A Comprehensive Atlas of Testicular lncRNAs Reveals Dynamic Changes and Regulatory Networks During Sexual Maturation in Tibetan Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Tissue Collection
2.2. RNA Extraction, Quality Control, and Library Construction
2.3. Transcriptome Assembly and LncRNA Identification
2.4. Differential Expression and Functional Enrichment Analysis
2.5. Co-Expression Network Construction
2.6. RT-qPCR Validation
2.7. Statistical Analysis
3. Results
3.1. Age-Dependent Morphological Changes, Hormone Levels, and LncRNA Characterization in Tibetan Sheep Testes
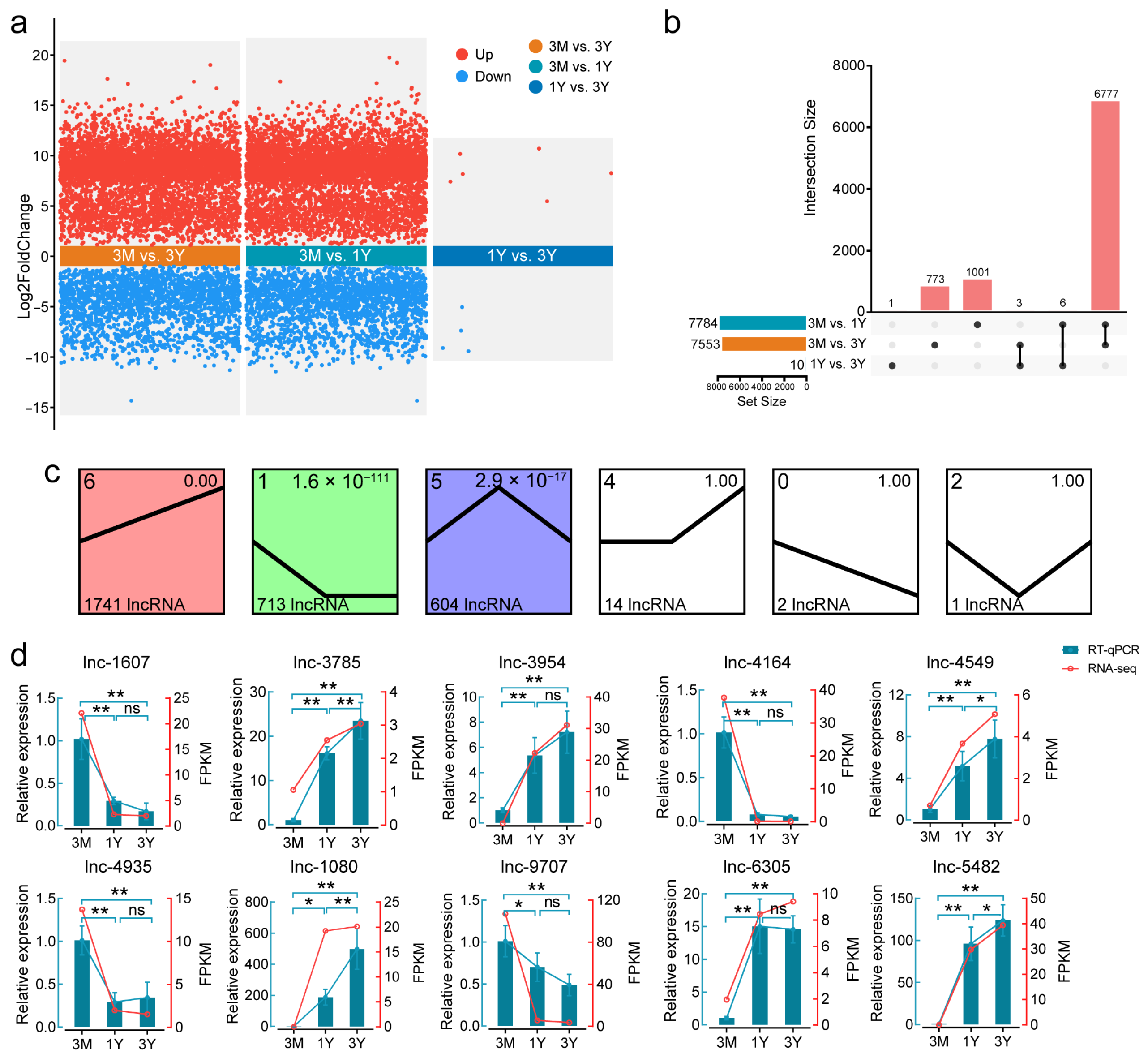
3.2. Differential Expression Analysis of LncRNAs
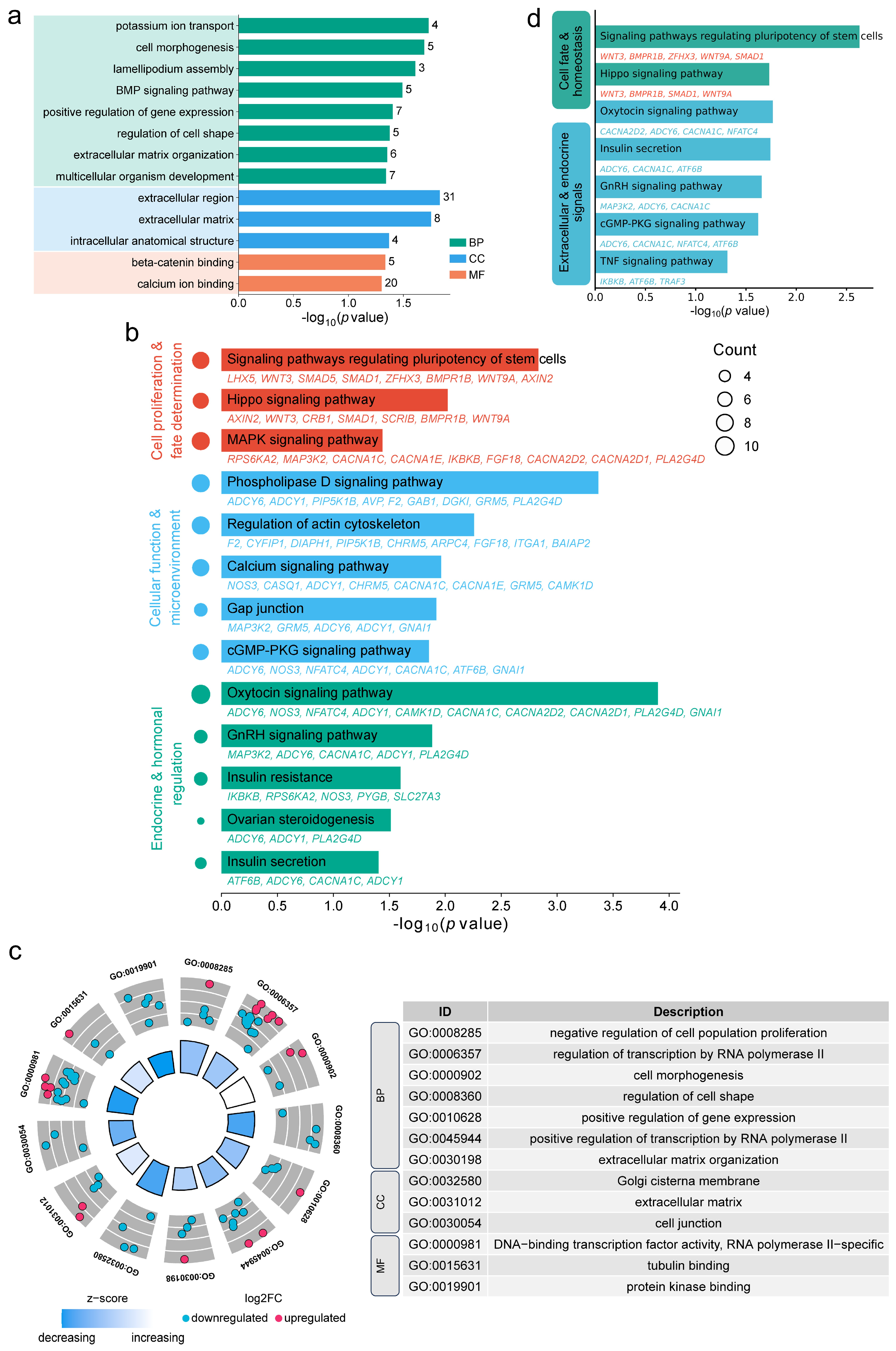
3.3. Prediction of Cis-Acting LncRNA Target Genes and Functional Enrichment
3.4. Functional Enrichment Analysis of Antisense LncRNA Target Genes
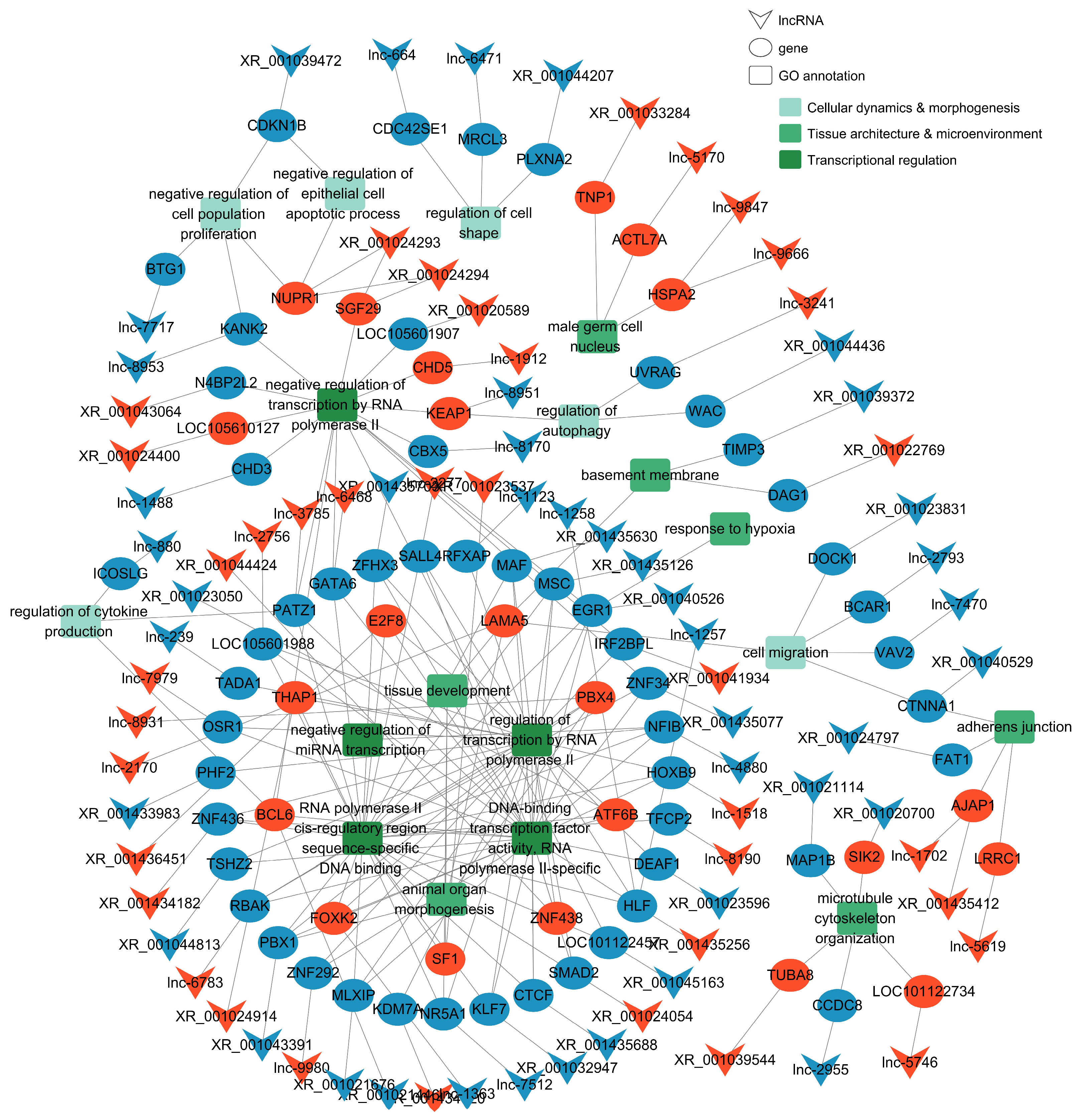
3.5. Target Gene Network and Functional Annotation of LncRNAs Upregulated Post-Puberty
4. Discussion
4.1. Histological and Endocrine Changes Underline Key Developmental Transitions
4.2. Dynamic LncRNA Expression Profiles Reflect Key Transitions in Testicular Development
4.3. Cis-Acting LncRNAs Modulate Core Pathways in Testicular Development and Spermatogenesis
4.4. Antisense LncRNAs Influence Testicular Maturation via Microenvironment and Transcriptional Regulation
4.5. Sexual Maturation Reshapes the lncRNA Regulatory Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1 Y | One year of age |
| 3 M | Three months of age |
| 3 Y | Three years of age |
| ACR | Acrosin |
| GATA6 | GATA-binding protein 6 |
| GO | Gene ontology |
| H&E | Hematoxylin and eosin |
| KEGG | Kyoto encyclopedia of gene and genomes |
| lncRNA | Long non-coding RNA |
| PCC | Pearson correlation coefficient |
| PFKP | Platelet-type phosphofructokinase |
| RNA-seq | RNA sequencing |
| RT-qPCR | Reverse transcription quantitative PCR |
| TNP1 | Transition nuclear protein 1 |
| TUBA8 | Tubulin alpha 8 |
References
- Yuan, L.; Song, X.; Yang, J.; Liang, X.; Chen, G. Comparison of distribution characteristics of TGF-β1 and its receptors in the testis of the Tibetan sheep before and after sexual mature. Acta Vet. Zootech. Sin. 2019, 50, 750–757. [Google Scholar] [CrossRef]
- Wang, W.; He, Y.; Han, Z.; Shi, J.; Huo, J.; Zhu, X. Expression of FSHR gene and LHR gene in the ovary of Ganjia Tibetan sheep in different stages of estrous cycle. Chin. Vet. Sci. 2016, 46, 97–103. [Google Scholar] [CrossRef]
- Joshi, M.; Rajender, S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod. Biol. Endocrinol. 2020, 18, 103. [Google Scholar] [CrossRef]
- Morgan, M.; Kabayama, Y.; Much, C.; Ivanova, I.; Di Giacomo, M.; Auchynnikava, T.; Monahan, J.M.; Vitsios, D.M.; Vasiliauskaitė, L.; Comazzetto, S.; et al. A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis. Cell Res. 2019, 29, 221–232. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, N.; Xu, J.; Ren, J.; Yang, X.; Li, X.; Yao, J.; Wu, S. Noncoding RNAs in host–microbiota interaction. Anim. Res. One Health 2025, 3, 358–367. [Google Scholar] [CrossRef]
- Si, J.; Su, X.; Jin, Z.; Duan, S. Uncovering essential lncRNAs through transcriptome-scale CRISPR-Cas13 screening. Adv. Biotechnol. 2025, 3, 27. [Google Scholar] [CrossRef]
- Lewandowski, J.P.; Dumbović, G.; Watson, A.R.; Hwang, T.; Jacobs-Palmer, E.; Chang, N.; Much, C.; Turner, K.M.; Kirby, C.; Rubinstein, N.D.; et al. The Tug1 lncRNA locus is essential for male fertility. Genome Biol. 2020, 21, 237. [Google Scholar] [CrossRef]
- Taylor, D.H.; Chu, E.T.-J.; Spektor, R.; Soloway, P.D. Long non-coding RNA regulation of reproduction and development. Mol. Reprod. Dev. 2015, 82, 932–956. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, K.; Matsubara, S.; Shiraishi, A.; Takei, N.; Satoh, Y.; Terao, M.; Takada, S.; Kotani, T.; Satake, H.; Kimura, A.P. A testis-specific long noncoding RNA, Start, is a regulator of steroidogenesis in mouse Leydig cells. Front. Endocrinol. 2021, 12, 665874. [Google Scholar] [CrossRef]
- Li, K.; Zhong, S.; Luo, Y.; Zou, D.; Li, M.; Li, Y.; Lu, Y.; Miao, S.; Wang, L.; Song, W. A long noncoding RNA binding to QKI-5 regulates germ cell apoptosis via p38 MAPK signaling pathway. Cell Death Dis. 2019, 10, 699. [Google Scholar] [CrossRef]
- Li, L.; Wang, M.; Wang, M.; Wu, X.; Geng, L.; Xue, Y.; Wei, X.; Jia, Y.; Wu, X. A long non-coding RNA interacts with Gfra1 and maintains survival of mouse spermatogonial stem cells. Cell Death Dis. 2016, 7, e2140. [Google Scholar] [CrossRef]
- Wichman, L.; Somasundaram, S.; Breindel, C.; Valerio, D.M.; McCarrey, J.R.; Hodges, C.A.; Khalil, A.M. Dynamic expression of long noncoding RNAs reveals their potential roles in spermatogenesis and fertility. Biol. Reprod. 2017, 97, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Qiao, L.; Dai, Z.; He, Q.; Lan, X.; Huang, S.; He, L. LncNONO-AS regulates AR expression by mediating NONO. Theriogenology 2019, 145, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, P.; Song, D.; Xiong, S.; Zhang, H.; Fu, J.; Gao, F.; Chen, H.; Zeng, X. Expression profiles and characteristics of human lncRNA in normal and asthenozoospermia sperm. Biol. Reprod. 2018, 100, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Ji, C.; Li, C.; Brand-Saberi, B.; Zhang, S. Multiple transcriptome analyses reveal mouse testis developmental dynamics. BMC Genom. 2024, 25, 395. [Google Scholar] [CrossRef]
- Weng, B.; Ran, M.; Chen, B.; He, C.; Dong, L.; Peng, F. Genome-wide analysis of long non-coding RNAs and their role in postnatal porcine testis development. Genomics 2017, 109, 446–456. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, H.; Hu, Z.; Sahlu, B.W.; Heng, N.; Gong, J.; Wang, H.; Zhu, H. Identification of spermatogenesis-related lncRNA in Holstein bull testis after sexual maturity based on transcriptome analysis. Anim. Reprod. Sci. 2022, 247, 107146. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Tafer, H.; Hofacker, I.L. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics 2008, 24, 2657–2663. [Google Scholar] [CrossRef]
- Volkova, O.A.; Kondrakhin, Y.V.; Kashapov, T.A.; Sharipov, R.N. Comparative analysis of protein-coding and long non-coding transcripts based on RNA sequence features. J. Bioinform. Comput. Biol. 2018, 16, 1840013. [Google Scholar] [CrossRef]
- Riquier, S.; Mathieu, M.; Bessiere, C.; Boureux, A.; Ruffle, F.; Lemaitre, J.M.; Djouad, F.; Gilbert, N.; Commes, T. Long non-coding RNA exploration for mesenchymal stem cell characterisation. BMC Genom. 2021, 22, 412. [Google Scholar] [CrossRef]
- Staub, C.; Johnson, L. Review: Spermatogenesis in the bull. Animal 2018, 12, s27–s35. [Google Scholar] [CrossRef] [PubMed]
- Xi, B.; Zhao, S.; Zhang, R.; Lu, Z.; Li, J.; An, X.; Yue, Y. Transcriptomic study of different stages of development in the testis of sheep. Animals 2024, 14, 2767. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, X.; Zhang, H.; Chen, Z.; Zhao, X.; Ma, Y. Histomorphological comparisons and expression patterns of BOLL gene in sheep testes at different development stages. Animals 2019, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Rey, R.A. The Role of androgen signaling in male sexual development at puberty. Endocrinology 2021, 162, bqaa215. [Google Scholar] [CrossRef]
- O’Shaughnessy, P.J. Hormonal control of germ cell development and spermatogenesis. Semin. Cell Dev. Biol. 2014, 29, 55–65. [Google Scholar] [CrossRef]
- Chojnacka, K.; Zarzycka, M.; Mruk, D.D. Biology of the Sertoli cell in the fetal, pubertal, and adult mammalian testis. Results Probl. Cell Differ. 2016, 58, 225–251. [Google Scholar] [CrossRef]
- Xu, H.; Sun, W.; Pei, S.; Li, W.; Li, F.; Yue, X. Identification of key genes related to postnatal testicular development based on transcriptomic data of testis in Hu sheep. Front. Genet. 2021, 12, 773695. [Google Scholar] [CrossRef]
- Yang, H.; Wang, F.; Li, F.; Ren, C.; Pang, J.; Wan, Y.; Wang, Z.; Feng, X.; Zhang, Y. Comprehensive analysis of long noncoding RNA and mRNA expression patterns in sheep testicular maturation. Biol. Reprod. 2018, 99, 650–661. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef]
- Li, T.; Wang, H.; Luo, R.; An, X.; Li, Q.; Su, M.; Shi, H.; Chen, H.; Zhang, Y.; Ma, Y. Proteome informatics in Tibetan sheep (Ovis aries) testes suggest the crucial proteins related to development and functionality. Front. Vet. Sci. 2022, 9, 923789. [Google Scholar] [CrossRef]
- Yang, H.; Ma, J.; Wan, Z.; Wang, Q.; Wang, Z.; Zhao, J.; Wang, F.; Zhang, Y. Characterization of sheep spermatogenesis through single-cell RNA sequencing. FASEB J. 2021, 35, e21187. [Google Scholar] [CrossRef]
- Lochab, A.K.; Extavour, C.G. Bone Morphogenetic Protein (BMP) signaling in animal reproductive system development and function. Dev. Biol. 2017, 427, 258–269. [Google Scholar] [CrossRef]
- Satoh, Y.; Takei, N.; Kawamura, S.; Takahashi, N.; Kotani, T.; Kimura, A.P. A novel testis-specific long noncoding RNA, Tesra, activates the Prss42/Tessp-2 gene during mouse spermatogenesis. Biol. Reprod. 2019, 100, 833–848. [Google Scholar] [CrossRef]
- Karpova, T.; Ravichandiran, K.; Insisienmay, L.; Rice, D.; Agbor, V.; Heckert, L.L. Steroidogenic factor 1 differentially regulates fetal and adult leydig cell development in male mice. Biol. Reprod. 2015, 93, 83. [Google Scholar] [CrossRef]
- Kato, T.; Esaki, M.; Matsuzawa, A.; Ikeda, Y. NR5A1 is required for functional maturation of Sertoli cells during postnatal development. Reproduction 2012, 143, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Zhang, Y.G.; Du, J.D. HIF-1α restricts proliferation and apoptosis of Tca8113 cells through up regulation of Hippo signaling pathway under hypoxic conditions. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6832–6837. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.L.; Batchelor, E. Coordination of MAPK and p53 dynamics in the cellular responses to DNA damage and oxidative stress. Mol. Syst. Biol. 2022, 18, e11401. [Google Scholar] [CrossRef]
- Wang, B.; He, J.; Cui, Y.; Yu, S.; Zhang, H.; Wei, P.; Zhang, Q. The HIF-1α/EGF/EGFR signaling pathway facilitates the proliferation of yak alveolar type ii epithelial cells in hypoxic conditions. Int. J. Mol. Sci. 2024, 25, 1442. [Google Scholar] [CrossRef]
- He, Z.; Li, S.; Zhao, F.; Sun, H.; Hu, J.; Wang, J.; Liu, X.; Li, M.; Zhao, Z.; Luo, Y. LncRNA and protein expression profiles reveal heart adaptation to high-altitude hypoxia in Tibetan sheep. Int. J. Mol. Sci. 2023, 25, 385. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, J.; Wang, D.; Wang, Y.; Zhang, F.; Jin, G.; Yuan, C.; Wang, X.; Qin, Q. Effect of silencing HIF-1α gene on testicle spermatogenesis function in varicocele rats. Cell Tissue Res. 2019, 378, 543–554. [Google Scholar] [CrossRef]
- Jiang, Q.; Di, Q.; Shan, D.; Xu, Q. Nonylphenol inhibited HIF-1alpha regulated aerobic glycolysis and induced ROS mediated apoptosis in rat Sertoli cells. Ecotoxicol. Environ. Saf. 2022, 241, 113822. [Google Scholar] [CrossRef]
- Cao, X.; Fang, X.; Guo, M.; Li, X.; He, Y.; Xie, M.; Xu, Y.; Liu, X. TRB3 mediates vascular remodeling by activating the MAPK signaling pathway in hypoxic pulmonary hypertension. Respir. Res. 2021, 22, 312. [Google Scholar] [CrossRef]
- Chen, H.; Fok, K.L.; Yu, S.; Jiang, J.; Chen, Z.; Gui, Y.; Cai, Z.; Chan, H.C. CD147 is required for matrix metalloproteinases-2 production and germ cell migration during spermatogenesis. Mol. Hum. Reprod. 2011, 17, 405–414. [Google Scholar] [CrossRef]
- Chang, Y.F.; Lee-Chang, J.S.; Harris, K.Y.; Sinha-Hikim, A.P.; Rao, M.K. Role of β-catenin in post-meiotic male germ cell differentiation. PLoS ONE 2011, 6, e28039. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Zhang, D.; Mi, J.; Du, W.; Yang, Y.; Chen, R.; Tian, C.; Zhao, X.; Zou, K. E-cadherin maintains the undifferentiated state of mouse spermatogonial progenitor cells via β-catenin. Cell Biosci. 2022, 12, 141. [Google Scholar] [CrossRef]
- Finkelstein, M.; Etkovitz, N.; Breitbart, H. Ca2+ signaling in mammalian spermatozoa. Mol. Cell. Endocrinol. 2020, 516, 110953. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Maiti, S.; Alam, N.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; Lécureuil, C.; Guillou, F.; Huff, V. The Wilms tumor gene, Wt1, is required for Sox9 expression and maintenance of tubular architecture in the developing testis. Proc. Natl. Acad. Sci. USA 2006, 103, 11987–11992. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, S.; He, C.; Chen, Z.; Lyu, T.; Zeng, L.; Wang, L.; Zhang, F.; Chen, H.; Zhao, R.C. Long non-coding RNA regulation of mesenchymal stem cell homeostasis and differentiation: Advances, challenges, and perspectives. Front. Cell Dev. Biol. 2021, 9, 711005. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Li, Q.; Chen, N.; Li, T.; Wang, H.; Su, M.; Shi, H.; Ma, Y. Effects of Pgam1-mediated glycolysis pathway in Sertoli cells on Spermatogonial stem cells based on transcriptomics and energy metabolomics. Front. Vet. Sci. 2022, 9, 992877. [Google Scholar] [CrossRef]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Su, L.; Cheng, C.Y.; Mruk, D.D. Adjudin-mediated Sertoli-germ cell junction disassembly affects Sertoli cell barrier function in vitro and in vivo. Int. J. Biochem. Cell Biol. 2010, 42, 1864–1875. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Jiang, Y.; Wang, Y.; Han, X.; Zhang, C.; Zhong, R.; Ge, W.; Han, B.; Ge, Z.; Huang, G.; et al. LncRNA8276 primes cell-cell adhesion for regulation of spermatogenesis. Andrology 2022, 10, 1687–1701. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, D.; Niu, T.; Sun, Z.; Wang, Y.; Han, X.; Xiong, B.; Shen, W.; Sun, Q.; Zhao, Y.; et al. LncRNA5251 inhibits spermatogenesis via modification of cell-cell junctions. Biol. Direct 2023, 18, 31. [Google Scholar] [CrossRef]
- Mirsanei, J.S.; Gholipour, H.; Zandieh, Z.; Jahromi, M.G.; Masroor, M.J.; Mehdizadeh, M.; Amjadi, F. Transition nuclear protein 1 as a novel biomarker in patients with fertilization failure. Clin. Exp. Reprod. Med. 2023, 50, 185–191. [Google Scholar] [CrossRef]
- Şahin, Y.; Aslan, E.S.; Aktuna, S.; Baltacı, V. Evaluation of TNP1 and PRM1 gene expression in male infertility patients with low or high sperm DNA fragmentation. J. Turk. Ger. Gynecol. Assoc. 2025, 26, 7–14. [Google Scholar] [CrossRef]
- Sen Sharma, S.; Vats, A.; Majumdar, S. Regulation of Hippo pathway components by FSH in testis. Reprod. Biol. 2019, 19, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C.; Wakitani, S.; Loveland, K.L. TGF-β superfamily signaling in testis formation and early male germline development. Semin. Cell Dev. Biol. 2015, 45, 94–103. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Li, T.; Wang, H.; Luo, R.; Song, J.; Wu, Y.; Jia, M.; Zhang, Y.; Ma, Y. A Comprehensive Atlas of Testicular lncRNAs Reveals Dynamic Changes and Regulatory Networks During Sexual Maturation in Tibetan Sheep. Animals 2026, 16, 176. https://doi.org/10.3390/ani16020176
Li T, Wang H, Luo R, Song J, Wu Y, Jia M, Zhang Y, Ma Y. A Comprehensive Atlas of Testicular lncRNAs Reveals Dynamic Changes and Regulatory Networks During Sexual Maturation in Tibetan Sheep. Animals. 2026; 16(2):176. https://doi.org/10.3390/ani16020176
Chicago/Turabian StyleLi, Taotao, Huihui Wang, Ruirui Luo, Juanjuan Song, Yi Wu, Meng Jia, Yong Zhang, and Youji Ma. 2026. "A Comprehensive Atlas of Testicular lncRNAs Reveals Dynamic Changes and Regulatory Networks During Sexual Maturation in Tibetan Sheep" Animals 16, no. 2: 176. https://doi.org/10.3390/ani16020176
APA StyleLi, T., Wang, H., Luo, R., Song, J., Wu, Y., Jia, M., Zhang, Y., & Ma, Y. (2026). A Comprehensive Atlas of Testicular lncRNAs Reveals Dynamic Changes and Regulatory Networks During Sexual Maturation in Tibetan Sheep. Animals, 16(2), 176. https://doi.org/10.3390/ani16020176





