Heat Acclimation Enhances Brain Resilience to Acute Thermal Stress in Clarias fuscus by Modulating Cell Adhesion, Anti-Apoptotic Pathways, and Intracellular Degradation Mechanisms
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
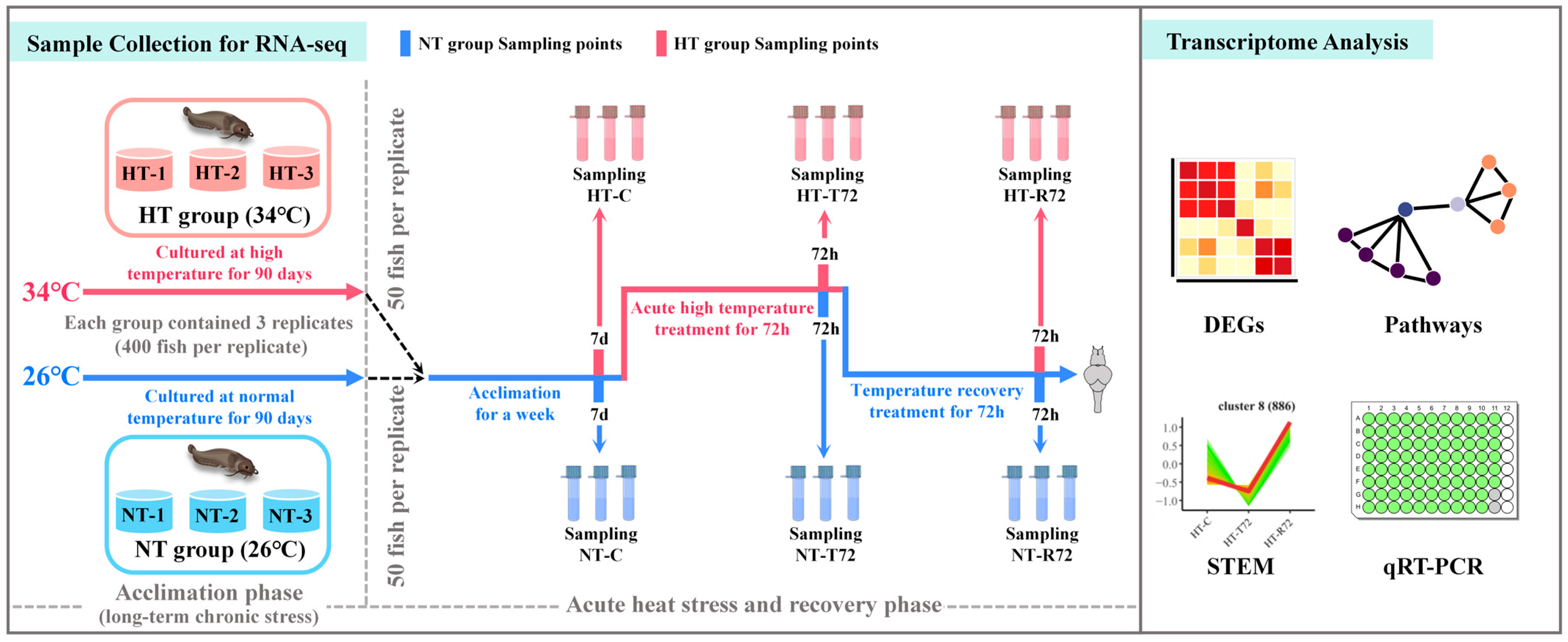
2.2. Animals
2.3. Brain Transcriptome Analysis
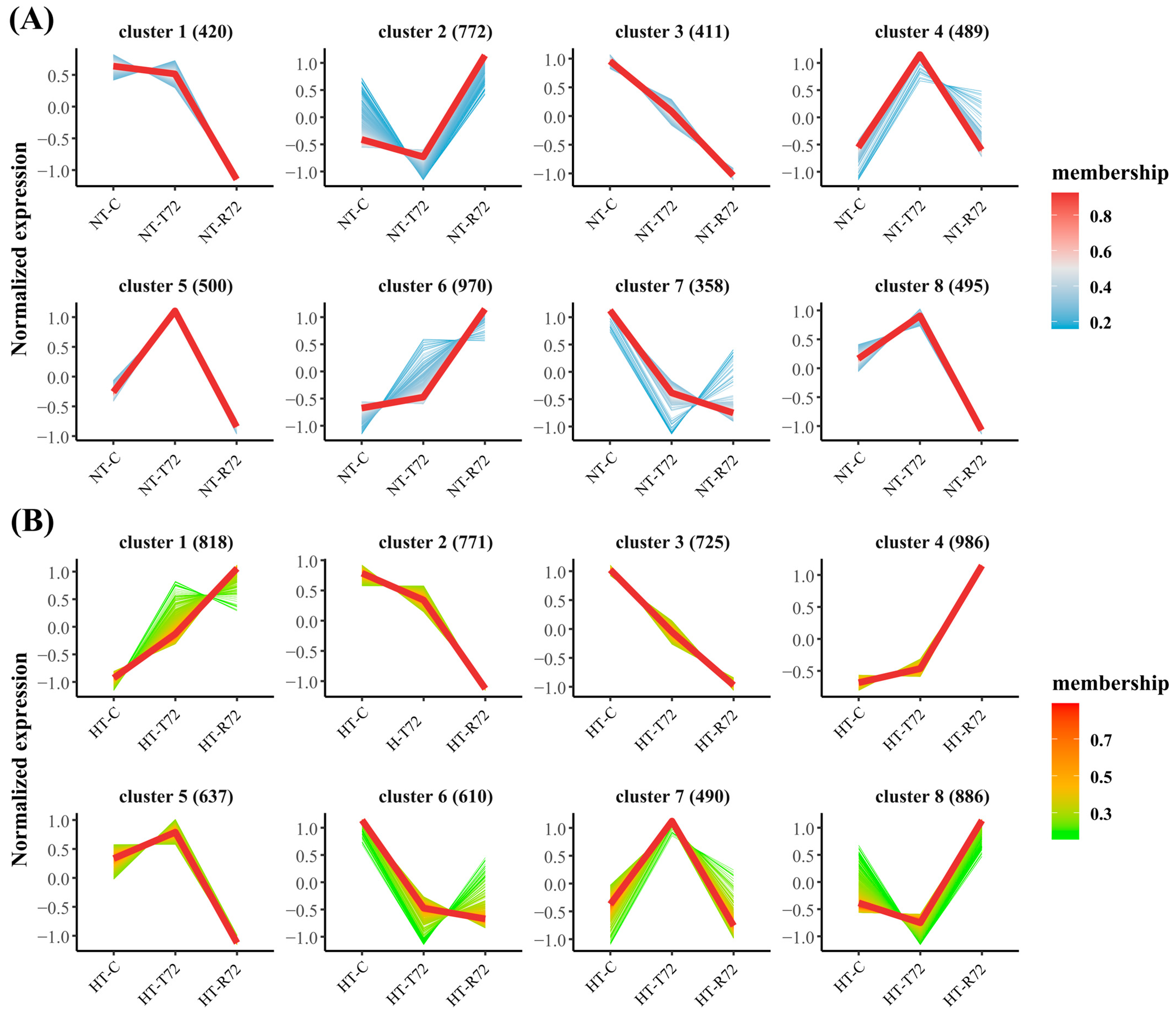
2.4. Trend Clustering
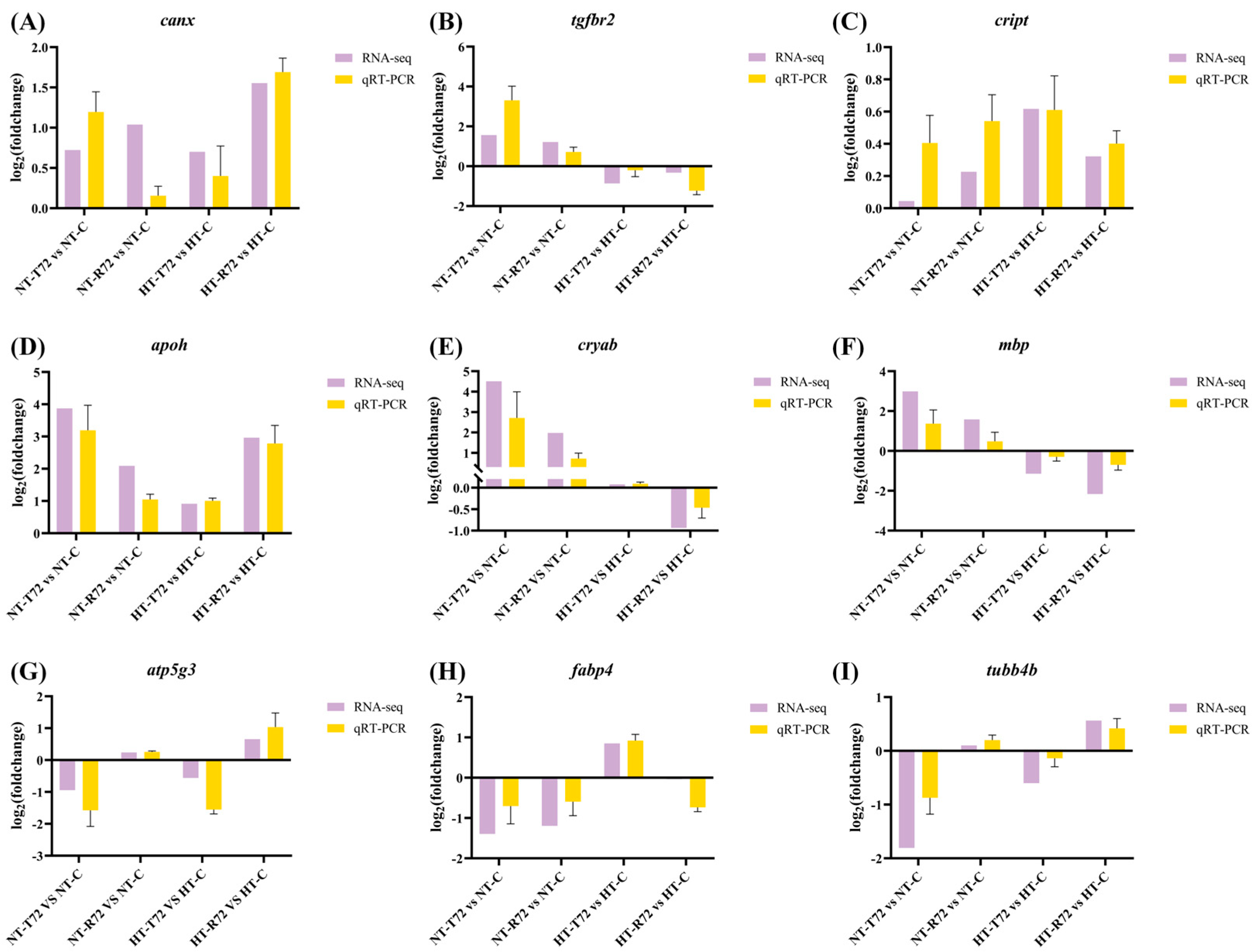
2.5. qRT-PCR Validation
3. Results
3.1. Quality of Transcriptome Sequencing
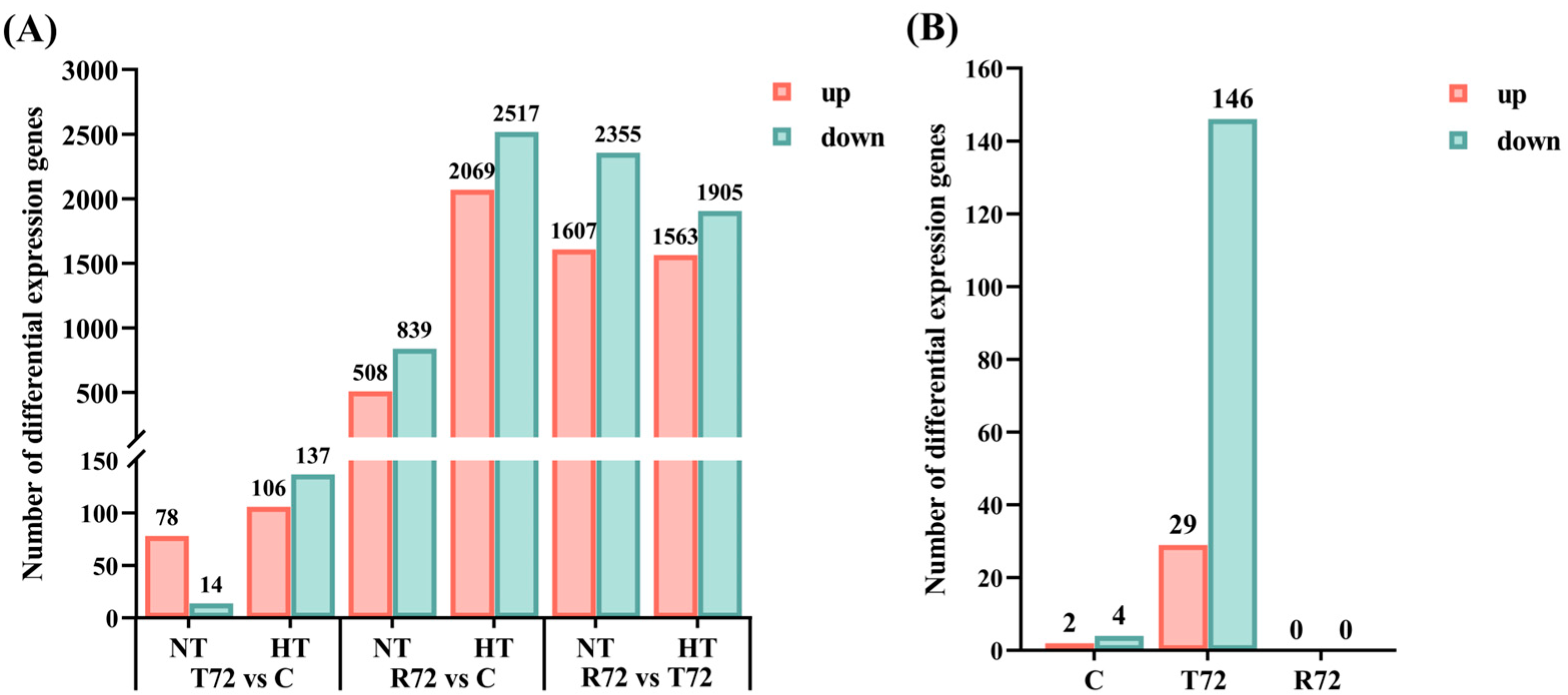
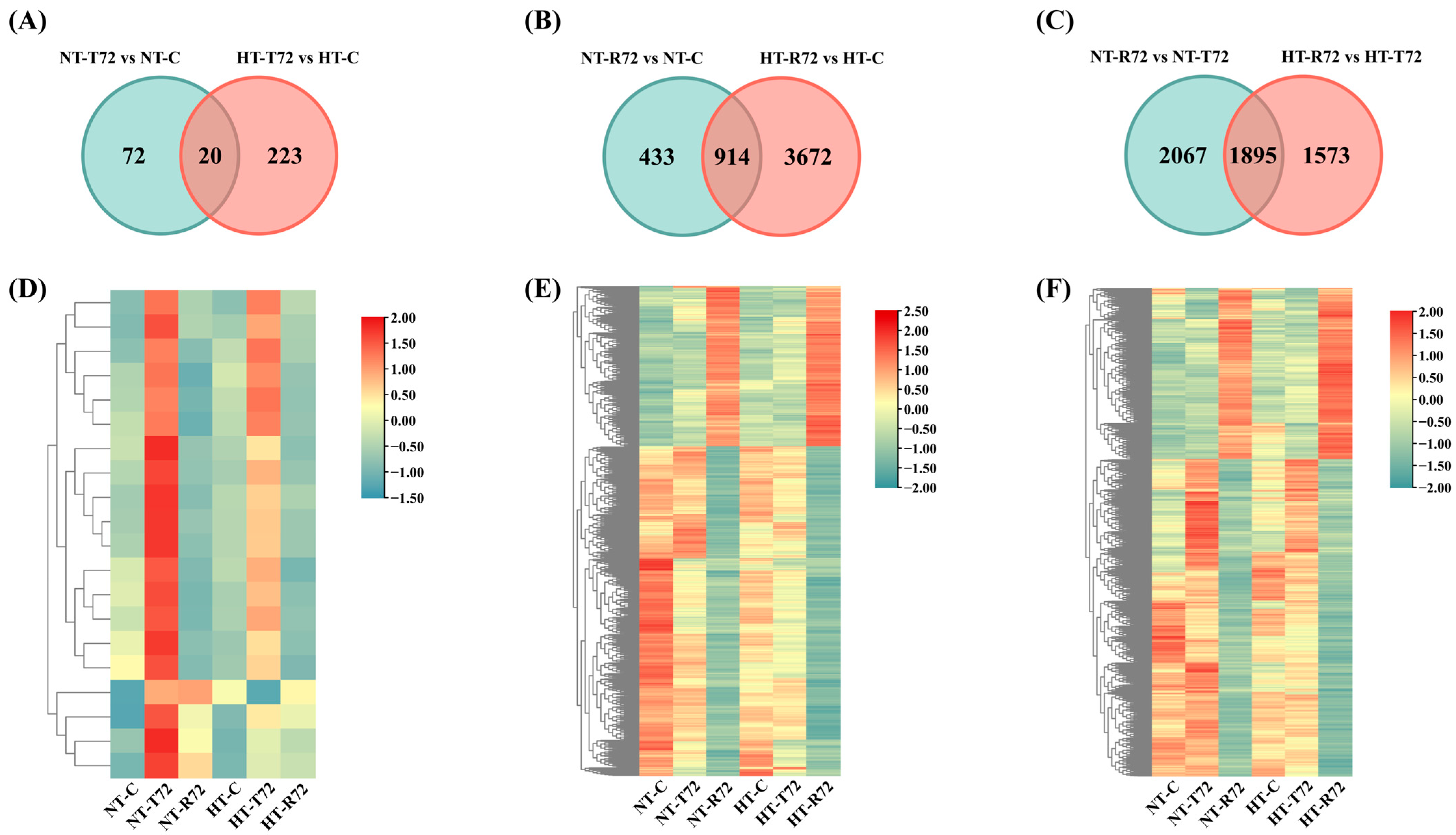
3.2. Analysis of DEGs
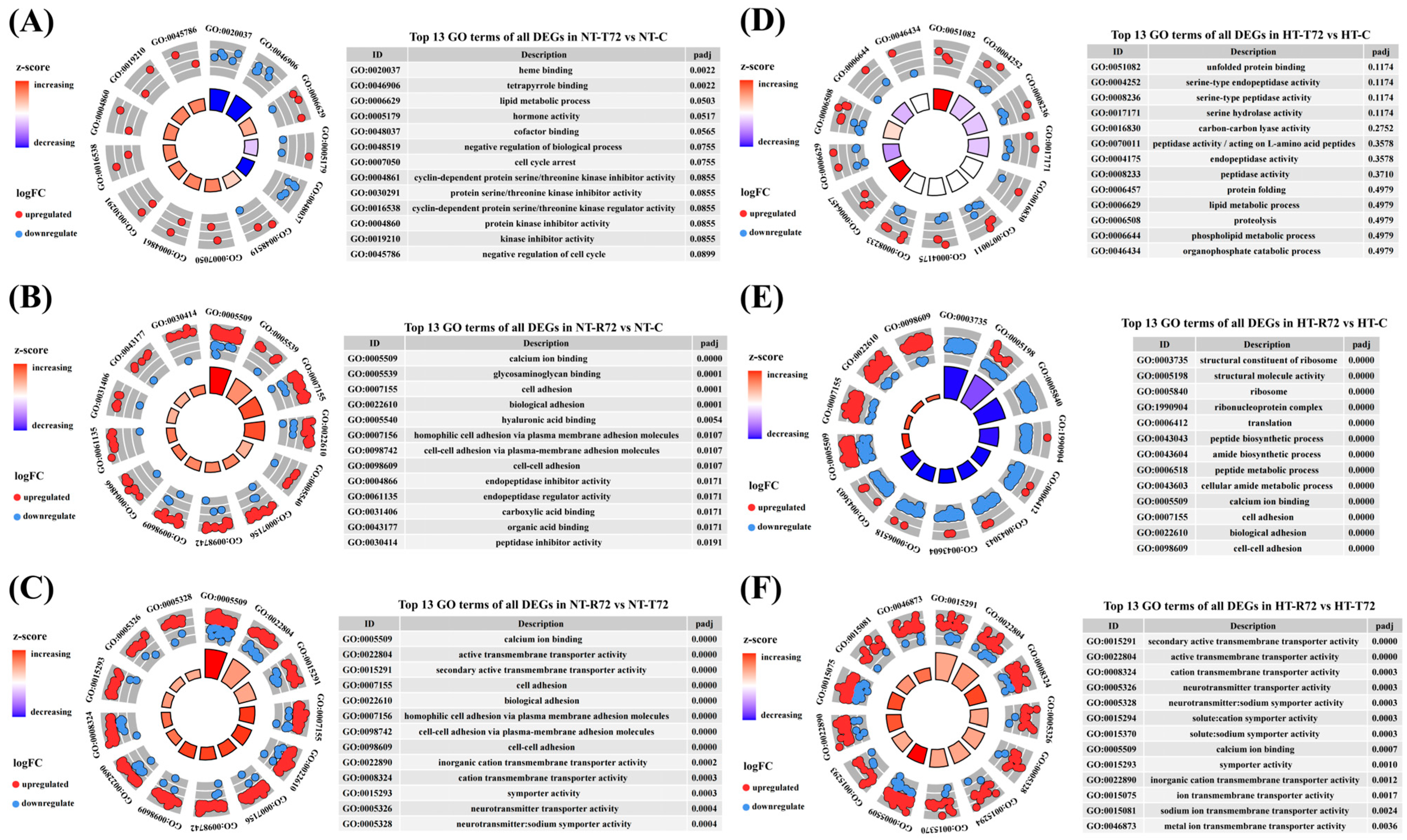
3.3. Functional and Pathway Enrichment Analysis of DEGs
3.4. Trend Clustering Analysis
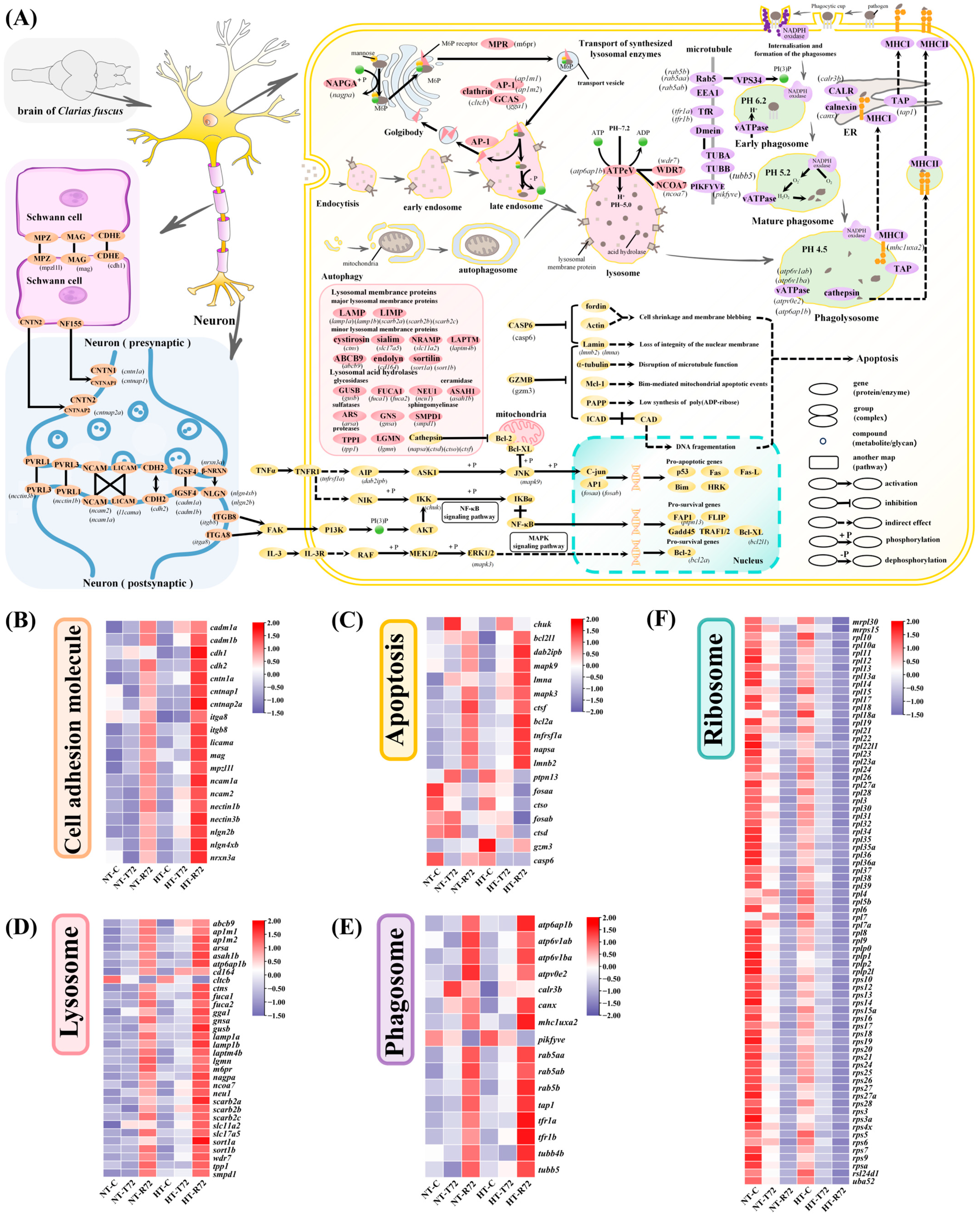
3.5. Molecular Pathway Alterations in the Brain Under Heat Stress
3.6. qRT-PCR Validation
4. Discussion
4.1. Enhanced Neural Resilience Through Accelerated Activation of Cell Adhesion and ECM Pathways
4.2. Dual Regulation of Cell Death and Degradation Pathways Supports Neuroprotection Under Heat Stress
4.3. Cellular Energy-Saving Mode Suppresses Ribosomal Gene Expression and Reduces Protein Synthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kiarsi, M.; Amiresmaili, M.; Mahmoodi, M.R.; Farahmandnia, H.; Nakhaee, N.; Zareiyan, A.; Aghababaeian, H. Heat Waves and Adaptation: A Global Systematic Review. J. Therm. Biol. 2023, 116, 103588. [Google Scholar] [CrossRef]
- Xu, W.; Wang, L. Editorial: The Physiological and Molecular Response of Aquatic Animals to Environmental Stresses. Front. Physiol. 2022, 13, 987004. [Google Scholar] [CrossRef] [PubMed]
- Ge, G.; Long, Y.; Song, G.; Li, Q.; Cui, Z.; Yan, H. Transcriptomic Profiling Revealed Signaling Pathways Associated with the Spawning of Female Zebrafish under Cold Stress. Int. J. Mol. Sci. 2022, 23, 7494. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhao, Z.; Zhang, R.; Guo, K.; Wang, S.; Xu, W.; Wang, C. The Effects of Temperature Changes on the Isozyme and Hsp70 Levels of the Amur Sturgeon, Acipenser Schrenckii, at Two Acclimation Temperatures. Aquaculture 2022, 551, 737743. [Google Scholar] [CrossRef]
- Li, S.; Liu, Y.; Li, B.; Ding, L.; Wei, X.; Wang, P.; Chen, Z.; Han, S.; Huang, T.; Wang, B.; et al. Physiological Responses to Heat Stress in the Liver of Rainbow Trout (Oncorhynchus mykiss) Revealed by UPLC-QTOF-MS Metabolomics and Biochemical Assays. Ecotoxicol. Environ. Saf. 2022, 242, 113949. [Google Scholar] [CrossRef]
- Maulu, S.; Hasimuna, O.J.; Haambiya, L.H.; Monde, C.; Musuka, C.G.; Makorwa, T.H.; Munganga, B.P.; Phiri, K.J.; Nsekanabo, J.D. Climate Change Effects on Aquaculture Production: Sustainability Implications, Mitigation, and Adaptations. Front. Sustain. Food Syst. 2021, 5, 609097. [Google Scholar] [CrossRef]
- Cheng, X.; Li, F.; Lu, J.; Wen, Y.; Li, Z.; Liao, J.; Cao, J.; He, X.; Sun, J.; Liu, Q. Transcriptome Analysis in Gill Reveals the Adaptive Mechanism of Domesticated Common Carp to the High Temperature in Shallow Rice Paddies. Aquaculture 2024, 578, 740107. [Google Scholar] [CrossRef]
- Nonnis, S.; Angiulli, E.; Maffioli, E.; Frabetti, F.; Negri, A.; Cioni, C.; Alleva, E.; Romeo, V.; Tedeschi, G.; Toni, M. Acute Environmental Temperature Variation Affects Brain Protein Expression, Anxiety and Explorative Behaviour in Adult Zebrafish. Sci. Rep. 2021, 11, 2521. [Google Scholar] [CrossRef]
- Haesemeyer, M.; Robson, D.N.; Li, J.M.; Schier, A.F.; Engert, F. A Brain-Wide Circuit Model of Heat-Evoked Swimming Behavior in Larval Zebrafish. Neuron 2018, 98, 817–831.e6. [Google Scholar] [CrossRef]
- Zhao, Y.; Duan, M.; Lin, X.; Li, W.; Liu, H.; Meng, K.; Liu, F.; Hu, W.; Luo, D. Molecular Underpinnings Underlying Behaviors Changes in the Brain of Juvenile Common Carp (Cyrinus carpio) in Response to Warming. J. Adv. Res. 2024, 63, 43–56. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, X.; Li, J.; Shen, Y. Transcriptomic Analysis of the Liver and Brain in Grass Carp (Ctenopharyngodon idella) Under Heat Stress. Mar. Biotechnol. 2022, 24, 856–870. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Dorta, V.; Caballero, M.J.; Betancor, M.B.; Manchado, M.; Tort, L.; Torrecillas, S.; Zamorano, M.J.; Izquierdo, M.; Montero, D. Effects of Thermal Stress on the Expression of Glucocorticoid Receptor Complex Linked Genes in Senegalese Sole (Solea senegalensis): Acute and Adaptive Stress Responses. Gen. Comp. Endocrinol. 2017, 252, 173–185. [Google Scholar] [CrossRef]
- Yang, Q.; Zhong, Y.; Yang, F.; Li, H.; Tran, N.T.; Zhang, M.; Wang, L.; He, L.; Zhang, Z.; Ge, H.; et al. Transcriptome Analysis of Sea Cucumber (Apostichopus japonicus) in Southern China under Heat Stress. Aquac. Rep. 2024, 36, 102036. [Google Scholar] [CrossRef]
- Yang, S.; Zhao, T.; Ma, A.; Huang, Z.; Liu, Z.; Cui, W.; Zhang, J.; Zhu, C.; Guo, X.; Yuan, C. Metabolic Responses in Scophthalmus Maximus Kidney Subjected to Thermal Stress. Fish Shellfish Immunol. 2020, 103, 37–46. [Google Scholar] [CrossRef]
- Zhao, T.; Ma, A.; Huang, Z.; Liu, Z.; Sun, Z.; Zhu, C.; Yang, J.; Li, Y.; Wang, Q.; Qiao, X.; et al. Transcriptome Analysis Reveals That High Temperatures Alter Modes of Lipid Metabolism in Juvenile Turbot (Scophthalmus maximus) Liver. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100887. [Google Scholar] [CrossRef]
- Li, Q.; Xiong, L.; Zhu, Y.; Zheng, A.; Zheng, S. Effects of Acute Temperature Stress on the Expression of Related Genes in the Brain of Opsariichthys bidens. Fishes 2024, 9, 248. [Google Scholar] [CrossRef]
- Lyu, L.; Wen, H.; Li, Y.; Li, J.; Zhao, J.; Zhang, S.; Song, M.; Wang, X. Deep Transcriptomic Analysis of Black Rockfish (Sebastes schlegelii) Provides New Insights on Responses to Acute Temperature Stress. Sci. Rep. 2018, 8, 9113. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.-X.; Lin, X.-H.; Zhou, D.-Y.; Chen, Y.; Shen, Y.-J.; Ye, M.-H.; Duan, C.-Y.; Zhang, Y.-L.; Yang, B.-L.; Deng, S.-P.; et al. A Chromosome-Level Genome Assembly of Hong Kong Catfish (Clarias fuscus) Uncovers a Sex-Determining Region. BMC Genom. 2023, 24, 291. [Google Scholar] [CrossRef]
- Anderson, M.J.; Fast, A.W. Temperature and Feed Rate Effects on Chinese Catfish, Clarias fuscus (Lacepède), Growth. Aquac. Res. 1991, 22, 435–442. [Google Scholar] [CrossRef]
- Liu, Y.; Tian, C.; Yang, Z.; Huang, C.; Jiao, K.; Yang, L.; Duan, C.; Zhang, Z.; Li, G. Effects of Chronic Heat Stress on Growth, Apoptosis, Antioxidant Enzymes, Transcriptomic Profiles, and Immune-Related Genes of Hong Kong Catfish (Clarias fuscus). Animals 2024, 14, 1006. [Google Scholar] [CrossRef]
- Duan, C.; Tian, C.; Guan, Y.; Xu, H.; Yang, L.; Chen, Y.; Liu, Y.; Shen, Y.; Zhang, Y.; Cao, S.; et al. Long-Term Thermal Stress Induces Hepatic Injury and Alters the Thermotolerance Response in Hong Kong Catfish (Clarias fuscus). Aquaculture 2024, 590, 741041. [Google Scholar] [CrossRef]
- Duan, C.; Zhou, D.; Feng, R.; Li, X.; Yang, L.; Li, X.; Li, G.; Chen, H.; Liao, Y.; Tian, C. Long-Term Thermal Acclimation Enhances Heat Resistance of Hong Kong Catfish (Clarias fuscus) by Modulating Gill Tissue Structure, Antioxidant Capacity and Immune Metabolic Pathways. Ecotoxicol. Environ. Saf. 2024, 284, 116930. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Yang, L.; Chen, W.; Zhou, D.; Cao, S.; Zhang, Y.; Li, G.; Chen, H.; Tian, C. Long-Term Thermal Stress Reshapes the Tolerance of Head Kidney of Hong Kong Catfish (Clarias fuscus) to Acute Heat Shock by Regulating Energy Metabolism and Immune Response. Comp. Biochem. Physiol. Part D Genom. Proteom. 2025, 54, 101437. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Tan, J.; Shen, Y.; Yang, B.; Zhang, Y.; Liao, Y.; Wang, P.; Zhou, D.; Li, G.; Tian, C. A High-Density Genetic Linkage Map and QTL Mapping for Sex in Clarias Fuscus. Aquaculture 2022, 561, 738723. [Google Scholar] [CrossRef]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational Methods for Transcriptome Annotation and Quantification Using RNA-Seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef]
- Shen, Y.; Zhu, Y.; Yang, Z.; Ye, M.; Zhou, D.; Li, G.; Tian, C. Screening for qRT-PCR Internal Reference Genes in Clarias Fuscus. Prog. Fish. Sci. 2024, 45, 144–154. [Google Scholar] [CrossRef]
- Takeichi, M. The Cadherin Superfamily in Neuronal Connections and Interactions. Nat. Rev. Neurosci. 2007, 8, 11–20. [Google Scholar] [CrossRef]
- Huang, Z.; Ma, A.; Yang, S.; Liu, X.; Zhao, T.; Zhang, J.; Wang, X.; Sun, Z.; Liu, Z.; Xu, R. Transcriptome Analysis and Weighted Gene Co-Expression Network Reveals Potential Genes Responses to Heat Stress in Turbot Scophthalmus Maximus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 33, 100632. [Google Scholar] [CrossRef]
- Kraitavin, W.; Yoshitake, K.; Igarashi, Y.; Mitsuyama, S.; Kinoshita, S.; Kambayashi, D.; Watabe, S.; Asakawa, S. Transcriptome Analysis of Yamame (Oncorhynchus masou) in Normal Conditions after Heat Stress. Biology 2019, 8, 21. [Google Scholar] [CrossRef]
- Brusés, J.L. N-Cadherin Signaling in Synapse Formation and Neuronal Physiology. Mol. Neurobiol. 2006, 33, 237–252. [Google Scholar] [CrossRef]
- Basu, R.; Taylor, M.R.; Williams, M.E. The Classic Cadherins in Synaptic Specificity. Cell Adhes. Migr. 2015, 9, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Dityatev, A.; Bukalo, O.; Schachner, M. Modulation of Synaptic Transmission and Plasticity by Cell Adhesion and Repulsion Molecules. Neuron Glia Biol. 2008, 4, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Faivre-Sarrailh, C.; Devaux, J.J. Neuro-Glial Interactions at the Nodes of Ranvier: Implication in Health and Diseases. Front. Cell. Neurosci. 2013, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Belluardo, N.; White, T.W.; Srinivas, M.; Trovato-Salinaro, A.; Ripps, H.; Mudò, G.; Bruzzone, R.; Condorelli, D.F. Identification and Functional Expression of HCx31.9, a Novel Gap Junction Gene. Cell Commun. Adhes. 2001, 8, 173–178. [Google Scholar] [CrossRef]
- López-Bendito, G.; Molnár, Z. Thalamocortical Development: How Are We Going to Get There? Nat. Rev. Neurosci. 2003, 4, 276–289. [Google Scholar] [CrossRef]
- Takai, Y.; Miyoshi, J.; Ikeda, W.; Ogita, H. Nectins and Nectin-like Molecules: Roles in Contact Inhibition of Cell Movement and Proliferation. Nat. Rev. Mol. Cell Biol. 2008, 9, 603–615. [Google Scholar] [CrossRef]
- Südhof, T.C. Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 2017, 171, 745–769. [Google Scholar] [CrossRef]
- Maness, P.F.; Schachner, M. Neural Recognition Molecules of the Immunoglobulin Superfamily: Signaling Transducers of Axon Guidance and Neuronal Migration. Nat. Neurosci. 2007, 10, 19–26. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: Bidirectional, Allosteric Signaling Machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, H.; Li, H.; Wang, A.; Yu, H. Effect of High Temperature on Immune Response of Grass Carp (Ctenopharyngodon idellus) by Transcriptome Analysis. Fish Shellfish Immunol. 2016, 58, 89–95. [Google Scholar] [CrossRef]
- Cheng, C.-H.; Yang, F.-F.; Ling, R.-Z.; Liao, S.-A.; Miao, Y.-T.; Ye, C.-X.; Wang, A.-L. Effects of Ammonia Exposure on Apoptosis, Oxidative Stress and Immune Response in Pufferfish (Takifugu obscurus). Aquat. Toxicol. 2015, 164, 61–71. [Google Scholar] [CrossRef]
- Liu, E.; Zhao, X.; Li, C.; Wang, Y.; Li, L.; Zhu, H.; Ling, Q. Effects of Acute Heat Stress on Liver Damage, Apoptosis and Inflammation of Pikeperch (Sander lucioperca). J. Therm. Biol. 2022, 106, 103251. [Google Scholar] [CrossRef] [PubMed]
- Fleisch, V.C.; Fraser, B.; Allison, W.T. Investigating Regeneration and Functional Integration of CNS Neurons: Lessons from Zebrafish Genetics and Other Fish Species. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2011, 1812, 364–380. [Google Scholar] [CrossRef]
- Tan, C.; Pang, X.; Zhang, J.; Yan, C.; Xu, Z.; Shao, W.; Wu, J.; Li, Y.; Du, X.; Yang, S. Effects of Chronic Heat Stress on Spleen Structure, Apoptosis and Immune Response in Siberian Sturgeon (Acipenser baerii). Isr. J. Aquac. Bamidgeh 2024, 76, 69–74. [Google Scholar] [CrossRef]
- Cavalcante, G.C.; Schaan, A.P.; Cabral, G.F.; Santana-da-Silva, M.N.; Pinto, P.; Vidal, A.F.; Ribeiro-dos-Santos, Â. A Cell’s Fate: An Overview of the Molecular Biology and Genetics of Apoptosis. Int. J. Mol. Sci. 2019, 20, 4133. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Chen, L.; Song, K.; Shu, T.; Fang, Z.; Ding, L.; Liu, J.; Jiang, L.; Zhang, G.; Zhang, B.; et al. Irreversible Electroporation Mediates Glioma Apoptosis via Upregulation of AP-1 and Bim: Transcriptome Evidence. Brain Sci. 2022, 12, 1465. [Google Scholar] [CrossRef]
- Yabu, T.; Shiba, H.; Shibasaki, Y.; Nakanishi, T.; Imamura, S.; Touhata, K.; Yamashita, M. Stress-Induced Ceramide Generation and Apoptosis via the Phosphorylation and Activation of nSMase1 by JNK Signaling. Cell Death Differ. 2015, 22, 258–273. [Google Scholar] [CrossRef]
- Wang, Y.; Su, C.; Liu, Q.; Hao, X.; Han, S.; Doretto, L.B.; Rosa, I.F.; Yang, Y.; Shao, C.; Wang, Q. Transcriptome Analysis Revealed the Early Heat Stress Response in the Brain of Chinese Tongue Sole (Cynoglossus semilaevis). Animals 2023, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhou, Y.; Guo, X.; Wei, W.; Li, Z.; Zhou, L.; Wang, Z.; Gui, J. Comparative Transcriptomes and Metabolomes Reveal Different Tolerance Mechanisms to Cold Stress in Two Different Catfish Species. Aquaculture 2022, 560, 738543. [Google Scholar] [CrossRef]
- Jing, Z.; Chen, Q.; Yan, C.; Zhang, C.; Xu, Z.; Huang, X.; Wu, J.; Li, Y.; Yang, S. Effects of Chronic Heat Stress on Kidney Damage, Apoptosis, Inflammation, and Heat Shock Proteins of Siberian Sturgeon (Acipenser baerii). Animals 2023, 13, 3733. [Google Scholar] [CrossRef]
- Han, P.; Yuan, M.; Sun, Z.; Xue, Y.; Liu, X.; Chen, J.; Yu, H.; Wang, X. Effect of Heat Exposure on Histology, Transcriptomics and Co-Expression Network: A Synthetic Study in Japanese Flounder (Paralichthys olivaceus). Aquaculture 2025, 595, 741490. [Google Scholar] [CrossRef]
- Logan, C.A.; Somero, G.N. Effects of Thermal Acclimation on Transcriptional Responses to Acute Heat Stress in the Eurythermal Fish Gillichthys mirabilis (Cooper). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1373–R1383. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Ao, H.; Guo, G.; Liu, M. The Role and Mechanism of Metformin in Inflammatory Diseases. J. Inflamm. Res. 2023, 16, 5545–5564. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Pan, C.; Liu, E.; Zhao, X.; Ling, Q. Alterations to Transcriptomic Profile, Histopathology, and Oxidative Stress in Liver of Pikeperch (Sander lucioperca) under Heat Stress. Fish Shellfish Immunol. 2019, 95, 659–669. [Google Scholar] [CrossRef]
- Beemelmanns, A.; Zanuzzo, F.S.; Xue, X.; Sandrelli, R.M.; Rise, M.L.; Gamperl, A.K. The Transcriptomic Responses of Atlantic Salmon (Salmo Salar) to High Temperature Stress Alone, and in Combination with Moderate Hypoxia. BMC Genom. 2021, 22, 261. [Google Scholar] [CrossRef]
- Mostowy, S.; Shenoy, A.R. The Cytoskeleton in Cell-Autonomous Immunity: Structural Determinants of Host Defence. Nat. Rev. Immunol. 2015, 15, 559–573. [Google Scholar] [CrossRef]
- Nair-Gupta, P.; Baccarini, A.; Tung, N.; Seyffer, F.; Florey, O.; Huang, Y.; Banerjee, M.; Overholtzer, M.; Roche, P.A.; Tampé, R.; et al. TLR Signals Induce Phagosomal MHC-I Delivery from the Endosomal Recycling Compartment to Allow Cross-Presentation. Cell 2014, 158, 506–521. [Google Scholar] [CrossRef] [PubMed]
- Münz, C. The Macroautophagy Machinery in MHC Restricted Antigen Presentation. Front. Immunol. 2021, 12, 628429. [Google Scholar] [CrossRef]
- Jiao, C.; Zou, J.; Chen, Z.; Zheng, F.; Xu, Z.; Lin, Y.-H.; Wang, Q. Dietary Glutamine Inclusion Regulates Immune and Antioxidant System, as Well as Programmed Cell Death in Fish to Protect against Flavobacterium Columnare Infection. Antioxidants 2021, 11, 44. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.; Sun, F.; Zhang, J.; Feng, J.; Liu, H.; Rajendran, K.V.; Sun, L.; Zhang, Y.; Jiang, Y.; et al. RNA-Seq Reveals Expression Signatures of Genes Involved in Oxygen Transport, Protein Synthesis, Folding, and Degradation in Response to Heat Stress in Catfish. Physiol. Genom. 2013, 45, 462–476. [Google Scholar] [CrossRef]
- Quan, J.; Kang, Y.; Luo, Z.; Zhao, G.; Ma, F.; Li, L.; Liu, Z. Identification and Characterization of Long Noncoding RNAs Provide Insight into the Regulation of Gene Expression in Response to Heat Stress in Rainbow Trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 36, 100707. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-Q.; Zhang, J.; Wang, H.-Y.; Niu, S.-F.; Wu, R.-X.; Tang, B.-G.; Wang, Q.-H.; Liang, Z.-B.; Liang, Y.-S. Transcriptomic Response of the Liver Tissue in Trachinotus ovatus to Acute Heat Stress. Animals 2023, 13, 2053. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.-Q.; Ka, W.; Zhang, H.-J.; Li, Y.-L.; Gao, P.; Long, R.-J.; Yang, S.-W.; Wang, J.-L. RNA-Seq Analysis of the Key Long Noncoding RNAs and mRNAs Related to the Regulation of Acute Heat Stress in Rainbow Trout. Animals 2022, 12, 325. [Google Scholar] [CrossRef] [PubMed]
- Ge, K.; Fan, Z.; Huang, T.; Gu, W.; Wang, G.; Liu, E.; Pan, R.; Li, D.; Sun, Y.; Yao, Z.; et al. Influence of Increasing Acclimation Temperature on Growth, Digestion, Antioxidant Capacity, Liver Transcriptome and Intestinal Microflora of Ussruri Whitefish Coregonus Ussuriensis Berg. Fish Shellfish Immunol. 2024, 151, 109667. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, Y.; Duan, C.; Xie, X.; Luo, Z.; Zhou, D.; Zhang, Y.; Li, G.; Liao, Y.; Tian, C. Heat Acclimation Enhances Brain Resilience to Acute Thermal Stress in Clarias fuscus by Modulating Cell Adhesion, Anti-Apoptotic Pathways, and Intracellular Degradation Mechanisms. Animals 2025, 15, 1220. https://doi.org/10.3390/ani15091220
Guan Y, Duan C, Xie X, Luo Z, Zhou D, Zhang Y, Li G, Liao Y, Tian C. Heat Acclimation Enhances Brain Resilience to Acute Thermal Stress in Clarias fuscus by Modulating Cell Adhesion, Anti-Apoptotic Pathways, and Intracellular Degradation Mechanisms. Animals. 2025; 15(9):1220. https://doi.org/10.3390/ani15091220
Chicago/Turabian StyleGuan, Yingyi, Cunyu Duan, Xinyu Xie, Zhuoying Luo, Dayan Zhou, Yulei Zhang, Guangli Li, Yu Liao, and Changxu Tian. 2025. "Heat Acclimation Enhances Brain Resilience to Acute Thermal Stress in Clarias fuscus by Modulating Cell Adhesion, Anti-Apoptotic Pathways, and Intracellular Degradation Mechanisms" Animals 15, no. 9: 1220. https://doi.org/10.3390/ani15091220
APA StyleGuan, Y., Duan, C., Xie, X., Luo, Z., Zhou, D., Zhang, Y., Li, G., Liao, Y., & Tian, C. (2025). Heat Acclimation Enhances Brain Resilience to Acute Thermal Stress in Clarias fuscus by Modulating Cell Adhesion, Anti-Apoptotic Pathways, and Intracellular Degradation Mechanisms. Animals, 15(9), 1220. https://doi.org/10.3390/ani15091220







