H.; Boehm, U.; Storch, K.F. Global but not gonadotrope-specific disruption of Bmal1 abolishes the luteinizing hormone surge without affecting ovulation. Endocrinology 2013, 154, 2924–2935. [Google Scholar] [CrossRef]
- Beymer, M.; Henningsen, J.; Bahougne, T.; Simonneaux, V. The role of kisspeptin and RFRP in the circadian control of female reproduction. Mol. Cell. Endocrinol. 2016, 438, 89–99. [Google Scholar] [CrossRef]
- Cardinet, G.H. Skeletal Muscle Function. In Clinical Biochemistry of Domestic Animals; Elsevier: Amsterdam, The Netherlands, 1997; pp. 407–440. [Google Scholar]
- Reinberg, A.; Bicakova-Rocher, A.; Nouguier, J.; Gorceix, A.; Mechkouri, M.; Touitou, Y.; Ashkenazi, I. Circadian rhythm period in reaction time to light signals: Difference between right- and left-hand side. Brain Res. Cogn. Brain Res. 1997, 6, 135–140. [Google Scholar] [CrossRef]
- Shub, Y.; Ashkenazi, I.E.; Reinberg, A. Differences between left-and right-hand reaction time rhythms: Indications of shifts in strategies of human brain activity. Cogn. Brain Res. 1997, 6, 141–146. [Google Scholar] [CrossRef]
- De Placido, G.; Alviggi, C.; Clarizia, R.; Mollo, A.; Alviggi, E.; Strina, I.; Fiore, E.; Wilding, M.; Pagano, T.; Matarese, G. Intra-follicular leptin concentration as a predictive factor for in vitro oocyte fertilization in assisted reproductive techniques. J. Endocrinol. Investig. 2006, 29, 719–726. [Google Scholar] [CrossRef]
- Elias, C.F.; Purohit, D. Leptin signaling and circuits in puberty and fertility. Cell. Mol. Life Sci. 2013, 70, 841–862. [Google Scholar] [CrossRef]
- Finn, P.D.; Cunningham, M.J.; Pau, K.Y.; Spies, H.G.; Clifton, D.K.; Steiner, R.A. The stimulatory effect of leptin on the neuroendocrine reproductive axis of the monkey. Endocrinology 1998, 139, 4652–4662. [Google Scholar] [CrossRef]
- Friedman, J.M. The function of leptin in nutrition, weight, and physiology. Nutr. Rev. 2002, 60, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Hu, S.; Xiao, Q.; Han, C.; Gan, C.; Gou, H.; Liu, H.; Li, L.; Xu, H.; He, H.; et al. Leptin exerts proliferative and anti-apoptotic effects on goose granulosa cells through the PI3K/Akt/mTOR signaling pathway. J. Steroid Biochem. Mol. Biol. 2015, 149, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Barash, I.A.; Cheung, C.C.; Weigle, D.S.; Ren, H.; Kabigting, E.B.; Kuijper, J.L.; Clifton, D.K.; Steiner, R.A. Leptin is a metabolic signal to the reproductive system. Endocrinology 1996, 137, 3144–3147. [Google Scholar] [CrossRef] [PubMed]
- Mounzih, K.; Lu, R.; Chehab, F.F. Leptin treatment rescues the sterility of genetically obese ob/ob males. Endocrinology 1997, 138, 1190–1193. [Google Scholar] [CrossRef]
- Craig, J.; Zhu, H.; Dyce, P.W.; Petrik, J.; Li, J. Leptin enhances oocyte nuclear and cytoplasmic maturation via the mitogen-activated protein kinase pathway. Endocrinology 2004, 145, 5355–5363. [Google Scholar] [CrossRef]
- Batista, A.M.; Silva, D.M.; Rego, M.J.; Silva, F.L.; Silva, E.C.; Beltrao, E.I.; Gomes Filho, M.A.; Wischral, A.; Guerra, M.M. The expression and localization of leptin and its receptor in goat ovarian follicles. Anim. Reprod. Sci. 2013, 141, 142–147. [Google Scholar] [CrossRef]
- Dupuis, L.; Schuermann, Y.; Cohen, T.; Siddappa, D.; Kalaiselvanraja, A.; Pansera, M.; Bordignon, V.; Duggavathi, R. Role of leptin receptors in granulosa cells during ovulation. Reprod. Camb. Engl. 2014, 147, 221–229. [Google Scholar] [CrossRef]
- Bjorbaek, C.; Uotani, S.; da Silva, B.; Flier, J.S. Divergent signaling capacities of the long and short isoforms of the leptin receptor. J. Biol. Chem. 1997, 272, 32686–32695. [Google Scholar] [CrossRef]
- Bates, S.H.; Stearns, W.H.; Dundon, T.A.; Schubert, M.; Tso, A.W.; Wang, Y.; Banks, A.S.; Lavery, H.J.; Haq, A.K.; Maratos-Flier, E.; et al. STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature 2003, 421, 856–859. [Google Scholar] [CrossRef]
- Harrath, A.H.; Ostrup, O.; Rafay, J.; Konickova Florkovicova, I.; Laurincik, J.; Sirotkin, A.V. Metabolic state defines the response of rabbit ovarian cells to leptin. Reprod. Biol. 2017, 17, 19–24. [Google Scholar] [CrossRef]
- Hu, S.; Gan, C.; Wen, R.; Xiao, Q.; Gou, H.; Liu, H.; Zhang, Y.; Li, L.; Wang, J. Role of leptin in the regulation of sterol/steroid biosynthesis in goose granulosa cells. Theriogenology 2014, 82, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, L.; Chu, G.; Kito, G.; Yamauchi, N.; Shigeyoshi, Y.; Hashimoto, S.; Hattori, M.A. FSH induces the development of circadian clockwork in rat granulosa cells via a gap junction protein Cx43-dependent pathway. Am. J. Physiol. Endocrinol. Metab. 2013, 304, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yin, L.; Bai, L.; Ma, G.; Zhao, C.; Xiang, A.; Pang, W.; Yang, G.; Chu, G. Bmal1 interference impairs hormone synthesis and promotes apoptosis in porcine granulosa cells. Theriogenology 2017, 99, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Mereness, A.L.; Murphy, Z.C.; Forrestel, A.C.; Butler, S.; Ko, C.; Richards, J.S.; Sellix, M.T. Conditional Deletion of Bmal1 in Ovarian Theca Cells Disrupts Ovulation in Female Mice. Endocrinology 2016, 157, 913–927. [Google Scholar] [CrossRef]
- Zachow, R.J.; Magoffin, D.A. Direct intraovarian effects of leptin: Impairment of the synergistic action of insulin-like growth factor-I on follicle-stimulating hormone-dependent estradiol-17 beta production by rat ovarian granulosa cells. Endocrinology 1997, 138, 847–850. [Google Scholar] [CrossRef]
- Liu, Y.; Johnson, B.P.; Shen, A.L.; Wallisser, J.A.; Krentz, K.J.; Moran, S.M.; Sullivan, R.; Glover, E.; Parlow, A.F.; Drinkwater, N.R.; et al. Loss of BMAL1 in ovarian steroidogenic cells results in implantation failure in female mice. Proc. Natl. Acad. Sci. USA 2014, 111, 14295–14300. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).