Effects of Short-Term Feeding with Diets Containing Insect Meal on the Gut Microbiota of African Catfish Hybrids
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Feeding and Rearing Conditions
2.2. Intestinal Sampling
2.3. DNA Purification and Amplicon Sequencing
2.4. Analyses of the Intestinal Microbiota Composition
2.5. Culturing and Characterization of Bacterial Isolates from BSL Feed
2.6. Whole Genome Sequencing of the Isolates Cultured from BSL Feed
2.7. Shotgun Metagenomic Sequencing and Assembly
2.8. Functional Annotation of Shotgun Metagenomic and WGS Data
2.9. Statistical Data Analyses
3. Results
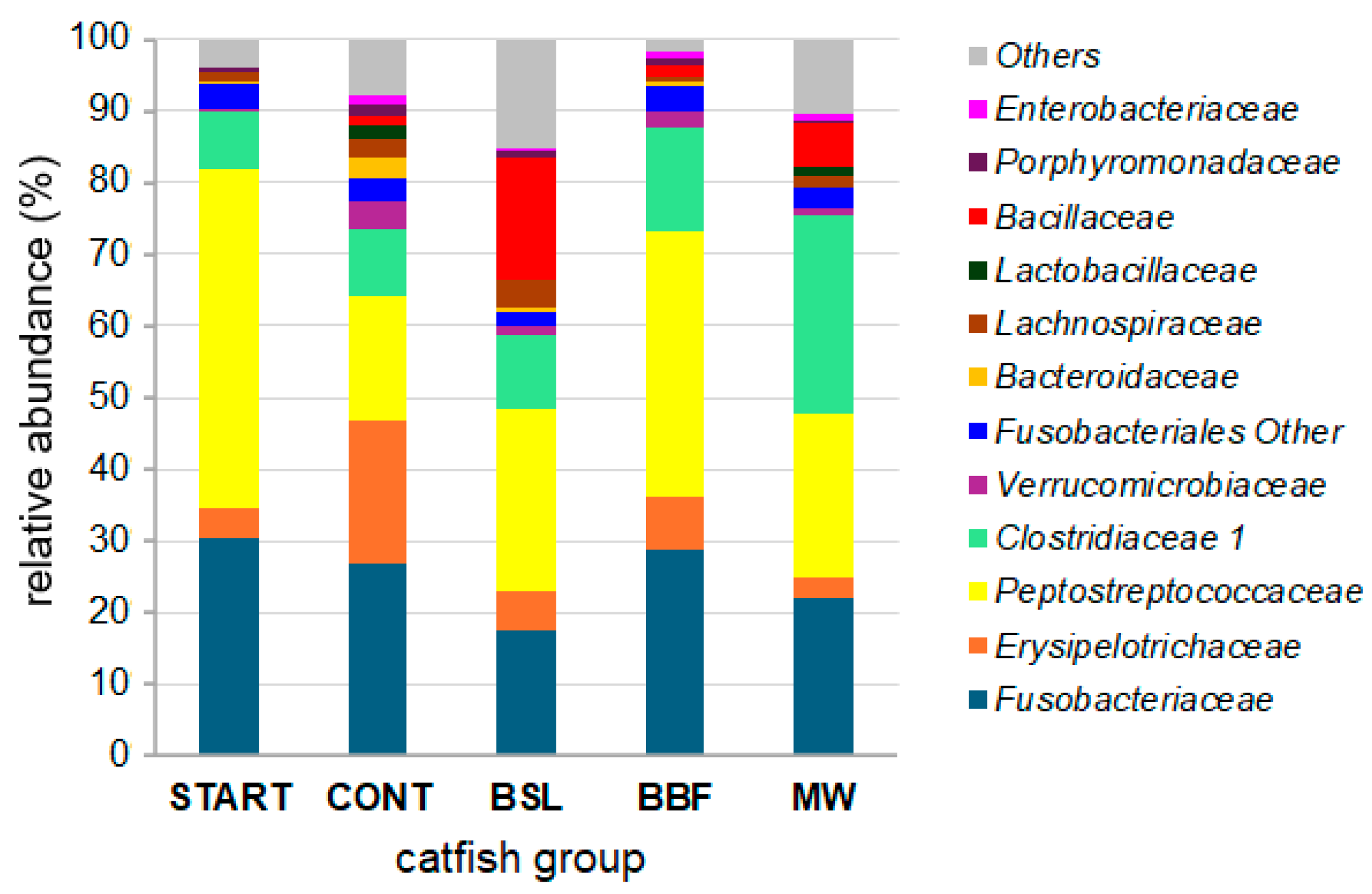
3.1. Bacterial Composition of the African Catfish Intestinal Microbiota
3.2. Changes in the Intestinal Microbiota Between Groups START and CONT
3.3. Effects of Short-Term Feeding with Diets Containing Insect Meal on the Intestinal Microbiota
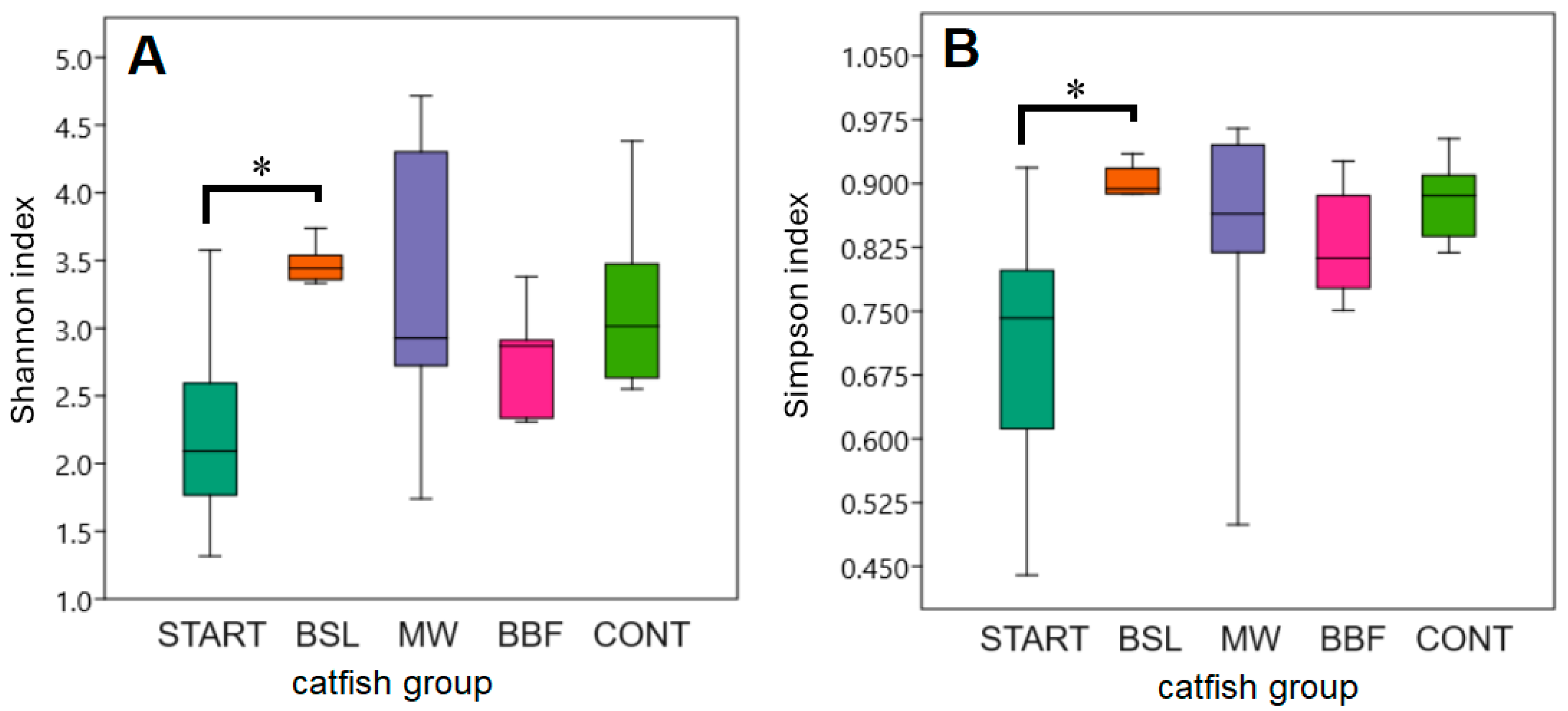
3.4. α-Diversity Analyses of the Intestinal Microbiota
3.5. β-Diversity Analyses of the Intestinal Microbiota
3.6. Identification of Bacterial Strains Cultured from BSL Feed
3.7. Screening for Acquired Antibiotic-Resistance and Chitinase Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Huis, A.; Oonincx, D.G.A.B. The environmental sustainability of insects as food and feed. A review. Agron. Sustain. Dev. 2017, 37, 43. [Google Scholar] [CrossRef]
- Sun-Waterhouse, D.; Waterhouse, G.I.N.; You, L.; Zhang, J.; Liu, Y.; Ma, L.; Gao, J.; Dong, Y. Transforming insect biomass into consumer wellness foods: A review. Food Res. Int. 2016, 89, 129–151. [Google Scholar] [CrossRef] [PubMed]
- Gasco, L.; Biancarosa, I.; Liland, N.S. From waste to feed: A review of recent knowledge on insects as producers of protein and fat for animal feeds. Curr. Opin. Green Sustain. Chem. 2020, 23, 67–79. [Google Scholar] [CrossRef]
- Sándor, Z.J.; Banjac, V.; Vidosavljević, S.; Káldy, J.; Egessa, R.; Lengyel-Kónya, É.; Tömösközi-Farkas, R.; Zalán, Z.; Adányi, N.; Libisch, B.; et al. Apparent digestibility coefficients of black soldier fly (Hermetia illucens), yellow mealworm (Tenebrio molitor), and blue bottle fly (Calliphora vicina) insects for juvenile African catfish hybrids (Clarias gariepinus × Heterobranchus longifilis). Aquac. Nutr. 2022, 2022, 4717014. [Google Scholar] [CrossRef] [PubMed]
- Dobermann, D.; Swift, J.A.; Field, L.M. Opportunities and hurdles of edible insects for food and feed. Nutr. Bull. 2017, 42, 293–308. [Google Scholar] [CrossRef]
- Charlton, A.J.; Dickinson, M.; Wakefield, M.E.; Fitches, E.; Kenis, M.; Han, R.; Zhu, F.; Kone, N.; Grant, M.; Devic, E.; et al. Exploring the chemical safety of fly larvae as a source of protein for animal feed. J. Insects Food Feed. 2015, 1, 7–16. [Google Scholar] [CrossRef]
- Catarino, M.M.R.S.; Fernandes, S.C.G.; de Freitas Ferreira, S.M. Feed for Rearing Omnivorous Fish. European Patent Application 17752492.3 WO2018/020395, 5 June 2019. Available online: https://patentimages.storage.googleapis.com/e1/d2/dc/e5127bb24050ba/EP3491932A1.pdf (accessed on 24 April 2025).
- Adah, P.M.; Onyia, L.U.; Obande, R.A. Fish hybridization in some catfishes: A review. Biotechnology 2014, 13, 248–251. [Google Scholar] [CrossRef]
- Keremah, R.I.; Deekae, S.N.; Akalokwu, U.A. Growth and feed utilization of catfish hybrid (Heterobranchus longifilis × Clarias gariepinus) fingerlings fed practical diets. Greener J. Agric. Sci. 2013, 3, 286–290. [Google Scholar] [CrossRef]
- Ózsvári, L.; Máté, M. Az akvakultúra-ágazat globális, európai és magyarországi fejlődése. Gazdálkodás 2021, 65, 289–309. [Google Scholar] [CrossRef]
- Federation of European Aquaculture Producers (FEAP) European Aquaculture Production Report 2014–2020 (2021). Available online: https://feap.info/wp-content/uploads/2020/10/20201007_feap-production-report-2020.pdf (accessed on 1 September 2024).
- European Market Observatory for Fisheries and Aquaculture (EUMOFA). Freshwater Aquaculture in the EU; Publications Office of the European Union: Luxembourg, 2021; ISBN 978-92-76-38492-2. [Google Scholar]
- Dadebo, E.; Aemro, D.; Tekle-Giorgis, Y. Food and feeding habits of the African catfish Clarias gariepinus (Burchell, 1822) (Pisces: Clariidae) in Lake Koka, Ethiopia. Afr. J. Ecol. 2014, 52, 471–478. [Google Scholar] [CrossRef]
- Dessie, A.; Mingist, M.; Mequanent, D.; Aemro, D. Food and feeding habits of the African catfish Clarias gariepinus (Burchell, 1822) (Pisces: Clariidae) in the newly built Ribb Reservoir, north-west Ethiopia. Indian J. Fish. 2024, 71, 37–42. [Google Scholar]
- Tesfahun, A. Feeding biology of the African catfish Clarias gariepinus (Burchell) in some of Ethiopian lakes: A review. Int. J. Fauna Biol. Stud. 2018, 5, 19–23. [Google Scholar]
- Effiong, B.N.; Mohammed, I. Effect of seasonal variation on the nutrient composition in selected fish species in Lake Kainji-Nigeria. Nat. Sci. 2008, 6, 1–5. [Google Scholar]
- Ballesteros, T.M.; Torres-Mejia, M.; Ramírez-Pinilla, M.P. How does diet influence the reproductive seasonality of tropical freshwater fish? A case study of a characin in a tropical mountain river. Neotrop. Ichthyol. 2009, 7, 693–700. [Google Scholar] [CrossRef]
- Kokou, F.; Gupta, S.; Kumar, V. Editorial: Understanding the interplay between diet, feed ingredients and gut microbiota for sustainable aquaculture. Front. Mar. Sci. 2022, 9, 853548. [Google Scholar] [CrossRef]
- Maulu, S.; Hasimuna, O.J.; Haambiya, L.H.; Monde, C.; Musuka, C.G.; Makorwa, T.H.; Munganga, B.P.; Phiri, K.J.; Nsekanabo, J.D. Climate change effects on aquaculture production: Sustainability implications, mitigation, and adaptations. Front. Sustain. Food Syst. 2021, 5, 609097. [Google Scholar] [CrossRef]
- Kamalam, B.S.; Pandey, P.K. Nutrition and environment interactions in aquaculture. In Outlook of Climate Change and Fish Nutrition; Springer Nature: Singapore, 2023; pp. 407–422. [Google Scholar]
- Mente, E.; Nikouli, E.; Antonopoulou, E.; Martin, S.A.M.; Kormas, K.A. Core versus diet-associated and postprandial bacterial communities of the rainbow trout (Oncorhynchus mykiss) midgut and faeces. Biol. Open 2018, 7, bio034397. [Google Scholar]
- Mes, W.; Lücker, S.; Jetten, M.S.M.; Siepel, H.; Gorissen, M.; van Kessel, M.A.H.J. Feeding strategy and feed protein level affect the gut microbiota of common carp (Cyprinus carpio). Environ. Microbiol. Rep. 2024, 16, e13262. [Google Scholar] [CrossRef]
- Parris, D.J.; Morgan, M.M.; Stewart, F.J. Feeding rapidly alters microbiome composition and gene transcription in the clownfish gut. Appl. Environ. Microbiol. 2019, 85, e02479-18. [Google Scholar] [CrossRef]
- Reid, C.E.; Bissett, A.; Huynh, C.; Bowman, J.P.; Taylor, R.S. Time from feeding impacts farmed Atlantic salmon (Salmo salar) gut microbiota and faecal score. Aquaculture 2024, 579, 740174. [Google Scholar] [CrossRef]
- Ji, B.W.; Sheth, R.U.; Dixit, P.D.; Tchourine, K.; Vitkup, D. Macroecological dynamics of gut microbiota. Nat. Microbiol. 2020, 5, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Bledsoe, J.W.; Waldbieser, G.C.; Swanson, K.S.; Peterson, B.C.; Small, B.C. Comparison of channel catfish and blue catfish gut microbiota assemblages shows minimal effects of host genetics on microbial structure and inferred function. Front. Microbiol. 2018, 9, 1073. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef] [PubMed]
- Gericke, S.J.; Salie, K.; de Wet, L.; Goosen, N.J. Effects of dietary supplementation of endo-(1,4)-β-xylanase in plant-based diets on growth performance, hindgut microbial diversity, and blood chemistry in large on-growing African catfish (Clarias gariepinus). J. Appl. Aquac. 2023, 35, 561–584. [Google Scholar] [CrossRef]
- Matuk, K.; Gulyás, T. New possibilities in fish anaesthesia process [A halak altatásának újabb lehetőségei]. Halászat 1987, 33, 11–13. (In Hungarian) [Google Scholar]
- Libisch, B.; Keresztény, T.; Kerényi, Z.; Kocsis, R.; Sipos, R.; Papp, P.P.; Olasz, F. Metagenomic analysis of acquired antibiotic resistance determinants in the gut microbiota of wild boars (Sus scrofa)–Preliminary Results. J. Vet. Res. 2020, 64, 111–118. [Google Scholar] [CrossRef]
- Libisch, B.; Abdulkadir, S.; Keresztény, T.; Papp, P.P.; Olasz, F.; Fébel, H.; Sándor, Z.J.; Rasschaert, G.; Lambrecht, E.; Heyndrickx, M.; et al. Detection of acquired antibiotic resistance genes in domestic pig (Sus scrofa) and common carp (Cyprinus carpio) intestinal samples by metagenomics analyses in Hungary. Antibiotics 2022, 11, 1441. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020, 48, W395–W402. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina paired-end read merger. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Weber, N.; Liou, D.; Dommer, J.; MacMenamin, P.; Quiñones, M.; Misner, I.; Oler, A.J.; Wan, J.; Kim, L.; Coakley McCarthy, M.; et al. Nephele: A cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 2018, 34, 1411–1413. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y.; et al. QIIME2 enables comprehensive end-to-end analysis of diverse microbiome data and comparative studies with publicly available data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Sun, J. Hypothesis testing and statistical analysis of microbiome. Genes Dis. 2017, 4, 138–148. [Google Scholar] [CrossRef]
- Zeebone, Y.Y.; Bóta, B.; Halas, V.; Libisch, B.; Olasz, F.; Papp, P.; Keresztény, T.; Gerőcs, A.; Ali, O.; Kovács, M.; et al. Gut-faecal microbial and health-marker response to dietary fumonisins in weaned pigs. Toxins 2023, 15, 328. [Google Scholar] [CrossRef]
- Klinsoda, J.; Vötterl, J.; Zebeli, Q.; Metzler-Zebeli, B.U. Alterations of the viable ileal microbiota of the gut mucosa-lymph node axis in pigs fed phytase and lactic acid-treated cereals. Appl. Environ Microbiol. 2020, 86, e02128-19. [Google Scholar] [CrossRef]
- Ruczizka, U.; Metzler-Zebeli, B.; Unterweger, C.; Mann, E.; Schwarz, L.; Knecht, C.; Hennig-Pauka, I. Early parenteral administration of ceftiofur has gender-specific short- and long-term effects on the fecal microbiota and growth in pigs from the suckling to growing phase. Animals 2019, 10, 17. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Libisch, B.; Picot, C.; Ceballos-Garzon, A.; Moravkova, M.; Klimesová, M.; Telkes, G.; Chuang, S.T.; Le Pape, P. Prototheca infections and ecology from a One Health perspective. Microorganisms 2022, 10, 938. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T.; Gladman, S. Fasta Statistics: Display Summary Statistics for a Fasta File. 2012. Available online: https://github.com/galaxyproject/tools-iuc (accessed on 1 September 2024).
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antibiotic Resistance Genes. 2016. Available online: https://github.com/tseemann/abricate (accessed on 1 September 2024).
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Ahsan, N.; Shimizu, M. Lysinibacillus species: Their potential as effective bioremediation, biostimulant, and biocontrol agents. Rev. Agric. Sci. 2021, 9, 103–116. [Google Scholar] [CrossRef]
- Hasan, I.; Gai, F.; Cirrincione, S.; Rimoldi, S.; Saroglia, G.; Terova, G. Chitinase and insect meal in aquaculture nutrition: A comprehensive overview of the latest achievements. Fishes 2023, 8, 607. [Google Scholar] [CrossRef]
- O’Brien, M.A.R.K.; Colwell, R.R. A rapid test for chitinase activity that uses 4-methylumbelliferyl-N-acetyl-beta-D-glucosaminide. Appl. Environ. Microbiol. 1987, 53, 1718–1720. [Google Scholar] [CrossRef]
- Swiontek Brzezinska, M.; Jankiewicz, U.; Burkowska, A.; Walczak, M. Chitinolytic microorganisms and their possible application in environmental protection. Curr. Microbiol. 2014, 68, 71–81. [Google Scholar] [CrossRef]
- Sehnal, L.; Brammer-Robbins, E.; Wormington, A.M.; Blaha, L.; Bisesi, J.; Larkin, I.; Martyniuk, C.J.; Simonin, M.; Adamovsky, O. Microbiome composition and function in aquatic vertebrates: Small organisms making big impacts on aquatic animal health. Front. Microbiol. 2021, 12, 567408. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Kumar, P.; Sivachandran, P.; Marimuthu, K.; Arshad, A.; Paray, B.A.; ArockiaRaj, J. Gut microbiota metagenomics in aquaculture: Factors influencing gut microbiome and its physiological role in fish. Rev. Aquac. 2020, 12, 1903–1927. [Google Scholar] [CrossRef]
- Rosenau, S.; Oertel, E.; Mott, A.C.; Tetens, J. The Effect of a Total Fishmeal Replacement by Arthrospira platensis on the microbiome of African catfish (Clarias gariepinus). Life 2021, 11, 558. [Google Scholar] [CrossRef]
- Kawasaki, K.; Kawasaki, T.; Hirayasu, H.; Matsumoto, Y.; Fujitani, Y. Evaluation of fertilizer value of residues obtained after processing household organic waste with black soldier fly larvae (Hermetia illucens). Sustainability 2020, 12, 4920. [Google Scholar] [CrossRef]
- Mazza, L.; Xiao, X.; Ur Rehman, K.; Cai, M.; Zhang, D.; Fasulo, S.; Tomberlin, J.K.; Zheng, L.; Soomro, A.A.; Yu, Z.; et al. Management of chicken manure using black soldier fly (Diptera: Stratiomyidae) larvae assisted by companion bacteria. Waste Manag. 2020, 102, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Villazana, J. Black Soldier Fly Larvae as Value-Added Feed for Aquaculture in Maine. Master’s Thesis, University of Maine, Orono, ME, USA, 2018. [Google Scholar]
- Li, Y.; Gajardo, K.; Jaramillo-Torres, A.; Kortner, T.M.; Krogdahl, Å. Consistent changes in the intestinal microbiota of Atlantic salmon fed insect meal diets. Anim. Microbiome 2022, 4, 8. [Google Scholar] [CrossRef]
- Cody, R.M. Distribution of chitinase and chitobiase in Bacillus. Curr. Microbiol. 1989, 19, 201–205. [Google Scholar] [CrossRef]
- Askarian, F.; Zhou, Z.; Olsen, R.E.; Sperstad, S.; Ringø, E. Culturable autochthonous gut bacteria in Atlantic salmon (Salmo salar L.) fed diets with or without chitin. Characterization by 16S rRNA gene sequencing, ability to produce enzymes and in vitro growth inhibition of four fish pathogens. Aquaculture 2012, 326–329, 1–8. [Google Scholar] [CrossRef]
- Li, Y.; Bruni, L.; Jaramillo-Torres, A.; Gajardo, K.; Kortner, T.M.; Krogdahl, Å. Differential response of digesta- and mucosa-associated intestinal microbiota to dietary insect meal during the seawater phase of Atlantic salmon. Anim. Microbiome 2021, 3, 8. [Google Scholar] [CrossRef]
- Huyben, D.; Vidaković, A.; Werner Hallgren, S.W.; Langeland, M. High-throughput sequencing of gut microbiota in rainbow trout (Oncorhynchus mykiss) fed larval and pre-pupae stages of black soldier fly (Hermetia illucens). Aquaculture 2019, 500, 485–491. [Google Scholar] [CrossRef]
- Rimoldi, S.; Antonini, M.; Gasco, L.; Moroni, F.; Terova, G. Intestinal microbial communities of rainbow trout (Oncorhynchus mykiss) may be improved by feeding a Hermetia illucens meal/low-fishmeal diet. Fish Physiol. Biochem. 2021, 47, 365–380. [Google Scholar] [CrossRef]
- Terova, G.; Rimoldi, S.; Ascione, C.; Gini, E.; Ceccotti, C.; Gasco, L. Rainbow trout (Oncorhynchus mykiss) gut microbiota is modulated by insect meal from Hermetia illucens prepupae in the diet. Rev. Fish Biol. Fish. 2019, 29, 465–486. [Google Scholar] [CrossRef]
- Xavier, R.; Severino, R.; Silva, S.M. Signatures of dysbiosis in fish microbiomes in the context of aquaculture. Rev. Aquac. 2024, 16, 706–731. [Google Scholar] [CrossRef]
- Fan, K.; Liu, H.; Pei, Z.; Brown, P.B.; Huang, Y. A study of the potential effect of dietary fishmeal replacement with cricket meal (Gryllus bimaculatus) on growth performance, blood health, liver antioxidant activities, intestinal microbiota and immune-related gene expression of juvenile channel catfish. Anim. Feed Sci. Technol. 2023, 295, 115542. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D.; Refaey, M.M.; Xu, W.; Tang, R.; Li, L. Host Age Affects the Development of Southern Catfish Gut Bacterial Community Divergent from That in the Food and Rearing Water. Front. Microbiol. 2018, 9, 495. [Google Scholar] [CrossRef] [PubMed]
- Skvortsova, E.; Filinskaya, O.; Postrash, I.; Bushkareva, A.; Mostofina, A. Biodiversity of gut microorganisms in aquacultured African catfish. E3S Web Conf. 2023, 463, 01039. [Google Scholar] [CrossRef]
- Ingerslev, H.C.; von Gersdorff Jørgensen, L.; Lenz Strube, M.; Larsen, N.; Dalsgaard, I.; Boye, M.; Madsen, L. The development of the gut microbiota in rainbow trout (Oncorhynchus mykiss) is affected by first feeding and diet type. Aquaculture 2014, 424–425, 24–34. [Google Scholar] [CrossRef]
- Bakke, I.; Coward, E.; Andersen, T.; Vadstein, O. Selection in the host structures the microbiota associated with developing cod larvae (Gadus morhua). Environ. Microbiol. 2015, 17, 3914–3924. [Google Scholar] [CrossRef]
- Bledsoe, J.W.; Peterson, B.C.; Swanson, K.S.; Small, B.C. Ontogenetic characterization of the intestinal microbiota of channel catfish through 16S rRNA gene sequencing reveals insights on temporal shifts and the influence of environmental microbes. PLoS ONE 2016, 11, e0166379. [Google Scholar] [CrossRef]
- Smith, C.C.R.; Snowberg, L.K.; Gregory Chit, J.; Knight, R.; Bolnick, D.I. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 2015, 9, 2515–2526. [Google Scholar] [CrossRef]
- Fontes, T.V.; de Oliveira, K.R.B.; Gomes Almeida, I.L.; Maria Orlando, T.M.; Rodrigues, P.B.; Costa, D.V.D.; Rosa, P.V.E. Digestibility of insect meals for Nile tilapia fingerlings. Animals 2019, 9, 181. [Google Scholar] [CrossRef]
- Nattabi, J.K. Aspects of the Digestive Physiology of Larvae of the North African Catfish, Clarias Gariepinus (Burchell 1822), During Early Development. Ph.D. Thesis, Institute of Aquaculture, University of Stirling, Stirling, UK, 2018. [Google Scholar]
- Rapatsa, M.M.; Moyo, N.A.G. Enzyme activity and histological analysis of Clarias gariepinus fed on Imbrasia belina meal used for partial replacement of fishmeal. Fish Physiol. Biochem. 2019, 45, 1309–1320. [Google Scholar] [CrossRef]
- Holen, M.M.; Sandve, S.R.; Harvey, T.N.; Jin, Y.; Angell, I.L.; Rudi, K.; Kent, M.P. The effect of dietary chitin on Atlantic salmon (Salmo salar) chitinase activity, gene expression, and microbial composition. Aquac. Fish. 2024. [Google Scholar] [CrossRef]
- Tan, H.Y.; Chen, S.W.; Hu, S.Y. Improvements in the growth performance, immunity, disease resistance, and gut microbiota by the probiotic Rummeliibacillus stabekisii in Nile tilapia (Oreochromis niloticus). Fish Shellfish. Immunol. 2019, 92, 265–275. [Google Scholar] [CrossRef]
- Chen, W.; Du, L.; Cai, C.; Huang, L.; Zheng, Q.; Chen, J.; Wang, L.; Zhang, X.; Fang, X.; Wang, L.; et al. Take chicks as an example: Rummeliibacillus stabekisii CY2 enhances immunity and regulates intestinal microbiota by degrading LPS to promote organism growth and development. J. Funct. Foods 2023, 105, 105583. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, S.H.; Noh, D.I.; Lee, Y.S.; Kim, T.R.; Hasan, M.T.; Lee, E.W.; Jang, W.J. Combination of host-associated Rummeliibacillus sp. and Microbacterium sp. positively modulated the growth, feed utilization, and intestinal microbial population of olive flounder (Paralichthys olivaceus). Biology 2023, 12, 1443. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Statement on how to interpret the QPS qualification on ‘acquired antimicrobial resistance genes’. EFSA J. 2023, 21, e08323. [Google Scholar] [PubMed]
- Algammal, A.M.; Mabrok, M.; Sivaramasamy, E.; Youssef, F.M.; Atwa, M.H.; El-Kholy, A.W.; Hetta, H.F.; Hozzein, W.N. Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci. Rep. 2020, 10, 15961. [Google Scholar] [CrossRef]
- Algammal, A.M.; Mabrok, M.; Ezzat, M.; Alfifi, K.J.; Esawy, A.M.; Elmasry, N.; El-Tarabili, R.M. Prevalence, antimicrobial resistance (AMR) pattern, virulence determinant and AMR genes of emerging multi-drug resistant Edwardsiella tarda in Nile tilapia and African catfish. Aquaculture 2022, 548, 737643. [Google Scholar] [CrossRef]
- Alotaibi, K.; Khan, A.; Nawaz, S.; Khan, S.; Nawaz, M. Characterization of β-lactam-resistant Escherichia coli Strains isolated from catfish (Ictalurus punctatus) raised without the drug. J. Bacteriol. Mycol. 2019, 6, 1115. [Google Scholar]
- Fauzi, N.N.F.N.M.; Hamdan, R.H.; Mohamed, M.; Ismail, A.; Mat Zin, A.A.M.; Mohamad, N.F.A. Prevalence, antibiotic susceptibility, and presence of drug resistance genes in Aeromonas spp. isolated from freshwater fish in Kelantan and Terengganu states, Malaysia. Vet. World 2021, 14, 2064–2072. [Google Scholar] [CrossRef]
- Kampouris, I.D.; Klümper, U.; Kramer, L.; Sorum, H.; Wedekind, H.; Berendonk, T.U. Dissemination of antibiotic resistance in antibiotic-free recirculating aquaculture systems. J. Hazard. Mater. Adv. 2022, 8, 100201. [Google Scholar] [CrossRef]
- Tekedar, H.C.; Arick, M.A.; Hsu, C.Y.; Thrash, A.; Blom, J.; Lawrence, M.L.; Abdelhamed, H. Identification of antimicrobial resistance determinants in Aeromonas veronii strain MS-17-88 recovered from channel catfish (Ictalurus punctatus). Front. Cell Infect. Microbiol. 2020, 10, 348. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Sung, K.; Khan, S.A.; Khan, A.A.; Steele, R. Biochemical and molecular characterization of tetracycline-resistant Aeromonas veronii isolates from catfish. Appl. Environ. Microbiol. 2006, 72, 6461–6466. [Google Scholar] [CrossRef]
- Nawaz, M.; Khan, A.A.; Khan, S.; Sung, K.; Steele, R. Isolation and characterization of tetracycline-resistant Citrobacter spp. from catfish. Food Microbiol. 2008, 25, 85–91. [Google Scholar] [CrossRef]
- Nawaz, M.; Khan, A.A.; Khan, S.; Sung, K.; Kerdahi, K.; Steele, R. Molecular characterization of tetracycline-resistant genes and integrons from avirulent strains of Escherichia coli isolated from catfish. Foodborne Pathog. Dis. 2009, 6, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Soku, Y.K.; Mohamed, A.; Samuel, T.; Dessai, U.; Walls, I.; Rockwell, C.; Fortenberry, G.; Berutti, T.; Nieves-Miranda, S.; Nawrocki, E.M.; et al. A comparative study on antimicrobial resistance in Escherichia coli isolated from channel catfish and related freshwater fish species. J. Food Prot. 2024, 87, 100192. [Google Scholar] [CrossRef]
- Zhou, A.; Tang, H.; Zhang, L.; Junaid, M.; Xie, S.; Zhang, Y.; Li, X.; Xu, G.; Zou, J. Dynamics of bacterial community and antibiotic resistance genes in the aquaculture ponds of channel catfish (Ictalurus punctaus): An association with the mobile genetic elements and environmental factors. Aquaculture 2023, 562, 738726. [Google Scholar] [CrossRef]
- Libisch, B.; Giske, C.G.; Kovács, B.; Tóth, T.G.; Füzi, M. Identification of the first VIM metallo-β-lactamase-producing multiresistant Aeromonas hydrophila strain. J. Clin. Microbiol. 2008, 46, 1878–1880. [Google Scholar] [CrossRef]
- Libisch, B.; Watine, J.; Balogh, B.; Gacs, M.; Muzslay, M.; Szabó, G.; Füzi, M. Molecular typing indicates an important role for two international clonal complexes in dissemination of VIM-producing Pseudomonas aeruginosa clinical isolates in Hungary. Res. Microbiol. 2008, 159, 162–168. [Google Scholar] [CrossRef]
- Libisch, B. Molecular typing methods for the genus Pseudomonas. In Molecular Typing in Bacterial Infections; Humana Press: Totowa, NJ, USA, 2013; pp. 407–429. [Google Scholar] [CrossRef]
- Szabó, O.; Gulyás, D.; Szabó, N.; Kristóf, K.; Kocsis, B.; Szabó, D. Plasmid-mediated quinolone resistance determinants in Enterobacteriaceae from urine clinical samples. Acta. Microbiol. Immunol. Hung. 2018, 65, 255–265. [Google Scholar] [CrossRef]
- de Jong, A.; Muggeo, A.; El Garch, F.; Moyaert, H.; de Champs, C.; Guillard, T. Characterization of quinolone resistance mechanisms in Enterobacteriaceae isolated from companion animals in Europe (ComPath II study). Vet. Microbiol. 2018, 216, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Colorado Gómez, M.A.; Melo-Bolívar, J.F.; Ruíz Pardo, R.Y.; Rodriguez, J.A.; Villamil, L.M. Unveiling the probiotic potential of the anaerobic bacterium Cetobacterium sp. nov. C33 for enhancing Nile tilapia (Oreochromis niloticus) cultures. Microorganisms 2023, 11, 2922. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; Van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Wang, X.; Zhou, X.; Jiang, S.; Li, Y.; Ahmad, O.; Qi, L.; Li, P.; Li, J. Taxonomic distribution of FosB in human-microbiota and activity comparison of fosfomycin resistance. Front. Microbiol. 2019, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Adeoye, A.A.; Akegbejo-Samsons, Y.; Fawole, F.J.; Davies, S.J. Preliminary assessment of black soldier fly (Hermetia illucens) larval meal in the diet of African catfish (Clarias gariepinus): Impact on growth, body index, and hematological parameters. J. World Aquac. Soc. 2020, 51, 1024–1033. [Google Scholar] [CrossRef]
- Bartucz, T.; Csókás, E.; Nagy, B.; Gyurcsák, M.P.; Bokor, Z.; Bernáth, G.; Molnár, J.; Urbányi, B.; Csorbai, B. Black soldier fly (Hermetia illucens) Meal as Direct Replacement of Complex Fish Feed for Rainbow Trout (Oncorhynchus mykiss) and African Catfish (Clarias gariepinus). Life 2023, 13, 1978. [Google Scholar] [CrossRef]





| Group 1 | Group 2 | Permutations | Pseudo-F | p-Value | q-Value |
|---|---|---|---|---|---|
| START | CONT | 999 | 3.51 | 0.001 | 0.01 |
| BSL | CONT | 999 | 3.91 | 0.002 | 0.01 |
| BSL | START | 999 | 3.84 | 0.006 | 0.02 |
| IM | CONT | 999 | 2.70 | 0.013 | 0.019 |
| Order | Family | Genus | Relative Abundance (%) a | Number of Isolates |
|---|---|---|---|---|
| Bacillales | Bacillaceae | Bacillus | 15.8 | 4 |
| Bacillales | Bacillaceae | Lysinibacillus | 2.8 | 3 |
| Bacillales | Planococcaceae | Rummeliibacillus | 0.073 | 2 |
| Micrococcales | Micrococcaceae | Glutamicibacter | 0.0036 | 1 |
| Acquired ARGs Detected with the Coverage (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibiotic Class | AMN | AMN | TRI | FOS | MAC | QNL | SUL | TET | TET |
| Fish Group/ARG | aadA9 | aph(3′)-Ia | dfrG | fosB | lnu(C) | qnrD1 | sul1 | tetA(P) | tetB(P) |
| BSL | 74.5% | 60.6% | 48.2% | 100% | 52.2% | 63.5% | 100% | 99.3% | |
| CONT | 100% | 100% | 100% | 99.3% | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Libisch, B.; Sándor, Z.J.; Keresztény, T.; Ozoaduche, C.L.; Papp, P.P.; Posta, K.; Biró, J.; Stojkov, V.; Banjac, V.; Adányi, N.; et al. Effects of Short-Term Feeding with Diets Containing Insect Meal on the Gut Microbiota of African Catfish Hybrids. Animals 2025, 15, 1338. https://doi.org/10.3390/ani15091338
Libisch B, Sándor ZJ, Keresztény T, Ozoaduche CL, Papp PP, Posta K, Biró J, Stojkov V, Banjac V, Adányi N, et al. Effects of Short-Term Feeding with Diets Containing Insect Meal on the Gut Microbiota of African Catfish Hybrids. Animals. 2025; 15(9):1338. https://doi.org/10.3390/ani15091338
Chicago/Turabian StyleLibisch, Balázs, Zsuzsanna J. Sándor, Tibor Keresztény, Chioma Lilian Ozoaduche, Péter P. Papp, Katalin Posta, Janka Biró, Viktor Stojkov, Vojislav Banjac, Nóra Adányi, and et al. 2025. "Effects of Short-Term Feeding with Diets Containing Insect Meal on the Gut Microbiota of African Catfish Hybrids" Animals 15, no. 9: 1338. https://doi.org/10.3390/ani15091338
APA StyleLibisch, B., Sándor, Z. J., Keresztény, T., Ozoaduche, C. L., Papp, P. P., Posta, K., Biró, J., Stojkov, V., Banjac, V., Adányi, N., Berki, M., Lengyel-Kónya, É., Tömösközi-Farkas, R., & Olasz, F. (2025). Effects of Short-Term Feeding with Diets Containing Insect Meal on the Gut Microbiota of African Catfish Hybrids. Animals, 15(9), 1338. https://doi.org/10.3390/ani15091338







