Genome-Wide Association Study Identifies Candidate Genes for Body Size Traits in Wanyue Black Pigs
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Experimental Animals
2.3. Genotyping, Imputation, and Quality Control
2.4. Genome-Wide Association Analysis of Body Size Traits
2.5. Transcriptome-Wide Association Study
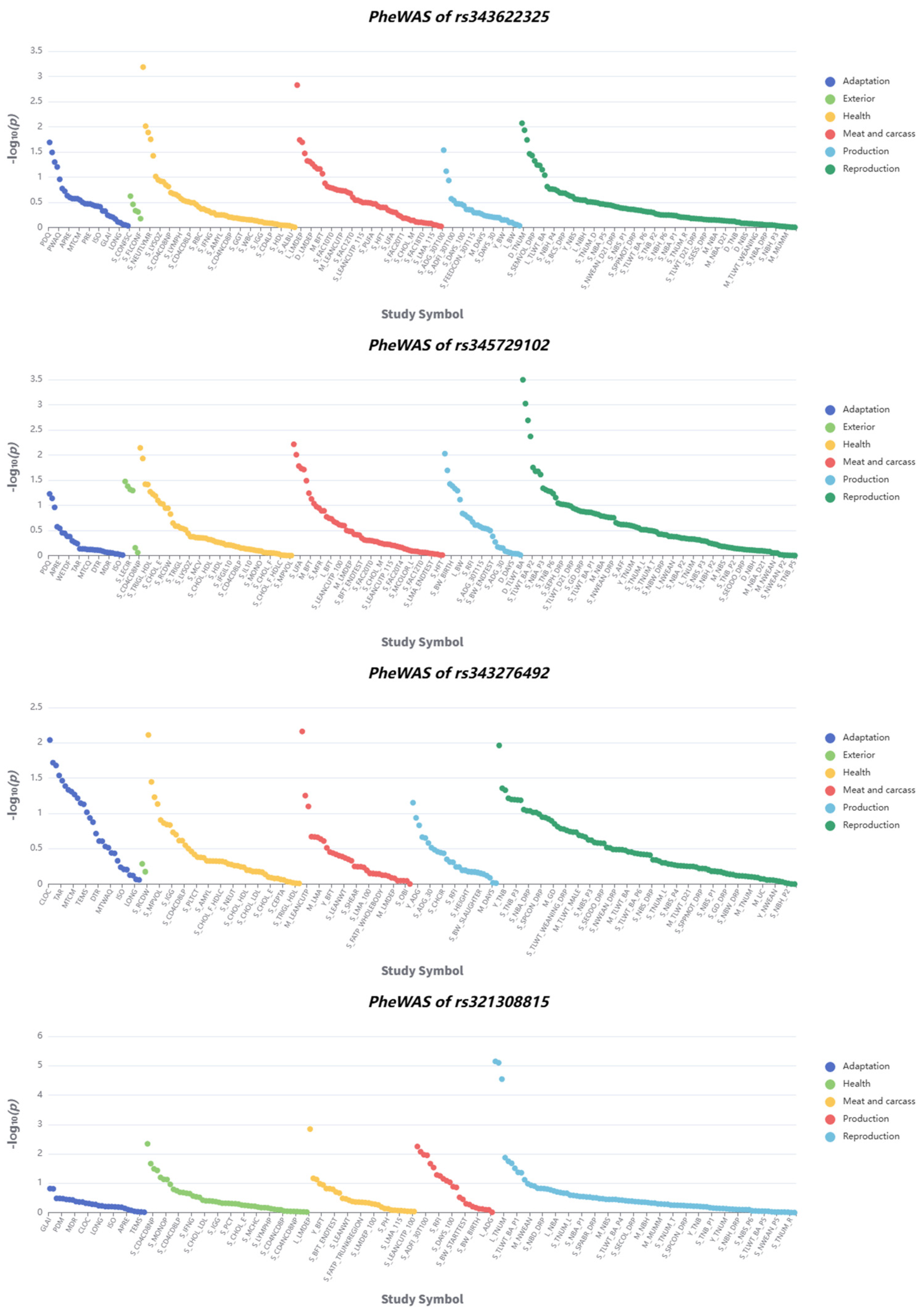
2.6. Functional Annotation and Phenome-Wide Association Study
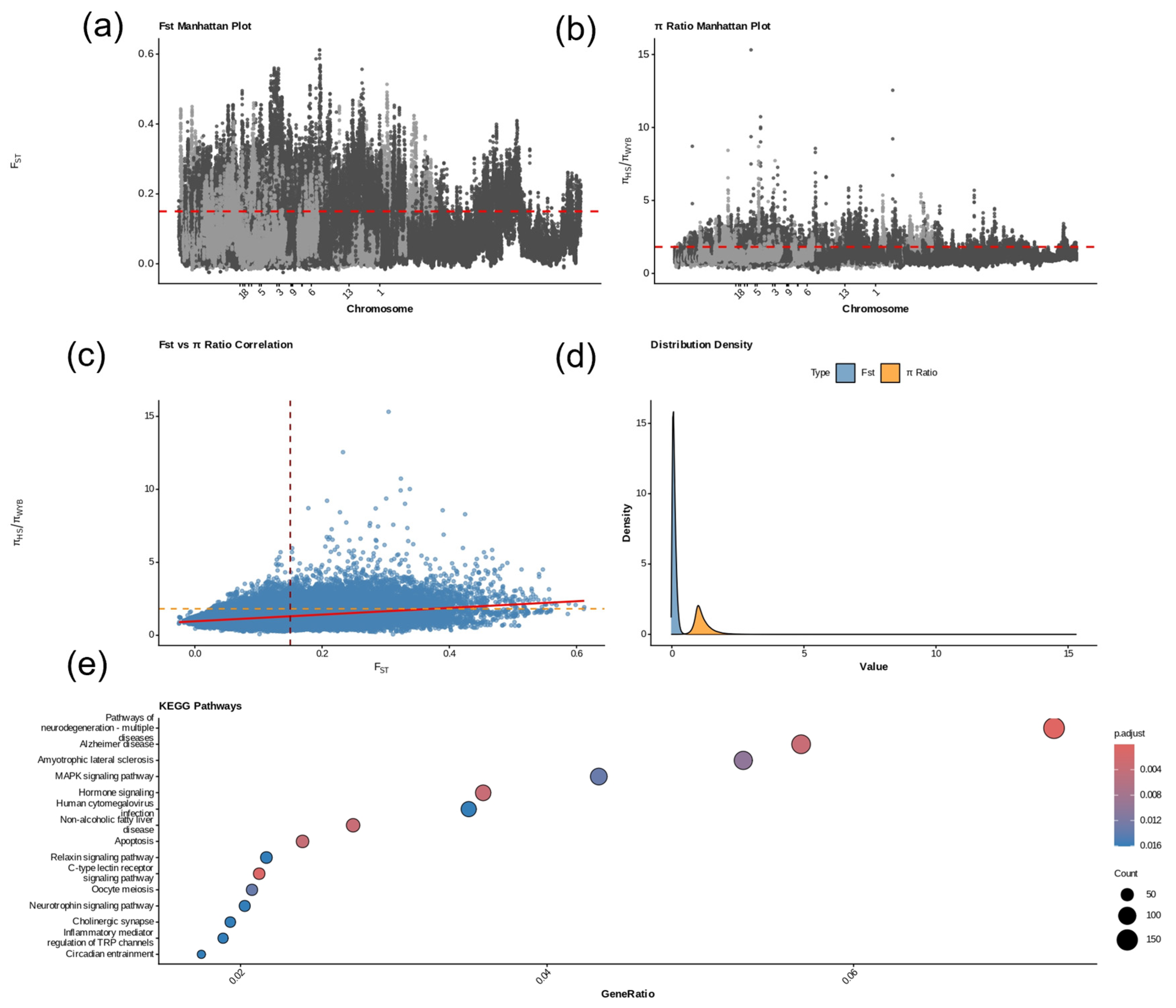
2.7. Selective Sweep Analysis Between Wanyue Black and Huoshou Black Pigs
3. Results
3.1. Statistical Characterization and Correlation Analysis of Phenotypic Traits
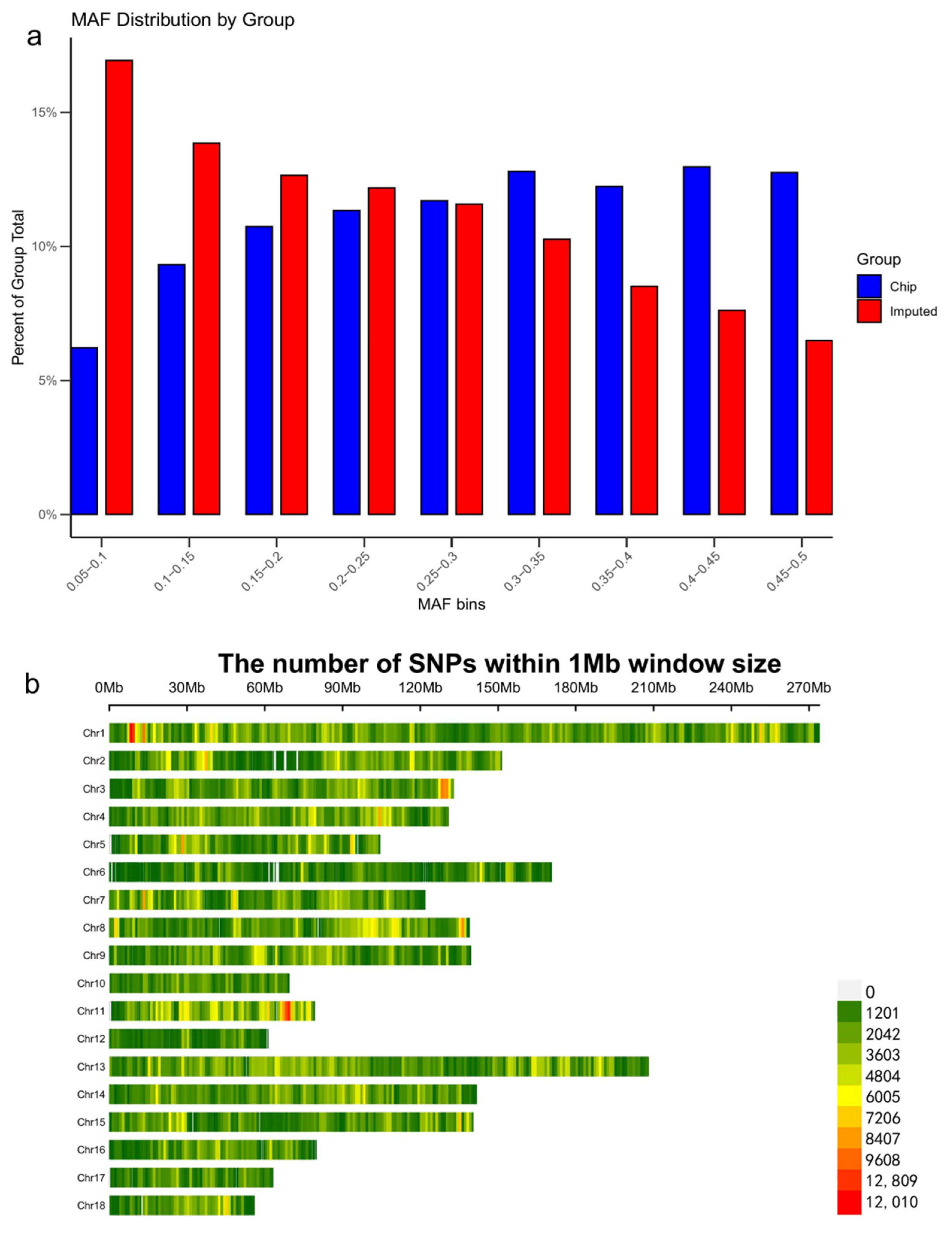
3.2. Imputation Accuracy
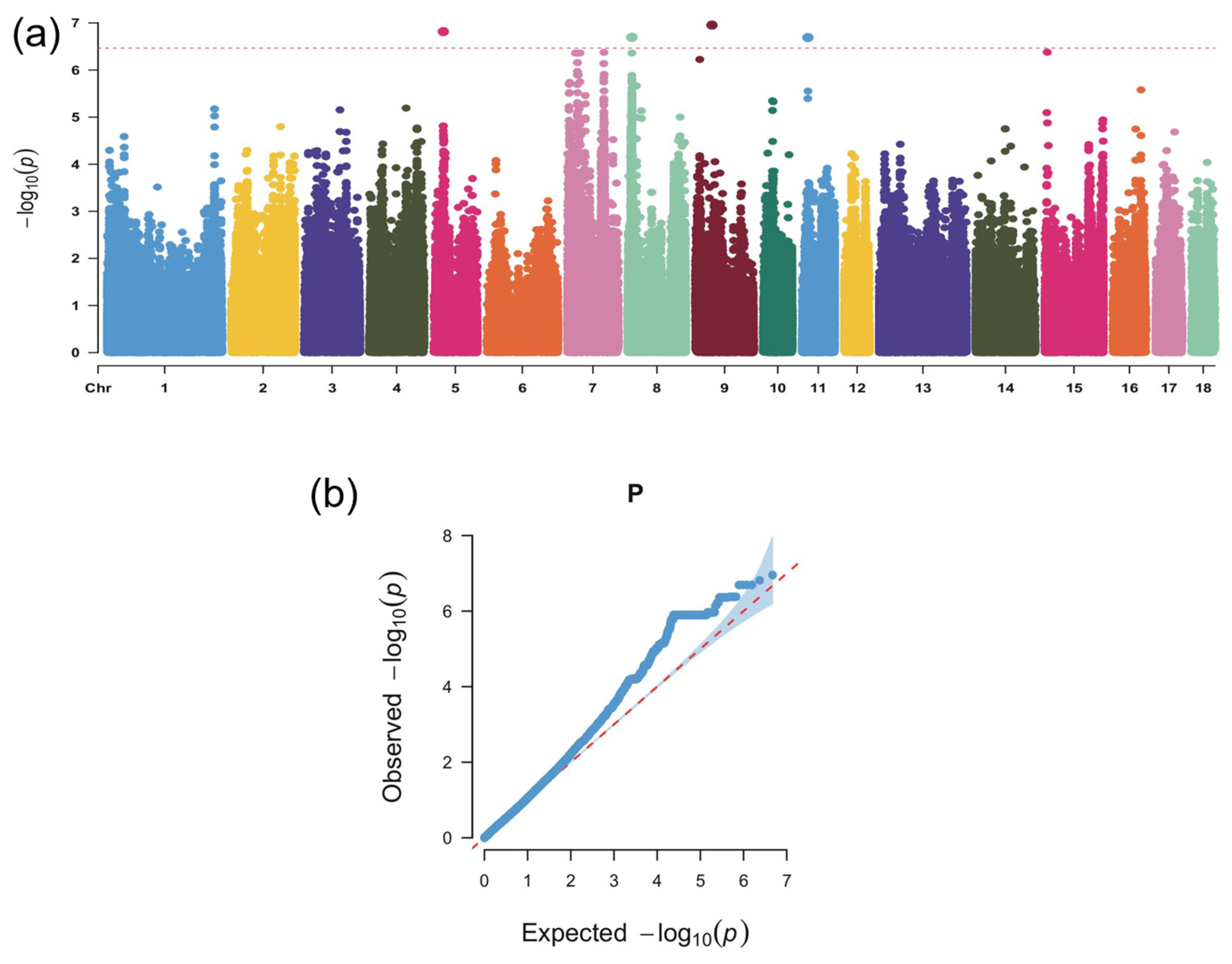
3.3. Genome-Wide Association Analysis Identifies Candidate Loci and Genes Linked to Body-Size-Related Traits in Wanyue Black Pigs
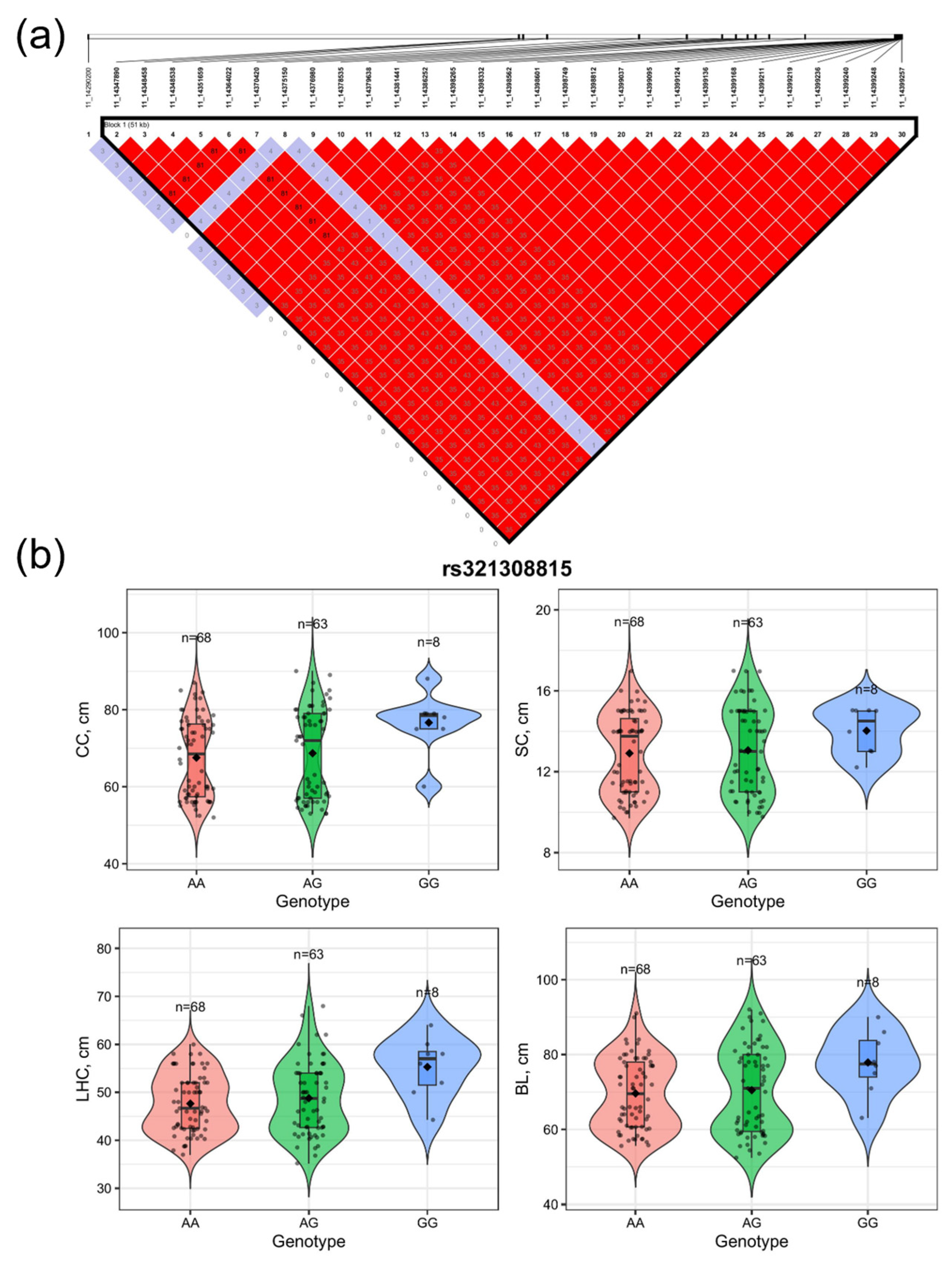
3.4. Linkage Disequilibrium and Genotypic Effect Analyses of Candidate SNPs Associated with Body Size Traits
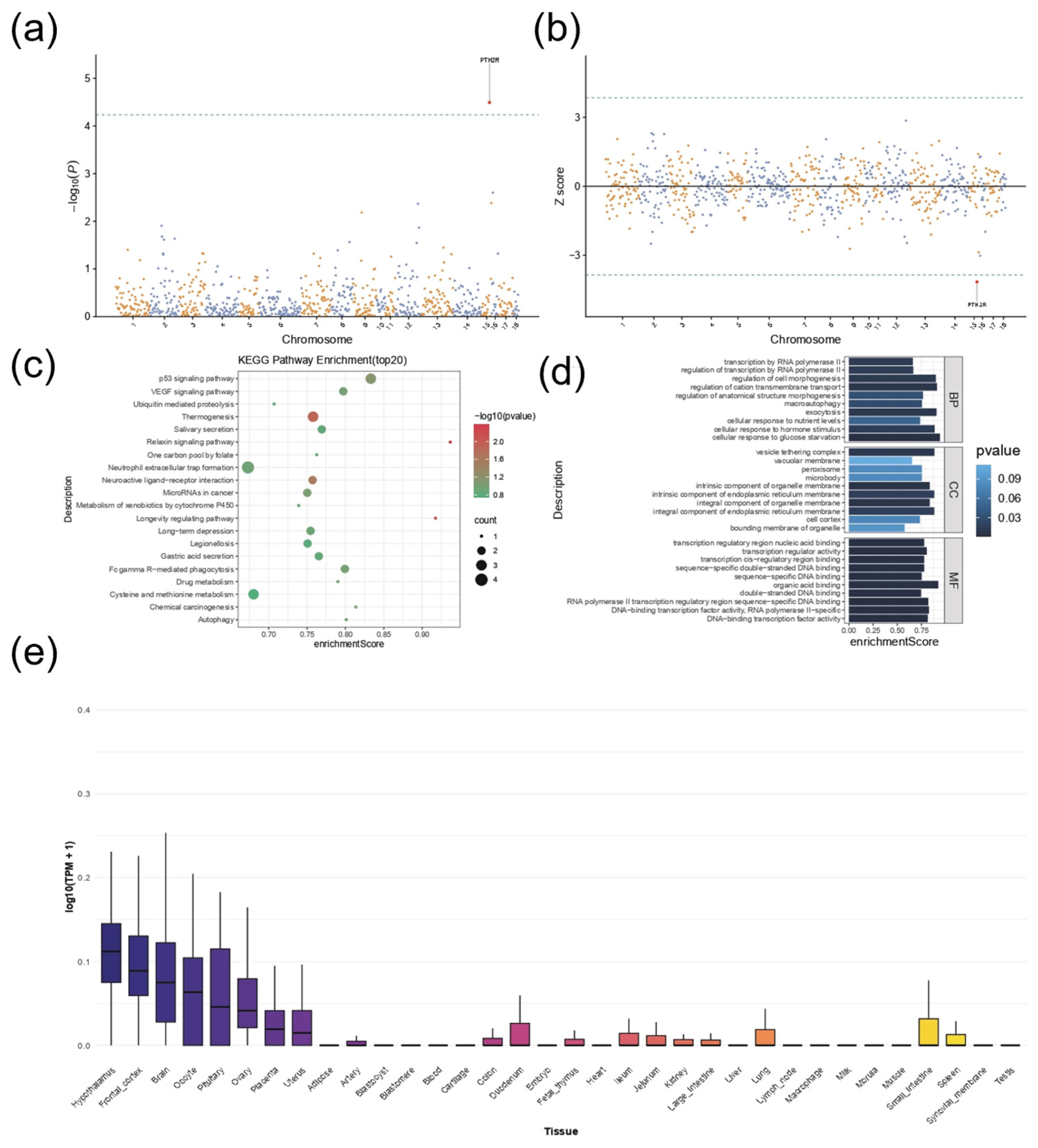
3.5. Transcriptome-Wide Association Study Reveals the Association Between Pituitary Traits and PTH2R Gene Expression
3.6. Functional Annotation and Phenome-Wide Association Study Reveal Candidate Genes Associated with Body Size Traits in Pigs
3.7. Genomic Selection Signals and Functional Pathway Analysis of Wanyue Black Pigs Relative to Huoshao Black Pigs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lebret, B.; Potokar, M.C. Review: Pork quality attributes from farm to fork. Part I. Carcass and fresh meat. Animal 2021, 16, 100402. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ortega, D.L.; Lin, W. Food values drive Chinese consumers’ demand for meat and milk substitutes. Appetite 2022, 181, 106392. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Liu, L.; Huang, M.; Ruan, D.; Ding, R.; Zhang, Z.; Zheng, E.; Wang, S.; Deng, S.; Meng, X.; et al. Dispersal, and Impact: Bidirectional Introgression Between Chinese and European Pig Populations. Adv. Sci. 2025, 12, 2570167. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, R.; Hu, X.; Shan, D.; Zhang, X.; Gou, Y.; Zheng, T.; Li, Y.; Fei, K.; Zhang, Q.; et al. Latitude-driven adaptive evolution of body length in diverse pig breeds across China. BMC Biol. 2025, 23, 150. [Google Scholar] [CrossRef]
- Yoo, C.K.; Park, H.B.; Lee, J.B.; Jung, E.J.; Kim, B.M.; Kim, H.I.; Ahn, S.J.; Ko, M.S.; Cho, I.C.; Lim, H.T. QTL analysis of body weight and carcass body length traits in an F-2 intercross between Landrace and Korean native pigs. Anim. Genet. 2014, 45, 589–592. [Google Scholar] [CrossRef]
- Gorssen, W.; Winters, C.; Meyermans, R.; D’hooge, R.; Janssens, S.; Buys, N. Estimating genetics of body dimensions and activity levels in pigs using automated pose estimation. Sci. Rep. 2022, 12, 15384. [Google Scholar] [CrossRef]
- Willson, H.E.; de Oliveira, H.R.; Schinckel, A.P.; Grossi, D.; Brito, L.F. Estimation of Genetic Parameters for Pork Quality, Novel Carcass, Primal-Cut and Growth Traits in Duroc Pigs. Animals 2020, 10, 779. [Google Scholar] [CrossRef]
- Martinsen, K.H.; Ødegård, J.; Aasmundstad, T.; Olsen, D.; Meuwissen, T.H. Genetic relationships between boar feed efficiency and sow piglet production, body condition score, and stayability in Norwegian Landrace pigs. J. Anim. Sci. 2016, 94, 3159. [Google Scholar] [CrossRef]
- Coyne, J.M.; Berry, D.P.; Matilainen, K.; Sevon-Aimonen, M.-L.; Mantysaari, E.A.; Juga, J.; Serenius, T.; McHugh, N. Genetic co-variance functions for live weight, feed intake, and efficiency measures in growing pigs. J. Anim. Sci. 2017, 95, 3822–3832. [Google Scholar] [CrossRef]
- Liu, H.; Song, H.; Jiang, Y.; Jiang, Y.; Zhang, F.; Liu, Y.; Shi, Y.; Ding, X.; Wang, C. A Single-Step Genome Wide Association Study on Body Size Traits Using Imputation-Based Whole-Genome Sequence Data in Yorkshire Pigs. Front. Genet. 2020, 12, 629049. [Google Scholar] [CrossRef]
- Zhou, S.; Ding, R.; Zhuang, Z.; Zeng, H.; Wen, S.; Ruan, D.; Wu, J.; Qiu, Y.; Zheng, E.; Cai, G.; et al. Genome-Wide Association Analysis Reveals Genetic Loci and Candidate Genes for Chest, Abdominal, and Waist Circumferences in Two Duroc Pig Populations. Front. Vet. Sci. 2022, 8, 807003. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Brown, M.A.; McCarthy, M.I.; Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012, 90, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Ling, C.; Xiao, H.; Zhang, Z. Identification of Gene Expression and Splicing QTLs in Porcine Muscle Associated with Meat Quality Traits. Animals 2025, 15, 1209. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, G.; Liu, Z.; Cheng, Y.; Ding, R.; Yang, G.; Yu, T. Whole genome and transcriptome analyses identify genetic markers associated with growth traits in Qinchuan black pig. BMC Genom. 2025, 26, 469. [Google Scholar] [CrossRef]
- Sun, K.; Hong, Y.; Zhang, W.; Dong, J.; Wen, Z.; Hu, Z.; Tan, X.; Li, H.; Zhao, A.; Huang, M.; et al. Single- and multiple-locus model genome-wide association study for growth traits in Dongliao Black pigs. Anim. Biosci. 2025, 38, 2312–2323. [Google Scholar] [CrossRef]
- Wu, Z.; Dou, T.; Wu, J.; Bai, L.; Zhang, Y.; Zan, S.; Yang, S.; Zhou, H.; Han, J.; Han, X.; et al. Single-step genome-wide association study reveals candidate genes for body mass index trait in Yunong-black pigs. Anim. Genet. 2025, 56, e13501. [Google Scholar] [CrossRef]
- Tian, J.; Niu, N.; Wang, X.; Shi, L.; Yang, L.; Li, M.; Shi, L.; Liu, X.; Gao, H.; Hou, X.; et al. Integration of multiomics data identifies candidate genes influencing pH levels in Beijing Black pigs. Anim. Res. One Health 2023, 2, 260–272. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, S.; Liu, Y.; Xie, F.; Wen, H.; Zhao, S.; Zheng, X.; Ding, Y.; Yin, Z.; Zhang, X. Comparative Analysis of the Ovary Transcriptome among Wanyue Black and Yorkshire Gilts Using RNA-Seq. Vet. Sci. 2024, 11, 115. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Wang, Q.; Zhang, Z.; Ye, X.; Gu, J.; Sun, J.; Cao, C.; Xiao, Q.; Chen, Q.; Xu, Z.; Wang, K.; et al. An updated Pig Haplotype Reference Panel (PHARP 4.0) comprising 13,298 haplotypes. Commun. Biol. 2025, 8, 1625. [Google Scholar] [CrossRef]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation, Contributed Packages; CRAN: Vienna, Austria, 2014. [CrossRef]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Teng, J.; Lin, Q.; Bai, Z.; Liu, S.; Guan, D.; Li, B.; Gao, Y.; Hou, Y.; Gong, M.; et al. The Farm Animal Genotype-Tissue Expression (FarmGTEx) Project. Nat. Genet. 2025, 57, 786–796. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Rui, Q.; Wang, J.; Li, Y.; Tan, X.; Bao, Y. Arabidopsis COG6 is essential for pollen tube growth and Golgi structure maintenance. Biochem. Biophys. Res. Commun. 2020, 528, 447–452. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, L.; Tan, L.; Luo, S.; Huang, Y.; Peng, H.; Cao, J.; He, X. Genetic analysis and prenatal diagnosis in a Chinese with growth retardation, abnormal liver function, and microcephaly. Mol. Genet. Genom. Med. 2021, 9, e1751. [Google Scholar] [CrossRef]
- Komlosi, K.; Gläser, S.; Kopp, J.; Hotz, A.; Alter, S.; Zimmer, A.D.; Beger, C.; Heinzel, S.; Schmidt, C.; Fischer, J. Neonatal presentation of COG6-DG with prominent skin phenotype. JIMD Rep. 2020, 55, 51–58. [Google Scholar] [CrossRef]
- Grigoriadis, D.; Sackey, E.; Riches, K.; van Zanten, M.; Brice, G.; England, R.; Mills, M.; Dobbins, S.E.; Lee, L.L. Lipoedema Consortium and Genomics England Research Consortium. Investigation of clinical characteristics and genome associations in the ‘UK Lipoedema’cohort. PLoS ONE 2022, 17, e0274867. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Wang, L.; Zhu, B.; Chen, Y.; Gao, H.; Gao, X.; Zhang, L.; Liu, G.E.; Li, J. Probe-based association analysis identifies several deletions associated with average daily gain in beef cattle. BMC Genom. 2019, 20, 31. [Google Scholar] [CrossRef]
- Yang, L.; Yang, T.; Wang, H.; Dou, T.; Fang, X.; Shi, L.; Li, X.; Feng, M. DNMBP-AS1 Regulates NHLRC3 Expression by Sponging miR-93-5p/17-5p to Inhibit Colon Cancer Progression. Front. Oncol. 2022, 12, 765163. [Google Scholar] [CrossRef]
- Sallam, A.M.; Abou-Souliman, I.; Reyer, H.; Wimmers, K.; Rabee, A.E. New insights into the genetic predisposition of brucellosis and its effect on the gut and vaginal microbiota in goats. Sci. Rep. 2023, 13, 20086. [Google Scholar] [CrossRef]
- Frank, C.G.; Aebi, M. ALG9 mannosyltransferase is involved in two different steps of lipid-linked oligosaccharide biosynthesis. Glycobiology 2005, 15, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Breitling, J.; Aebi, M. N-Linked Protein Glycosylation in the Endoplasmic Reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013359. [Google Scholar] [CrossRef] [PubMed]
- Tham, E.; Eklund, E.A.; Hammarsjö, A.; Bengtson, P.; Geiberger, S.; Lagerstedt-Robinson, K.; Malmgren, H.; Nilsson, D.; Grigelionis, G.; Conner, P.; et al. A novel phenotype in N-glycosylation disorders: Gillessen-Kaesbach-Nishimura skeletal dysplasia due to pathogenic variants in ALG9. Eur. J. Hum. Genet. 2016, 24, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Neupert, W.; Tzagoloff, A. The Metalloprotease Encoded by ATP23 Has a Dual Function in Processing and Assembly of Subunit 6 of Mitochondrial ATPase. Mol. Biol. Cell 2006, 18, 617–626. [Google Scholar] [CrossRef]
- Osman, C.; Wilmes, C.; Tatsuta, T.; Langer, T. Prohibitins Interact Genetically with Atp23, a Novel Processing Peptidase and Chaperone for the F1FO-ATP Synthase. Mol. Biol. Cell 2007, 18, 627–635. [Google Scholar] [CrossRef]
- Zhou, Z.; Menzel, F.; Benninghoff, T.; Chadt, A.; Du, C.; Holman, G.D.; Al-Hasani, H. Rab28 is a TBC1D1/TBC1D4 substrate involved in GLUT4 trafficking. Febs Lett. 2017, 591, 88–96. [Google Scholar] [CrossRef]
- Akella, J.S.; Carter, S.P.; Rizvi, F.; Nguyen, K.C.; Tsiropoulou, S.; Moran, A.L.; Silva, M.; Kennedy, B.N.; Hall, D.H.; Barr, M.M.; et al. A ciliary BBSome-ARL-6-PDE6D pathway trafficks RAB-28, a negative regulator of extracellular vesicle biogenesis. bioRxiv 2019, 715730. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, X.; Zhao, L.; Wang, Y.; Ye, C.; Zou, X.; Dai, A.; Cong, Z.; Chen, J.; Zhou, Q.; et al. Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2. Proc. Natl. Acad. Sci. USA 2021, 118, e2101279118. [Google Scholar] [CrossRef]
- Reyes, M.; Firat, D.; Hanna, P.; Khan, M.; Bruce, M.; Shvedova, M.; Kobayashi, T.; Schipani, E.; Gardella, T.J.; Jüppner, H. Substantially Delayed Maturation of Growth Plate Chondrocytes in “Humanized” PTH1R Mice with the H223R Mutation of Jansen’s Disease. JBMR PLUS 2023, 7, e10802. [Google Scholar] [CrossRef]
- Dobolyi, A.; Dimitrov, E.; Palkovits, M.; Usdin, T.B. The neuroendocrine functions of the parathyroid hormone 2 receptor. Front. Endocrinol. 2012, 3, 121. [Google Scholar] [CrossRef]
- Panda, D.K.; Goltzman, D.; Karaplis, A.C. Defective postnatal endochondral bone development by chondrocyte-specific targeted expression of parathyroid hormone type 2 receptor. Am. J. Physiol. Metab. 2012, 303, E1489–E1501. [Google Scholar] [CrossRef]
| Trait | N | Max | Min | Mean (±SD) | CV |
|---|---|---|---|---|---|
| BL | 139 | 89.62 | 65.38 | 70.48 ± 9.51 | 13.49% |
| CC | 139 | 87.27 | 62.52 | 68.57 ± 8.08 | 11.78% |
| SC | 139 | 16.14 | 12.65 | 14.24 ± 2.03 | 14.25% |
| LHC | 139 | 68.54 | 45.82 | 58.60 ± 4.96 | 8.46% |
| Trait | BL | CC | SC | LHC |
|---|---|---|---|---|
| BL | 1 | - | - | - |
| CC | 0.956 ** | 1 | - | - |
| SC | 0.914 ** | 0.935 ** | 1 | - |
| LHC | 0.898 ** | 0.893 ** | 0.873 ** | 1 |
| Chromosome | SNPID | Position | p_wald | Gene |
|---|---|---|---|---|
| 5 | rs343622325 | 23613371 | 1.528592 × 10−7 | ATP23 |
| 8 | rs345729102 | 9521800 | 2.020554 × 10−7 | RAB28 |
| 9 | rs343276492 | 39557329 | 1.111061 × 10−7 | ALG9 |
| 11 | rs321308815 | 14774102 | 2.042810 × 10−7 | LHFPL6, GOC6, NHLRC3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Ye, H.; Li, W.; Tian, F.; Wang, Q.; Ma, Z.; Guan, J.; Ding, Y.; Zheng, X.; Yin, Z.; Zhang, X. Genome-Wide Association Study Identifies Candidate Genes for Body Size Traits in Wanyue Black Pigs. Animals 2026, 16, 117. https://doi.org/10.3390/ani16010117
Ye H, Li W, Tian F, Wang Q, Ma Z, Guan J, Ding Y, Zheng X, Yin Z, Zhang X. Genome-Wide Association Study Identifies Candidate Genes for Body Size Traits in Wanyue Black Pigs. Animals. 2026; 16(1):117. https://doi.org/10.3390/ani16010117
Chicago/Turabian StyleYe, Haibo, Wei Li, Fang Tian, Qianqian Wang, Zhonghua Ma, Jinyu Guan, Yueyun Ding, Xianrui Zheng, Zongjun Yin, and Xiaodong Zhang. 2026. "Genome-Wide Association Study Identifies Candidate Genes for Body Size Traits in Wanyue Black Pigs" Animals 16, no. 1: 117. https://doi.org/10.3390/ani16010117
APA StyleYe, H., Li, W., Tian, F., Wang, Q., Ma, Z., Guan, J., Ding, Y., Zheng, X., Yin, Z., & Zhang, X. (2026). Genome-Wide Association Study Identifies Candidate Genes for Body Size Traits in Wanyue Black Pigs. Animals, 16(1), 117. https://doi.org/10.3390/ani16010117






