Functional Polymorphisms in the Neuropeptide Y (NPY) Gene Associated with Egg Production in Thai Native, Black-Bone, and Commercial Laying Hens Using SNP Markers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
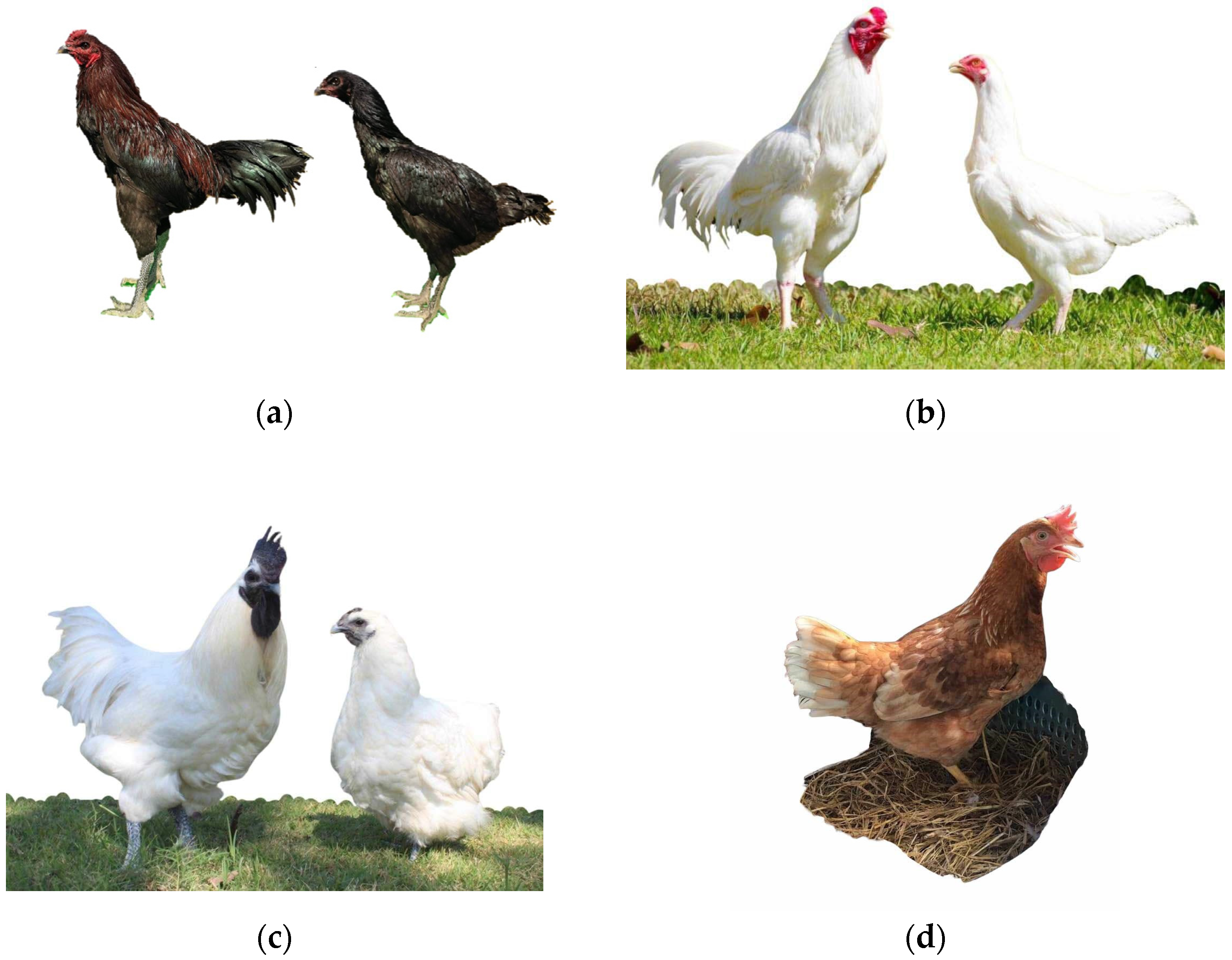
2.1. Experimental Animals and Phenotypic Data Collection
2.2. Single Nucleotide Polymorphisms (SNPs) Genotype
2.2.1. DNA Samples, Primer Designing, PCR Amplification
2.2.2. DNA Sequencing and Bioinformatics Analysis
2.3. Statistical Analysis
3. Results
3.1. Egg Production Traits in Thai Native Chicken, Black-Bone, and Commercial Laying Hens
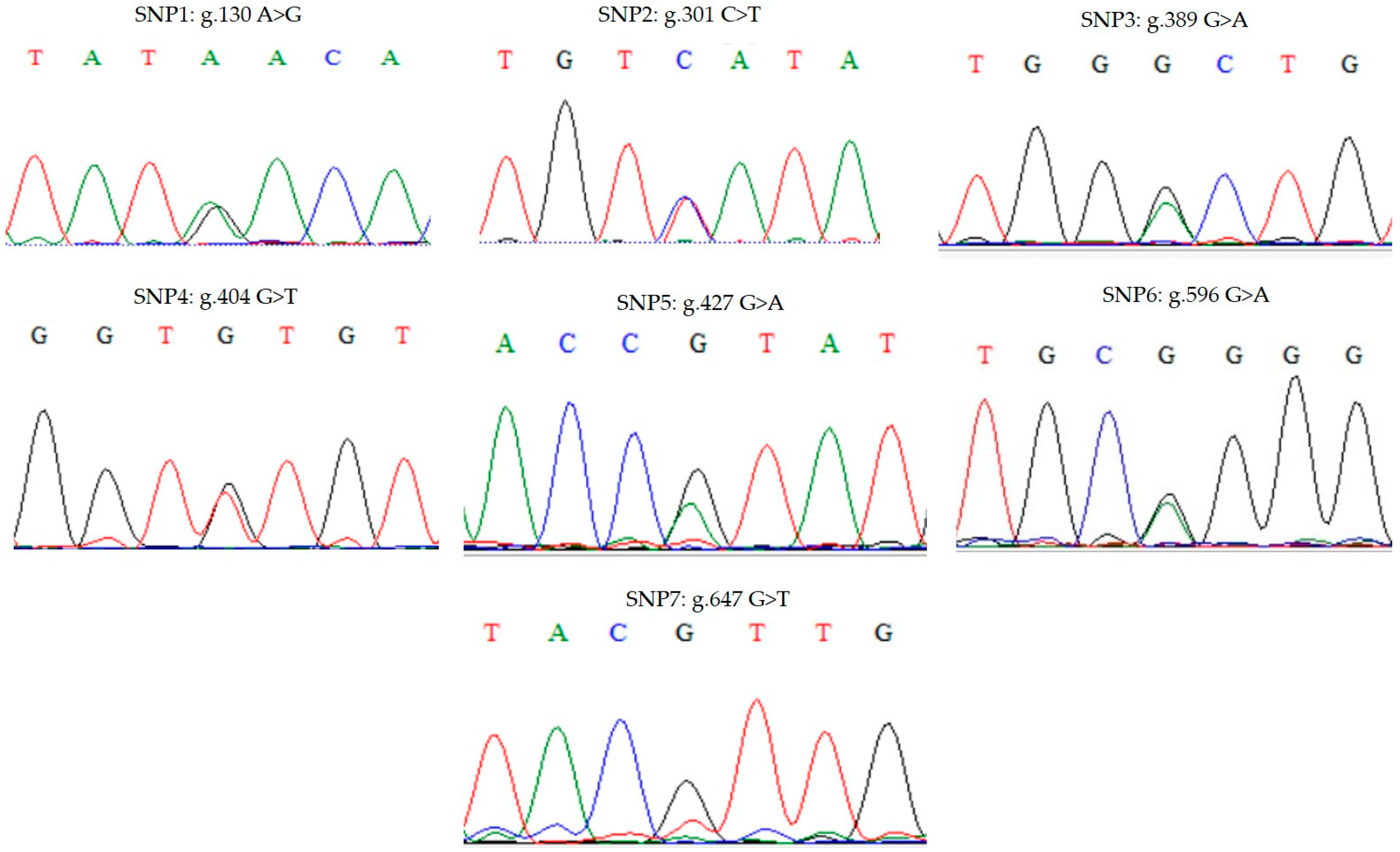
3.2. SNP Identification in the Coding Region of the NPY Gene
3.3. Genotype and Alleles Frequency
3.4. Genetic Diversity
3.5. Associations Between Seven SNP Polymorphisms and Chicken Egg Production
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Padhi, M.K. Importance of indigenous breeds of chicken for rural economy and their improvements for higher production performance. Scientifica 2016, 2016, 2604685. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, I.A.; Shamim, A.; Shad, M.K.; Ashari, H.; Yusuf, I. Circular economy-based integrated farming system for indigenous chicken: Fostering food security and sustainability. J. Clean. Prod. 2024, 436, 140368. [Google Scholar] [CrossRef]
- Loengbudnark, W.; Chankitisakul, V.; Duangjinda, M.; Boonkum, W. Sustainable growth through thai native chicken farming: Lessons from rural communities. Sustainability 2024, 16, 7811. [Google Scholar] [CrossRef]
- Kim, M.; Munyaneza, J.P.; Cho, E.; Jang, A.; Jo, C.; Nam, K.C.; Choo, H.J.; Lee, J.H. Genome-wide association study on the content of nucleotide-related compounds in Korean native chicken breast meat. Animals 2023, 13, 2966. [Google Scholar] [CrossRef]
- Jaturasitha, S.; Chaiwang, N.; Kreuzer, M. Thai native chicken meat: An option to meet the demands for specific meat quality by certain groups of consumers; a review. Anim Prod Sci. 2017, 57, 1582–1587. [Google Scholar] [CrossRef]
- Teinlek, P.; Siripattarapravat, K.; Tirawattanawanich, C. Genetic diversity analysis of Thai indigenous chickens based on complete sequences of mitochondrial DNA D-loop region. Asian-Australas. J. Anim. Sci. 2018, 31, 804–811. [Google Scholar] [CrossRef]
- Bungsrisawat, P.; Tumwasorn, S.; Loongyai, W.; Nakthong, S.; Sopannaratha, P. Genetic parameters of some carcass and meat quality traits in Betong chicken (KU line). Agric. Nat. Resour. 2018, 52, 274–279. [Google Scholar] [CrossRef]
- Kammongkun, J.; Promket, D. Growth performance and morphology traits associated with neuropeptide y (NPY) genes expression in native chickens. Adv. Anim. Vet. Sci. 2024, 12, 2263–2274. [Google Scholar] [CrossRef]
- Loengbudnark, W.; Chankitisakul, V.; Boonkum, W. The genetic impact of heat stress on the egg production of Thai native chickens (Pradu Hang dum). PLoS ONE 2023, 18, e0281328. [Google Scholar] [CrossRef]
- Li, J.-J.; Zhang, L.; Ren, P.; Wang, Y.; Yin, L.-Q.; Ran, J.-S.; Zhang, X.-X.; Liu, Y.-P. Genotype frequency distributions of 28 SNP markers in two commercial lines and five Chinese native chicken populations. BMC Genet. 2020, 21, 12. [Google Scholar] [CrossRef]
- Boonkum, W.; Chankitisakul, V.; Kananit, S.; Tuntiyasawasdikul, V.; Sirisan, V.; Kenchaiwong, W. Genetic profiles of purine, uric acid, superoxide dismutase, and growth in Thai slow-growing chickens. Animals 2024, 14, 3658. [Google Scholar] [CrossRef] [PubMed]
- Chomchuen, K.; Tuntiyasawasdikul, V.; Chankitisakul, V.; Boonkum, W. Genetic Evaluation of Body Weights and Egg Production Traits Using a Multi-Trait Animal Model and Selection Index in Thai Native Synthetic Chickens (Kaimook e-san2). Animals 2022, 12, 335. [Google Scholar] [CrossRef] [PubMed]
- Mengesha, Y.; Kebede, E.; Getachew, A. Review of chicken productive and reproductive performance and its challenges in Ethiopia. All Life 2022, 15, 118–125. [Google Scholar] [CrossRef]
- Hosnedlova, B.; Vernerova, K.; Kizek, R.; Bozzi, R.; Kadlec, J.; Curn, V.; Kouba, F.; Fernandez, C.; Machander, V.; Horna, H. Associations between IGF1, IGFBP2 and TGFß3 genes polymorphisms and growth performance of broiler chicken lines. Animals 2020, 10, 800. [Google Scholar] [CrossRef]
- AL-Jaryan, I.L.H.; Hassan, W.S.; AL-Rekabi, M.M. Association of the melatonin receptor c gene with egg production traits in local Iraqi chicken. Sys. Rev. Pharm. 2021, 12, 1406–1413. [Google Scholar]
- Cho, S.; Manjula, P.; Kim, M.; Cho, E.; Lee, D.; Lee, S.H.; Lee, J.H.; Seo, D. Comparison of selection signatures between Korean native and commercial chickens using 600k SNP array data. Genes 2021, 12, 824. [Google Scholar] [CrossRef]
- Machete, J.B.; Kgwatalala, P.M.; Nsoso, S.J.; Hlongwane, N.L.; Moreki, J.C. Genetic diversity and population structure of three strains of indigenous Tswana chickens and commercial broiler using single nucleotide polymormophic (SNP) markers. Open J. Anim. Sci. 2021, 11, 515–531. [Google Scholar] [CrossRef]
- Ali, M.Y.; Faruque, S.; Ahmadi, S.; Ohkubo, T. Genetic analysis of HSP70 and HSF3 polymorphisms and their associations with the egg production traits of Bangladeshi hilly chickens. Animals 2024, 14, 3552. [Google Scholar] [CrossRef]
- Velasco, V.V.; Tsudzuki, M.; Hashimoto, N.; Goto, N.; Ishikawa, A. Genetic diversity, runs of homozygosity, and selection signatures in native Japanese chickens: Insights from single-nucleotide polymorphisms. Animals 2024, 14, 3341. [Google Scholar] [CrossRef]
- Seo, D.; Cho, S.; Manjula, P.; Choi, N.; Kim, Y.-K.; Koh, Y.J.; Lee, S.H.; Kim, H.-Y.; Lee, J.H. Identification of target chicken populations by machine learning models using the minimum number of SNPs. Animals 2021, 11, 241. [Google Scholar] [CrossRef]
- Cai, K.; Liu, R.; Wei, L.; Wang, X.; Cui, H.; Luo, N.; Wen, J.; Chang, Y.; Zhao, G. Genome-wide association analysis identify candidate genes for feed efficiency and growth traits in Wenchang chickens. BMC Genom. 2024, 25, 645. [Google Scholar] [CrossRef]
- Sartsoongnoen, N.; Kamkrathok, B.; Songserm, T.; Chaiseha, Y. Distribution and variation of neuropeptide Y in the brain of native Thai chicken. Avian Biol Res. 2021, 14, 27–36. [Google Scholar] [CrossRef]
- Chen, W.C.; Liu, Y.B.; Liu, W.F.; Zhou, Y.Y.; He, H.F.; Lin, S. Neuropeptide Y is an immunomodulatory factor: Direct and indirect. Front. Immunol. 2020, 11, 580378. [Google Scholar] [CrossRef]
- Al-Zubaidi, K.S.O.; Al-Rekabi, M.M.J.; Allaw, A.A. Effect of polymorphism of the Neuropeptide Y (NPY) gene on some productive traits of Iraqi local white chickens. IOP Conf. Ser. Earth Environ. Sci. 2023, 1252, 012121. [Google Scholar] [CrossRef]
- Padwar, P.; Thakur, M.S. Association of neuropeptide-Y gene polymorphic variants with quantitative traits in Jabalpur colour and Kadaknath chicken. Indian J. Anim. Sci. 2021, 91, 729–732. [Google Scholar] [CrossRef]
- Ngu, N.T.; Xuan, N.H.; Vu, C.T.; An, N.T.; Dung, T.N.; Nhan, N.T.H. Effects of genetic polymorphisms on egg production in indigenous Noi chicken. J. Exp. Biol. Agric. Sci. 2015, 3, 487–493. [Google Scholar]
- Goodwin, W.; Linacre, A.; Hadi, S. An Introduction to Forensic Genetics, 2nd ed.; Wiley-Blackwell: Chichester, UK, 2011. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- SAS. SAS/STAT User’s Guide, Version 9.4; SAS Inst. Inc.: Cary, NC, USA, 2019.
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longmans Green: Harlow, Essex, UK, 1996. [Google Scholar]
- Tunim, S.; Phasuk, Y.; Aggrey, S.E.; Duangjinda, M. Increasing fat deposition via upregulates the transcription of peroxisome proliferator-activated receptor gamma in native crossbred chickens. Animals 2021, 11, 90. [Google Scholar] [CrossRef]
- Anh, N.T.L.; Kunhareang, S.; Duangjinda, M. Association of chicken growth hormones and insulin-like growth factor gene polymorphisms with growth performance and carcass traits in Thai broilers. Asian-Australas. J. Anim. Sci. 2015, 18, 1686–1695. [Google Scholar] [CrossRef]
- Ghaderi-Zefrehei, M.; Rafeie, F.; Zakizadeh, S.; Torshizi, M.E.; Peters, S.O.; Smith, J. Genetic variance components of the growth curve for Isfahan indigenous chicken. Vet. Med. Sci. 2024, 10, e1388. [Google Scholar] [CrossRef]
- Tongsiri, S.; Jeyaruban, G.M.; Hermesch, S.; van der Werf, J.H.J.; Li, L.; Chormai, T. Genetic parameters and inbreeding effects for production traits of Thai native chickens. Asian-Australas. J. Anim. Sci. 2019, 32, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Faruque, S.; Islam, M.S.; Afroz, M.A.; Rahman, M.M. Evaluation of the performance of native chicken and estimation of heritability for body weight. J. Bangladesh Acad. Sci. 2013, 37, 93–101. [Google Scholar] [CrossRef]
- Promket, D.; Pengmeesri, K.; Kammongkun, J.; Somchan, T. Identification of melatonin receptors type c (MTNR1C) and neuropeptide y (NPY) genes related to egg production in Thai indigenous chickens. Adv. Anim. Vet. Sci. 2024, 12, 206–215. [Google Scholar] [CrossRef]
- Ding, F.-X.; Zhang, G.-X.; Wang, J.-Y.; Yuan, L.; Zhang, L.-J.; Wei, Y.; Wang, H.-H.; Zhang, L.; Hou, Q.-R. Genetic diversity of a Chinese native chicken breed, Bian chicken, based on twenty-nine microsatellite markers. Asian-Australas. J. Anim. Sci. 2010, 23, 154–161. [Google Scholar] [CrossRef]
- Kubota, S.; Vandee, A.; Keawnakient, P.; Molee, W.; Yongsawatdikul, J.; Molee, A. Effects of the MC4R, CAPN1, and ADSL genes on body weight and purine content in slow-growing chickens. Poult. Sci. 2019, 98, 4327–4337. [Google Scholar] [CrossRef]
- Şener, K.; Alver, E.N.; Cevher, Ş.C. An overview of appetite regulation mechanisms. Koc. J. Sci. Eng. 2022, 5, 178–193. [Google Scholar] [CrossRef]
- Chen, A.; Zhao, X.; Wen, J.; Zhao, X.; Wang, G.; Zhang, X.; Ren, X.; Zhang, Y.; Cheng, X.; Yu, X.; et al. Genetic parameter estimation and molecular foundation of chicken egg-laying trait. Poult. Sci. 2024, 103, 103627. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Wu, Y.; Shen, J.; Pan, A.; Zhang, H.; Sun, J.; Liang, Z.; Huang, T.; Du, J.; Pi, J. Genome-wide association study of egg production traits in shuanglian chickens using whole genome sequencing. Genes 2023, 14, 2129. [Google Scholar] [CrossRef]
- Zhou, J.; Yu, J.-Z.; Zhu, M.-Y.; Yang, F.-X.; Hao, J.-P.; He, Y.; Zhu, X.-L.; Hou, Z.-C.; Zhu, F. Genome-wide association analysis and genetic parameters for egg production traits in Peking ducks. Animals 2024, 14, 1891. [Google Scholar] [CrossRef]
| Traits | Chicken Breeds | p-Value | |||
|---|---|---|---|---|---|
| PH | C | BB | LC | ||
| AFEP, day | 151.89 ± 9.59 c | 188.05 ± 24.14 a | 170.60 ± 12.41 b | 140.44 ± 5.18 d | <0.001 |
| FEW, g | 33.29 ± 6.07 b | 35.08 ± 3.86 b | 41.55 ± 1.05 a | 43.77 ± 7.05 a | <0.001 |
| EW_9M, g | 44.81 ± 3.10 b | 41.15 ± 2.94 c | 44.45 ± 1.09 b | 51.48 ± 6.91 a | <0.001 |
| EW_12M, g | 44.95 ± 3.06 c | 43.30 ± 3.16 c | 48.55 ± 1.05 b | 53.50 ± 6.91 a | <0.001 |
| NE_9M, egg | 159.61 ± 35.49 b | 55.30 ± 23.01 c | 52.85 ± 19.59 c | 195.61 ± 15.79 a | <0.001 |
| NE_12M, egg | 196.90 ± 43.32 b | 73.70 ± 30.70 c | 87.55 ± 28.78 c | 254.44 ± 14.08 a | <0.001 |
| EperM, egg | 16.40 ± 3.61 b | 6.14 ± 2.55 c | 7.29 ± 2.39 c | 21.20 ± 1.17 a | <0.001 |
| EMs, g/hen/day | 42.63 ± 9.82 b | 17.95 ± 5.76 c | 22.06 ± 6.03 c | 60.67 ± 8.76 a | <0.001 |
| SNPs | Chicken Breeds | Genotype Frequency (n) | Allele Frequency | PIC | HE | Chi-Square | |||
|---|---|---|---|---|---|---|---|---|---|
| SNP1: g.130 A>G | AA | AG | GG | A | G | ||||
| PH | 0.69 (41) | 0.14 (8) | 0.17 (10) | 0.76 | 0.24 | 0.30 | 0.36 | 23.08 | |
| C | 0.70 (14) | 0.25 (5) | 0.05 (1) | 0.82 | 0.175 | 0.25 | 0.29 | 0.36 | |
| BB | 0.75 (15) | 0.10 (2) | 0.15 (3) | 0.80 | 0.20 | 0.27 | 0.32 | 9.45 | |
| LC | 0.61 (11) | 0.11 (2) | 0.28 (5) | 0.67 | 0.33 | 0.35 | 0.44 | 10.13 | |
| All | 0.69 (81) | 0.15 (17) | 0.16 (19) | 0.76 | 0.24 | 0.29 | 0.36 | 41.55 | |
| SNP2: g.301 C>T | CC | CT | TT | C | T | ||||
| PH | 0.63 (37) | 0.12 (7) | 0.25 (15) | 0.69 | 0.31 | 0.34 | 0.43 | 30.96 | |
| C | 0.50 (10) | 0.35 (7) | 0.15 (3) | 0.68 | 0.32 | 0.34 | 0.44 | 0.82 | |
| BB | 0.65 (13) | 0.05 (1) | 0.30 (6) | 0.68 | 0.32 | 0.34 | 0.44 | 15.70 | |
| LC | 0.78 (14) | 0.06 (1) | 0.17 (3) | 0.81 | 0.19 | 0.26 | 0.31 | 12.18 | |
| All | 0.63 (74) | 0.14 (16) | 0.23 (27) | 0.70 | 0.30 | 0.33 | 0.42 | 53.13 | |
| SNP3: g.389 G>A | GG | GA | AA | G | A | ||||
| PH | 0.46 (27) | 0.15 (9) | 0.39 (23) | 0.53 | 0.47 | 0.37 | 0.50 | 28.38 | |
| C | 0.35 (7) | 0.35 (7) | 0.30 (6) | 0.52 | 0.48 | 0.37 | 0.50 | 1.78 | |
| BB | 0.35 (7) | 0.10 (2) | 0.55 (11) | 0.40 | 0.60 | 0.36 | 0.48 | 12.53 | |
| LC | 0.44 (8) | 0.22 (4) | 0.33 (6) | 0.56 | 0.44 | 0.37 | 0.49 | 5.45 | |
| All | 0.42 (49) | 0.19 (22) | 0.39 (46) | 0.51 | 0.49 | 0.37 | 0.50 | 45.51 | |
| SNP4: g.404 G>T | GG | GT | TT | G | T | ||||
| PH | 0.53 (31) | 0.03 (2) | 0.44 (26) | 0.54 | 0.46 | 0.37 | 0.50 | 30.35 | |
| C | 0.70 (14) | 0.15 (3) | 0.15 (3) | 0.78 | 0.23 | 0.29 | 0.35 | 6.50 | |
| BB | 0.65 (13) | 0.10 (2) | 0.25 (5) | 0.70 | 0.3 | 0.33 | 0.42 | 11.61 | |
| LC | 0.56 (10) | 0.22 (4) | 0.22 (4) | 0.67 | 0.33 | 0.35 | 0.44 | 4.50 | |
| All | 0.58 (68) | 0.09 (11) | 0.32 (38) | 0.63 | 0.37 | 0.36 | 0.47 | 74.64 | |
| SNP5: g.427 G>A | GG | GA | AA | G | A | ||||
| PH | 0.42 (25) | 0.08 (5) | 0.49 (29) | 0.47 | 0.53 | 0.37 | 0.50 | 40.62 | |
| C | 0.30 (6) | 0.35 (7) | 0.35 (7) | 0.47 | 0.53 | 0.37 | 0.50 | 1.78 | |
| BB | 0.45 (9) | 0.15 (3) | 0.40 (8) | 0.52 | 0.48 | 0.37 | 0.50 | 9.78 | |
| LC | 0.22 (4) | 0.28 (5) | 0.50 (9) | 0.36 | 0.64 | 0.35 | 0.46 | 2.85 | |
| All | 0.38 (44) | 0.17 (20) | 0.45 (53) | 0.46 | 0.54 | 0.50 | 0.50 | 50.36 | |
| SNP6: g.596 G>A | GG | GA | AA | G | A | ||||
| PH | 0.36 (21) | 0.19 (11) | 0.46 (27) | 0.45 | 0.55 | 0.37 | 0.49 | 22.92 | |
| C | 0.10 (2) | 0.40 (8) | 0.50 (10) | 0.30 | 0.70 | 0.33 | 0.42 | 0.05 | |
| BB | 0.05 (1) | 0.20 (4) | 0.75 (15) | 0.15 | 0.85 | 0.22 | 0.26 | 0.93 | |
| LC | 0.50 (9) | 0.22 (4) | 0.28 (5) | 0.61 | 0.39 | 0.36 | 0.48 | 5.10 | |
| All | 0.28 (33) | 0.23 (27) | 0.49 (57) | 0.40 | 0.60 | 0.36 | 0.48 | 31.42 | |
| SNP7: g.647 G>T | GG | GT | TT | G | T | ||||
| PH | 0.58 (34) | 0.24 (14) | 0.19 (11) | 0.69 | 0.31 | 0.33 | 0.42 | 11.44 | |
| C | 0.65 (13) | 0.20 (4) | 0.15 (3) | 0.75 | 0.25 | 0.30 | 0.38 | 4.36 | |
| BB | 0.70 (14) | 0.25 (5) | 0.05 (1) | 0.82 | 0.18 | 0.25 | 0.29 | 0.36 | |
| LC | 0.56 (10) | 0.39 (7) | 0.06 (1) | 0.75 | 0.25 | 0.30 | 0.38 | 0.02 | |
| All | 0.61 (71) | 0.26 (30) | 0.14 (16) | 0.74 | 0.26 | 0.31 | 0.39 | 13.66 | |
| SNPs | Genotype | AFEP | FEW | EW_9M | EW_12M | NE_9M | NE_12M | EperM | EMs |
|---|---|---|---|---|---|---|---|---|---|
| SNP1: g.130 A>G | AA | 161.84 a | 36.56 | 45.12 | 46.60 | 123.26 | 160.54 | 13.38 | 36.70 |
| AG | 158.82 ab | 37.14 | 45.64 | 47.05 | 132.47 | 164.59 | 13.71 | 37.97 | |
| GG | 150.21 b | 37.17 | 44.84 | 46.26 | 150.79 | 190.58 | 15.88 | 41.52 | |
| p-value | 0.02 | 0.88 | 0.64 | 0.61 | 0.32 | 0.57 | 0.57 | 0.89 | |
| SNP2: g.301 C>T | CC | 157.10 b | 37.04 | 45.56 | 47.06 | 132.41 | 168.05 | 14.00 | 38.45 |
| CT | 169.93 a | 37.13 | 44.25 | 45.54 | 120.38 | 150.81 | 12.56 | 34.75 | |
| TT | 159.92 ab | 35.69 | 44.55 | 46.01 | 125.07 | 169.41 | 14.11 | 37.27 | |
| p-value | 0.04 | 0.35 | 0.17 | 0.06 | 0.95 | 0.96 | 0.96 | 0.85 | |
| SNP3: g.389 G>A | GG | 157.04 | 36.47 | 44.53 | 45.89 | 140.47 | 183.51 a | 15.29 a | 40.40 |
| GA | 163.63 | 37.12 | 45.52 | 46.92 | 127.23 | 162.32 ab | 13.52 ab | 38.19 | |
| AA | 160.17 | 36.85 | 45.63 | 47.23 | 117.80 | 149.13 b | 12.42 b | 34.52 | |
| p-value | 0.35 | 0.40 | 0.38 | 0.07 | 0.06 | 0.03 | 0.03 | 0.21 | |
| SNP4: g.404 G>T | GG | 159.86 ab | 37.37 ab | 44.71 | 46.40 | 126.99 | 165.66 | 13.80 | 37.31 |
| GT | 168.63 b | 40.63 a | 46.76 | 48.39 | 115.00 | 152.45 | 12.70 | 36.56 | |
| TT | 156.23 a | 34.48 b | 45.47 | 46.46 | 136.87 | 170.55 | 14.21 | 38.64 | |
| p-value | 0.02 | <0.01 | 0.18 | 0.09 | 0.32 | 0.32 | 0.32 | 0.41 | |
| SNP5: g.427 G>A | GG | 161.18 | 35.67 | 44.16 b | 45.47 b | 118.93 b | 150.39 b | 12.53 b | 33.34 b |
| GA | 161.50 | 38.40 | 46.78 a | 48.76 a | 123.55 ab | 162.65 ab | 13.55 ab | 19.20 c | |
| AA | 157.37 | 37.00 | 45.35 ab | 46.74 ab | 139.57 a | 180.25 a | 15.02 a | 40.69 a | |
| p-value | 0.51 | 0.17 | <0.01 | <0.01 | 0.03 | 0.01 | 0.01 | <0.01 | |
| SNP6: g.596 G>A | GG | 151.21 b | 36.54 | 46.44 a | 47.34 a | 152.33 a | 189.42 a | 15.78 a | 42.86 a |
| GA | 162.88 a | 36.68 | 45.07 ab | 46.59 ab | 123.48 b | 158.81 ab | 13.23 ab | 36.51 ab | |
| AA | 162.71 a | 36.89 | 44.44 b | 46.19 b | 118.25 b | 155.86 b | 12.98 b | 35.21 b | |
| p-value | <0.01 | 0.33 | <0.01 | 0.03 | <0.01 | <0.01 | <0.01 | <0.01 | |
| SNP7: g.647 G>T | GG | 160.59 | 37.10 | 45.10 | 46.63 | 120.04 | 153.21 | 12.76 | 34.90 |
| GT | 156.93 | 37.19 | 46.03 | 47.42 | 143.20 | 187.73 | 15.64 | 42.90 | |
| TT | 155.56 | 34.31 | 43.69 | 45.00 | 142.63 | 182.06 | 15.17 | 40.14 | |
| p-value | 0.63 | 0.28 | 0.31 | 0.34 | 0.36 | 0.24 | 0.23 | 0.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Promket, D.; Kammongkun, J.; Insee, J.; Kenchaiwong, W.; Pengmeesri, K.; Somchan, T.; Boonkum, W. Functional Polymorphisms in the Neuropeptide Y (NPY) Gene Associated with Egg Production in Thai Native, Black-Bone, and Commercial Laying Hens Using SNP Markers. Animals 2025, 15, 744. https://doi.org/10.3390/ani15050744
Promket D, Kammongkun J, Insee J, Kenchaiwong W, Pengmeesri K, Somchan T, Boonkum W. Functional Polymorphisms in the Neuropeptide Y (NPY) Gene Associated with Egg Production in Thai Native, Black-Bone, and Commercial Laying Hens Using SNP Markers. Animals. 2025; 15(5):744. https://doi.org/10.3390/ani15050744
Chicago/Turabian StylePromket, Doungnapa, Jennarong Kammongkun, Jiranan Insee, Wootichai Kenchaiwong, Khanitta Pengmeesri, Thassawan Somchan, and Wuttigrai Boonkum. 2025. "Functional Polymorphisms in the Neuropeptide Y (NPY) Gene Associated with Egg Production in Thai Native, Black-Bone, and Commercial Laying Hens Using SNP Markers" Animals 15, no. 5: 744. https://doi.org/10.3390/ani15050744
APA StylePromket, D., Kammongkun, J., Insee, J., Kenchaiwong, W., Pengmeesri, K., Somchan, T., & Boonkum, W. (2025). Functional Polymorphisms in the Neuropeptide Y (NPY) Gene Associated with Egg Production in Thai Native, Black-Bone, and Commercial Laying Hens Using SNP Markers. Animals, 15(5), 744. https://doi.org/10.3390/ani15050744








