Variability Survey at Different Genetic Markers as Effective Tools for the Management of the Endangered Breeds: The Case of the Sicilian Native Donkeys
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Donkey Genetic Diversity Investigated by Microsatellite Markers (STRs)
2.3. Donkey Genetic Diversity Investigated by D-Loop Sequencing
3. Results
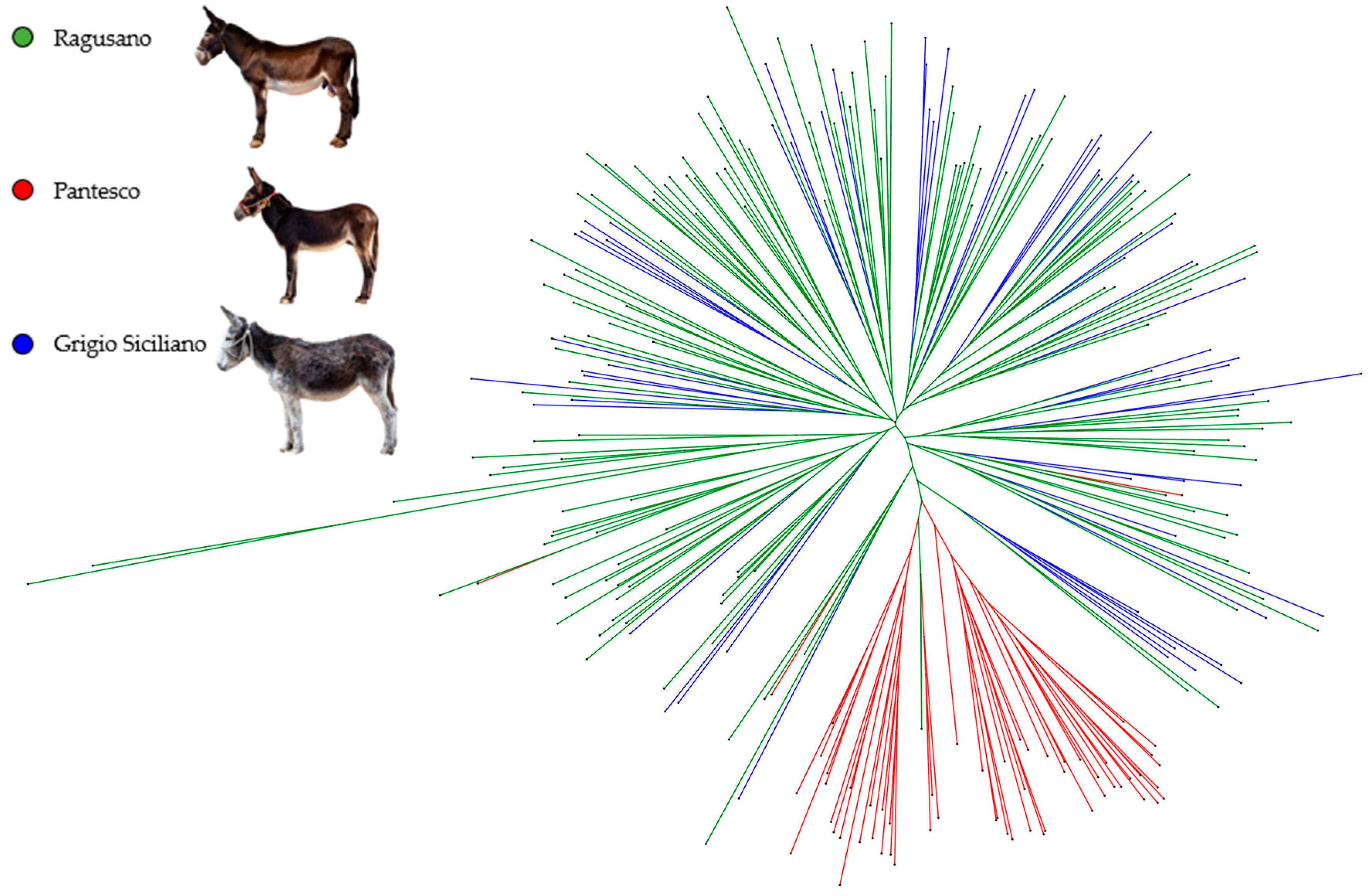
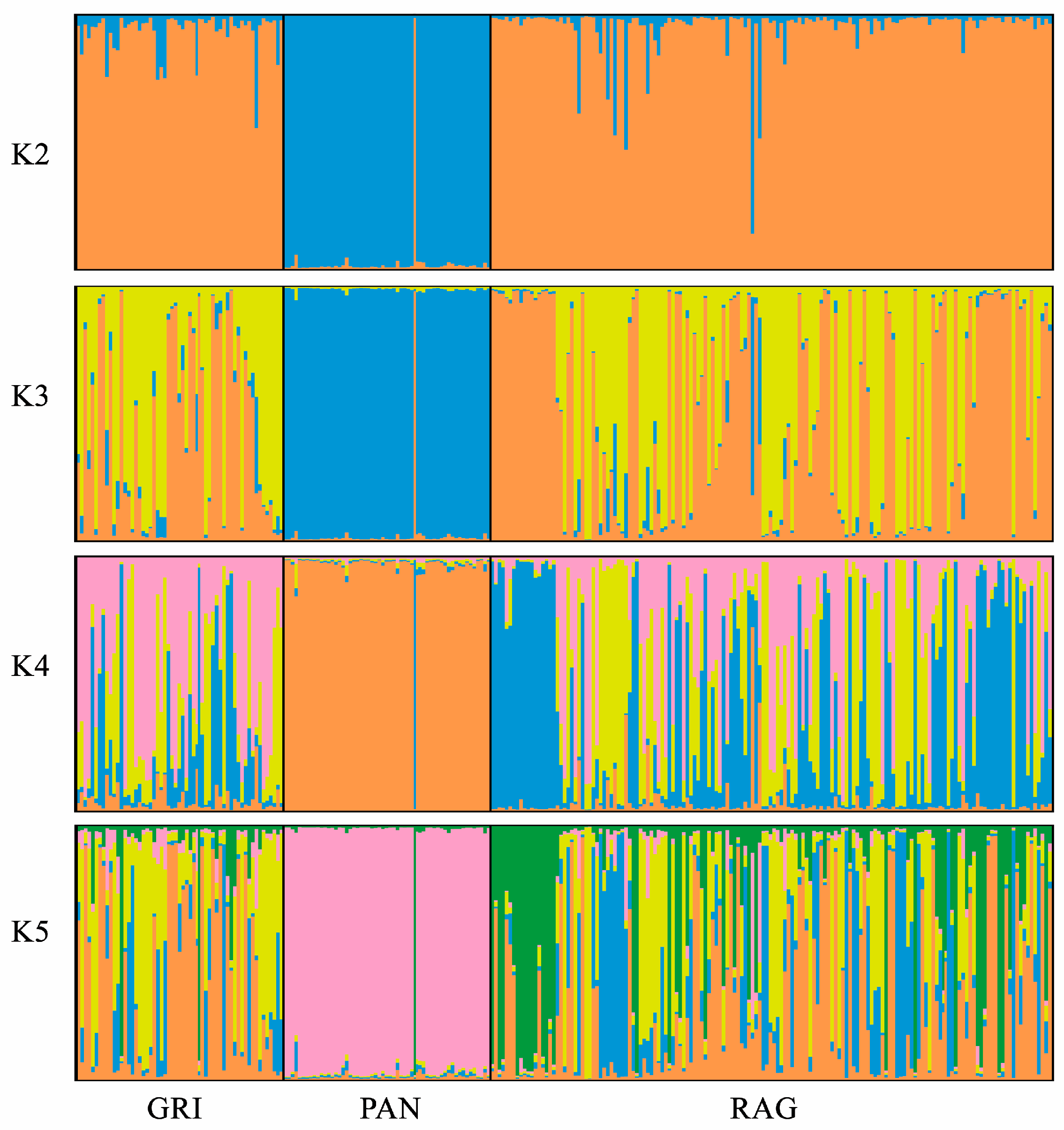
3.1. Genetic Diversity, Relationships, and Admixture Analysis by Means of STRs
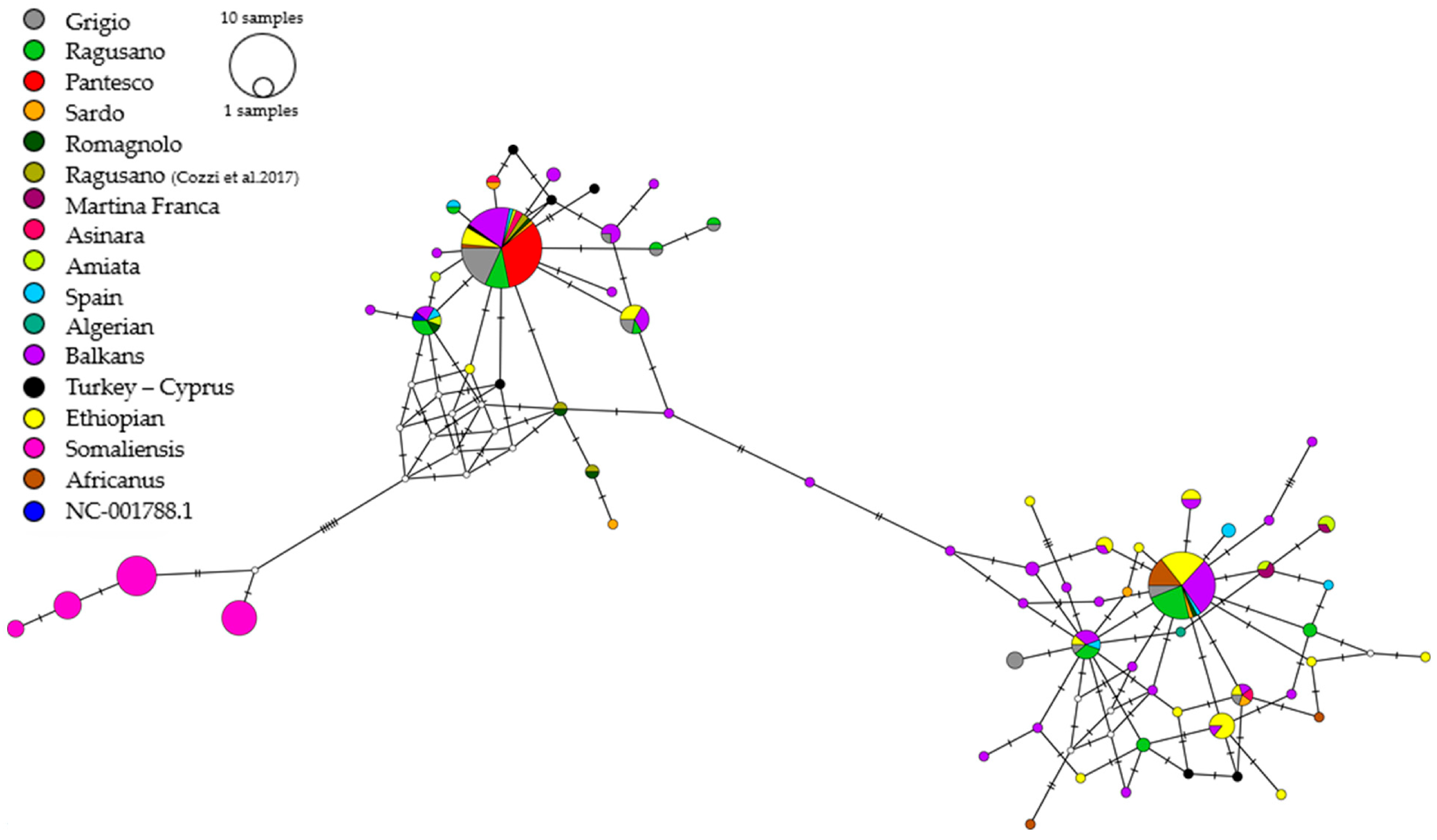
3.2. Donkey Genetic Diversity Investigated by Uniparental, Maternal Markers (mtDNA)
4. Discussion
4.1. Genetic Diversity, Relationships, and Admixture Analysis by Means of STRs
4.2. Donkey Genetic Diversity Investigated by Uniparental, Maternal Markers (mtDNA)
4.3. Main Practical Implication in the Use of Molecular Information in the Management of the Endangered Sicilian Donkey Breeds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Todd, E.T.; Tonasso-Calvière, L.; Chauvey, L.; Schiavinato, S.; Fages, A.; Seguin-Orlando, A.; Clavel, P.; Khan, N.; Pérez Pardal, L.; Patterson Rosa, L.; et al. The Genomic History and Global Expansion of Domestic Donkeys. Science 2022, 377, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Bordonaro, S.; Guastella, A.M.; Criscione, A.; Zuccaro, A.; Marletta, D. Genetic Diversity and Variability in Endangered Pantesco and Two Other Sicilian Donkey Breeds Assessed by Microsatellite Markers. Sci. World J. 2012, 2012, 648427. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.; Altomonte, I.; Licitra, R.; Salari, F. Nutritional and Nutraceutical Quality of Donkey Milk. J. Equine Vet. Sci. 2018, 65, 33–37. [Google Scholar] [CrossRef]
- Ivanković, A.; Šubara, G.; Bittante, G.; Šuran, E.; Amalfitano, N.; Aladrović, J.; Kelava Ugarković, N.; Pađen, L.; Pećina, M.; Konjačić, M. Potential of Endangered Local Donkey Breeds in Meat and Milk Production. Animals 2023, 13, 2146. [Google Scholar] [CrossRef]
- Galardi, M.; Contalbrigo, L.; Toson, M.; Bortolotti, L.; Lorenzetto, M.; Riccioli, F.; Moruzzo, R. Donkey Assisted Interventions: A Pilot Survey on Service Providers in North-Eastern Italy. Explore 2022, 18, 10–16. [Google Scholar] [CrossRef]
- Pécsek, B.; Beke, S. A Szamár Potenciális Szerepe Az Egészségturizmusban. Recreation 2023, 13, 16–20. [Google Scholar] [CrossRef]
- Camillo, F.; Rota, A.; Biagini, L.; Tesi, M.; Fanelli, D.; Panzani, D. The Current Situation and Trend of Donkey Industry in Europe. J. Equine Vet. Sci. 2018, 65, 44–49. [Google Scholar] [CrossRef]
- Colli, L.; Perrotta, G.; Negrini, R.; Bomba, L.; Bigi, D.; Zambonelli, P.; Verini Supplizi, A.; Liotta, L.; Ajmone-Marsan, P. Detecting Population Structure and Recent Demographic History in Endangered Livestock Breeds: The Case of the Italian Autochthonous Donkeys. Anim. Genet. 2013, 44, 69–78. [Google Scholar] [CrossRef]
- Matassino, D.; Cecchi, F.; Ciani, F.; Incoronato, C.; Occidente, M.; Santoro, L.; Ciampolini, R. Genetic Diversity and Variability in Two Italian Autochthonous Donkey Genetic Types Assessed by Microsatellite Markers. Ital. J. Anim. Sci. 2014, 13, 3028. [Google Scholar] [CrossRef]
- Jordana, J.; Ferrando, A.; Miró, J.; Goyache, F.; Loarca, A.; Martínez López, O.R.; Canelón, J.L.; Stemmer, A.; Aguirre, L.; Lara, M.A.C.; et al. Genetic Relationships among A Merican Donkey Populations: Insights into the Process of Colonization. J. Anim. Breed. Genet. 2016, 133, 155–164. [Google Scholar] [CrossRef]
- Nayak, S.S.; Rajawat, D.; Jain, K.; Sharma, A.; Gondro, C.; Tarafdar, A.; Dutt, T.; Panigrahi, M. A Comprehensive Review of Livestock Development: Insights into Domestication, Phylogenetics, Diversity, and Genomic Advances. Mamm. Genome 2024, 35, 577–599. [Google Scholar] [CrossRef]
- Sharma, R.; Sharma, H.; Ahlawat, S.; Panchal, P.; Pal, Y.; Behl, R.; Tantia, M.S. Simple Sequence Repeat (SSR) Genotypic Data Reveal High Genetic Diversity in Rajasthan Donkey of India. Indian J. Anim. Sci. 2018, 87, 79860. [Google Scholar] [CrossRef]
- Kim, S.M.; Yun, S.W.; Cho, G.J. Assessment of Genetic Diversity Using Microsatellite Markers to Compare Donkeys (Equus asinus) with Horses (Equus caballus). Anim. Biosci. 2021, 34, 1460–1465. [Google Scholar] [CrossRef]
- Xu, X.; Gullberg, A.; Arnason, U. The Complete Mitochondrial DNA (mtDNA) of the Donkey and mtDNA Comparisons among Four Closely Related Mammalian Species-Pairs. J. Mol. Evol. 1996, 43, 438–446. [Google Scholar] [CrossRef]
- Beja-Pereira, A.; England, P.R.; Ferrand, N.; Jordan, S.; Bakhiet, A.O.; Abdalla, M.A.; Mashkour, M.; Jordana, J.; Taberlet, P.; Luikart, G. African Origins of the Domestic Donkey. Science 2004, 304, 1781. [Google Scholar] [CrossRef]
- Rambaldi Migliore, N.; Bigi, D.; Milanesi, M.; Zambonelli, P.; Negrini, R.; Morabito, S.; Verini-Supplizi, A.; Liotta, L.; Chegdani, F.; Agha, S.; et al. Mitochondrial DNA Control-Region and Coding-Region Data Highlight Geographically Structured Diversity and Post-Domestication Population Dynamics in Worldwide Donkeys. PLoS ONE 2024, 19, e0307511. [Google Scholar] [CrossRef] [PubMed]
- Ivanković, A.; Bittante, G.; Šubara, G.; Šuran, E.; Ivkić, Z.; Pećina, M.; Konjačić, M.; Kos, I.; Kelava Ugarković, N.; Ramljak, J. Genetic and Population Structure of Croatian Local Donkey Breeds. Diversity 2022, 14, 322. [Google Scholar] [CrossRef]
- Cozzi, M.C.; Valiati, P.; Cherchi, R.; Gorla, E.; Prinsen, R.T.M.M.; Longeri, M.; Bagnato, A.; Strillacci, M.G. Mitochondrial DNA Genetic Diversity in Six Italian Donkey Breeds (Equus asinus). Mitochondrial DNA Part A 2018, 29, 409–418. [Google Scholar] [CrossRef]
- Mazzatenta, A.; Vignoli, M.; Caputo, M.; Vignola, G.; Tamburro, R.; De Sanctis, F.; Roig, J.M.; Bucci, R.; Robbe, D.; Carluccio, A. Maternal Phylogenetic Relationships and Genetic Variation among Rare, Phenotypically Similar Donkey Breeds. Genes 2021, 12, 1109. [Google Scholar] [CrossRef]
- Guastella, A.M.; Zuccaro, A.; Criscione, A.; Marletta, D.; Bordonaro, S. Genetic Analysis of Sicilian Autochthonous Horse Breeds Using Nuclear and Mitochondrial DNA Markers. J. Hered. 2011, 102, 753–758. [Google Scholar] [CrossRef]
- Criscione, A.; Moltisanti, V.; Chies, L.; Marletta, D.; Bordonaro, S. A Genetic Analysis of the Italian Salernitano Horse. Animal 2015, 9, 1610–1616. [Google Scholar] [CrossRef]
- Stanisic, L.J.; Aleksic, J.M.; Dimitrijevic, V.; Simeunovic, P.; Glavinic, U.; Stevanovic, J.; Stanimirovic, Z. New Insights into the Origin and the Genetic Status of the Balkan Donkey from Serbia. Anim. Genet. 2017, 48, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; ISBN 978-0-231-88671-0. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a Genetic Linkage Map in Man Using Restriction Fragment Length Polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- El Mousadik, A.; Petit, R.J. High Level of Genetic Differentiation for Allelic Richness among Populations of the Argan Tree [Argania spinosa (L.) Skeels] Endemic to Morocco. Theor. Appl. Genet. 1996, 92, 832–839. [Google Scholar] [CrossRef]
- Bowcock, A.M.; Ruiz-Linares, A.; Tomfohrde, J.; Minch, E.; Kidd, J.R.; Cavalli-Sforza, L.L. High Resolution of Human Evolutionary Trees with Polymorphic Microsatellites. Nature 1994, 368, 455–457. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. The SplitsTree App: Interactive Analysis and Visualization Using Phylogenetic Trees and Networks. Nat. Methods 2024, 21, 1773–1774. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Aranguren-Mendez, J.; Beja-Pereira, A.; Avellanet, R.; Dzama, K.; Jordana, J. Mitochondrial DNA Variation and Genetic Relationships in Spanish Donkey Breeds (Equus asinus). J. Anim. Breed. Genet. 2004, 121, 319–330. [Google Scholar] [CrossRef]
- de la Pena, A. Molecular Variability of Horse and Donkey Sex Chromosomes. 2015. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 1 April 2024).
- Ivankovic, A.; Kavar, T.; Caput, P.; Mioc, B.; Pavic, V.; Dovc, P. Genetic Diversity of Three Donkey Populations in the Croatian Coastal Region. Anim. Genet. 2002, 33, 169–177. [Google Scholar] [CrossRef]
- Pérez-Pardal, L.; Grizelj, J.; Traoré, A.; Cubric-Curik, V.; Arsenos, G.; Dovenski, T.; Marković, B.; Fernández, I.; Cuervo, M.; Álvarez, I.; et al. Lack of Mitochondrial DNA Structure in B Alkan Donkey Is Consistent with a Quick Spread of the Species after Domestication. Anim. Genet. 2014, 45, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Kul, C.; Bilgen, B.; Akyuz, N.; Ertugrul, B.; Okan, O. Molecular phylogeny of Anatolian and Cypriot donkey populations based on mitochondrial DNA and Y-chromosomal STRs. Ank. Üniv. Vet. Fakültesi Derg. 2016, 63, 143–149. [Google Scholar] [CrossRef]
- Kefena, E.; Dessie, T.; Tegegne, A.; Beja-Pereira, A.; Yusuf Kurtu, M.; Rosenbom, S.; Han, J.L. Genetic Diversity and Matrilineal Genetic Signature of Native Ethiopian Donkeys (Equus asinus) Inferred from Mitochondrial DNA Sequence Polymorphism. Livest. Sci. 2014, 167, 73–79. [Google Scholar] [CrossRef]
- Kimura, B.; Marshall, F.B.; Chen, S.; Rosenbom, S.; Moehlman, P.D.; Tuross, N.; Sabin, R.C.; Peters, J.; Barich, B.; Yohannes, H.; et al. Ancient DNA from Nubian and Somali Wild Ass Provides Insights into Donkey Ancestry and Domestication. Proc. R. Soc. B Biol. Sci. 2011, 278, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Criscione, A.; Chessari, G.; Cesarani, A.; Ablondi, M.; Asti, V.; Bigi, D.; Bordonaro, S.; Ciampolini, R.; Cipolat-Gotet, C.; Congiu, M.; et al. Analysis of ddRAD-Seq Data Provides New Insights into the Genomic Structure and Patterns of Diversity in Italian Donkey Populations. J. Anim. Sci. 2024, 102, skae165. [Google Scholar] [CrossRef]
- Bonadonna, T. Etnologia Zootecnica—Trattato Di Scienza e Tecnica Delle Produzioni Animali; UTeT: Torino, Italy, 1976; Volume VI. [Google Scholar]
- Dell, A.C.; Curry, M.C.; Yarnell, K.M.; Starbuck, G.R.; Wilson, P.B. Mitochondrial D-Loop Sequence Variation and Maternal Lineage in the Endangered Cleveland Bay Horse. PLoS ONE 2020, 15, e0243247. [Google Scholar] [CrossRef]
- Radoslavov, G.; Kalaydzhiev, G.; Yordanov, G.; Palova, N.; Salkova, D.; Hristov, P. Genetic Diversity of Native Bulgarian Sheep Breeds Based on the Mitochondrial D-Loop Sequence Analysis. Small Rumin. Res. 2025, 249, 107527. [Google Scholar] [CrossRef]
- Huang, B.; Khan, M.Z.; Chai, W.; Ullah, Q.; Wang, C. Exploring Genetic Markers: Mitochondrial DNA and Genomic Screening for Biodiversity and Production Traits in Donkeys. Animals 2023, 13, 2725. [Google Scholar] [CrossRef] [PubMed]
| Macro-Area | Code | Country | Sequences | References |
|---|---|---|---|---|
| Mediterranea and Balkan area | SIC | Italy | 81 | Present study |
| ITY | Italy | 28 | [18] | |
| SPA | Spain | 9 | [33,34] | |
| BAL | Croatia, Albania, Bulgaria, Greece, Kosovo, Montenegro, Romania, Serbia, Ukraine | 65 | [35,36] | |
| TRK-CYR | Turkey, Cyprus | 7 | [37] | |
| Africa | ETH | Ethiopia | 39 | [38] |
| SOM | Ethiopia | 41 | [15,38,39] | |
| NUB | Ethiopia | 10 | [15,38,39] |
| Locus | Ar | N | Ho | He | PIC |
|---|---|---|---|---|---|
| HTG10 | 8.6 | 10 | 0.657 | 0.752 | 0.714 |
| VHL20 | 4.4 | 6 | 0.611 | 0.587 | 0.509 |
| HTG07 | 10.4 | 11 | 0.789 | 0.769 | 0.732 |
| AHT04 | 6.0 | 10 | 0.589 | 0.622 | 0.566 |
| HMS3 | 5.6 | 7 | 0.460 | 0.504 | 0.448 |
| HMS06 | 3.2 | 4 | 0.349 | 0.348 | 0.281 |
| HMS07 | 6.2 | 8 | 0.257 | 0.253 | 0.237 |
| HMS02 | 5.2 | 6 | 0.574 | 0.597 | 0.538 |
| HTG06 | 4.2 | 5 | 0.570 | 0.585 | 0.527 |
| ASB23 | 5.6 | 6 | 0.689 | 0.702 | 0.655 |
| HMS18 | 7.3 | 10 | 0.565 | 0.645 | 0.577 |
| TKY297 | 6.2 | 7 | 0.629 | 0.697 | 0.637 |
| TKY312 | 4.6 | 6 | 0.551 | 0.601 | 0.522 |
| TKY337 | 5.4 | 7 | 0.437 | 0.440 | 0.395 |
| TKY343 | 3.5 | 4 | 0.562 | 0.534 | 0.427 |
| Mean | 5.69 | 7.13 | 0.553 | 0.576 | 0.518 |
| Breed | Size | MNA | Ar | Private | Ho ± SD | He ± SD | FIS (95% C.I.) | Fst | PIC |
|---|---|---|---|---|---|---|---|---|---|
| GRI | 58 | 5.27 ± 1.79 | 5.3 | 1 | 0.584 ± 0.017 | 0.617 ± 0.040 (−0.009–0.097) | 0.044 | 0.041 | 0.559 |
| PAN | 57 | 3.00 ± 1.00 | 3.0 | - | 0.495 ± 0.017 | 0.498 ± 0.052 (−0.064–0.084) | 0.010 | 0.081 | 0.435 |
| RAG | 157 | 7.07 ± 2.15 | 6.0 | 26 | 0.580 ± 0.010 | 0.612 ± 0.037 (0.020–0.079) | 0.050 | 0.050 | 0.559 |
| Haplotype | PAN | RAG | GRI | 1 5 4 8 4 | 1 5 4 9 0 | 1 5 5 0 3 | 1 5 5 2 7 | 1 5 5 3 2 | 1 5 5 6 9 | 1 5 5 8 0 | 1 5 5 9 2 | 1 5 5 9 8 | 1 5 5 9 9 | 1 5 6 2 1 | 1 5 6 2 6 | 1 5 6 4 4 | 1 5 6 4 5 | 1 5 6 5 2 | 1 5 6 6 2 | 1 5 6 9 8 | 1 5 7 1 3 | 1 5 7 1 8 | 1 5 7 4 6 | 1 5 7 4 8 | 1 5 7 7 0 | 1 5 8 0 1 | 1 5 8 0 2 | 1 5 8 0 6 | 1 5 8 2 0 | 1 5 8 2 1 | 1 5 8 2 2 |
| NC_001788.1 | G | C | T | C | A | A | A | A | C | A | A | A | G | A | C | A | C | C | C | G | G | T | C | T | C | C | G | G | |||
| H1 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||
| H2 | 3 | 1 | A | T | C | . | . | G | G | . | T | G | . | . | A | . | T | G | T | . | . | . | . | C | T | . | T | T | A | A | |
| H3 | 1 | . | . | . | . | . | G | G | . | . | G | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | A | . | ||
| H4 | 11 | 3 | A | T | C | . | . | G | G | . | T | G | G | . | A | . | T | G | T | . | . | . | . | C | T | . | T | T | A | A | |
| H5 | 19 | 4 | 9 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| H6 | 1 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | ||
| H7 | 1 | A | T | C | . | . | G | G | . | T | G | . | G | A | . | T | G | T | . | . | C | . | . | T | . | T | T | A | A | ||
| H8 | 1 | 2 | . | . | . | . | . | . | G | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| H9 | 2 | A | T | C | . | . | G | G | . | T | G | . | G | A | . | T | G | T | . | . | . | . | . | T | . | T | T | A | A | ||
| H10 | 1 | 1 | . | . | . | G | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| H11 | 1 | A | T | C | . | . | G | . | . | T | G | G | . | A | . | T | G | T | . | . | . | . | C | T | . | T | T | A | A | ||
| H12 | 2 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | ||
| H13 | 1 | 1 | . | . | . | G | G | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| H14 | 1 | 1 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | |
| H15 | 4 | 2 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | |
| H16 | 2 | A | T | C | . | . | G | G | G | T | G | G | . | A | . | T | G | T | . | . | . | . | C | T | . | T | T | A | A | ||
| H17 | 2 | A | T | C | . | . | G | G | . | T | G | . | . | A | G | T | G | T | . | . | . | . | C | T | . | T | T | A | A | ||
| H18 | 1 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . |
| Breed | n | S | SpI | Ss | NH | PH | π ± s.d. | Hd ± s.d. | k |
|---|---|---|---|---|---|---|---|---|---|
| Grigio | 26 | 25 | 20 | 5 | 13 | 8 | 0.0220 ± 0.0030 | 0.871 ± 0.056 | 8.237 |
| Pantesco | 23 | 1 | 1 | 0 | 2 | 0 | 0.0008 ± 0.0003 | 0.300 ± 0.105 | 0.300 |
| Ragusano | 32 | 23 | 21 | 2 | 12 | 5 | 0.0250 ± 0.0015 | 0.859 ± 0.048 | 9.246 |
| Whole sample | 82 | 28 | 25 | 3 | 18 | 13 | 0.0218 ± 0.0017 | 0.812 ± 0.036 | 7.981 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Carlentini, M.; Tumino, S.; Chessari, G.; Antoci, A.; Criscione, A.; Marletta, D.; Mastrangelo, S.; Bordonaro, S. Variability Survey at Different Genetic Markers as Effective Tools for the Management of the Endangered Breeds: The Case of the Sicilian Native Donkeys. Animals 2026, 16, 90. https://doi.org/10.3390/ani16010090
Carlentini M, Tumino S, Chessari G, Antoci A, Criscione A, Marletta D, Mastrangelo S, Bordonaro S. Variability Survey at Different Genetic Markers as Effective Tools for the Management of the Endangered Breeds: The Case of the Sicilian Native Donkeys. Animals. 2026; 16(1):90. https://doi.org/10.3390/ani16010090
Chicago/Turabian StyleCarlentini, Morena, Serena Tumino, Giorgio Chessari, Aurora Antoci, Andrea Criscione, Donata Marletta, Salvatore Mastrangelo, and Salvatore Bordonaro. 2026. "Variability Survey at Different Genetic Markers as Effective Tools for the Management of the Endangered Breeds: The Case of the Sicilian Native Donkeys" Animals 16, no. 1: 90. https://doi.org/10.3390/ani16010090
APA StyleCarlentini, M., Tumino, S., Chessari, G., Antoci, A., Criscione, A., Marletta, D., Mastrangelo, S., & Bordonaro, S. (2026). Variability Survey at Different Genetic Markers as Effective Tools for the Management of the Endangered Breeds: The Case of the Sicilian Native Donkeys. Animals, 16(1), 90. https://doi.org/10.3390/ani16010090






