Alterations in Whey Protein Abundance Correlated with the Somatic Cell Count Identified via Label-Free and Selected Reaction Monitoring Proteomic Approaches
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Milk Whey Separation
2.3. Protein Digestion
2.4. Liquid Chromatography–Tandem Mass Spectrometry Analysis
2.5. Protein Identification and Quantitation
2.6. Selected Reaction Monitoring (SRM) Validation
2.7. Data Analysis
3. Results
3.1. Proteomics Analysis of Milk Whey Among the Five Different Milk Groups
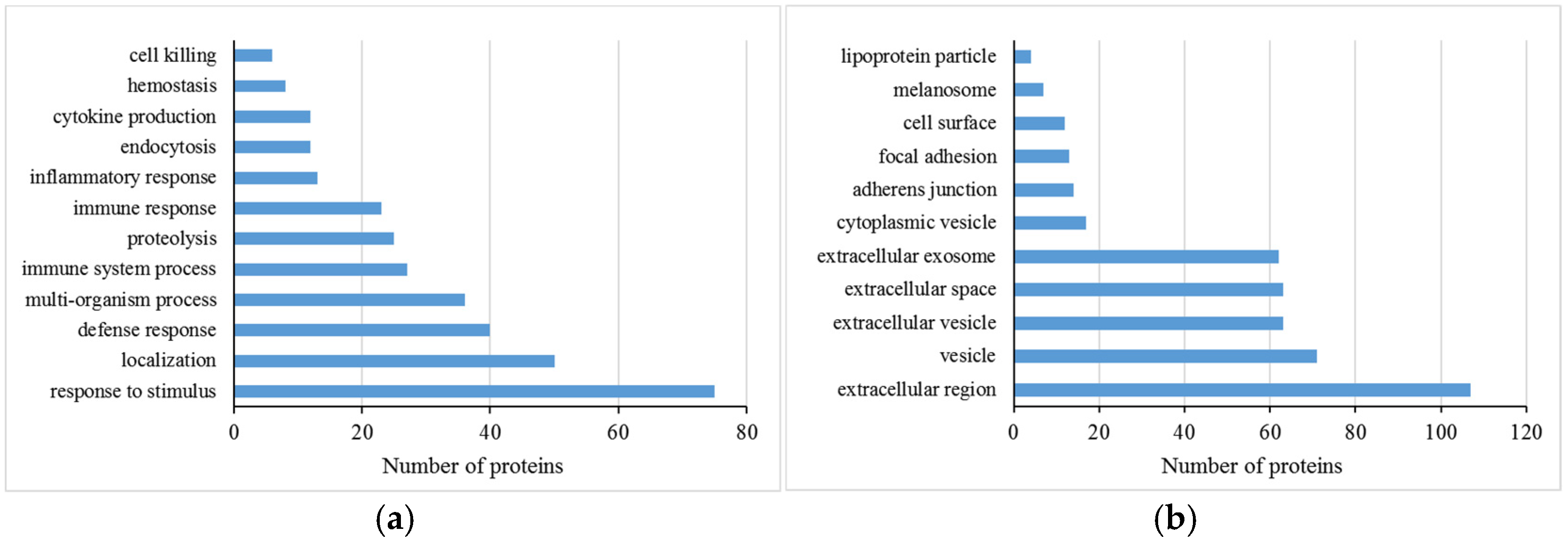
3.2. Functional Analysis of Differentially Abundant Proteins
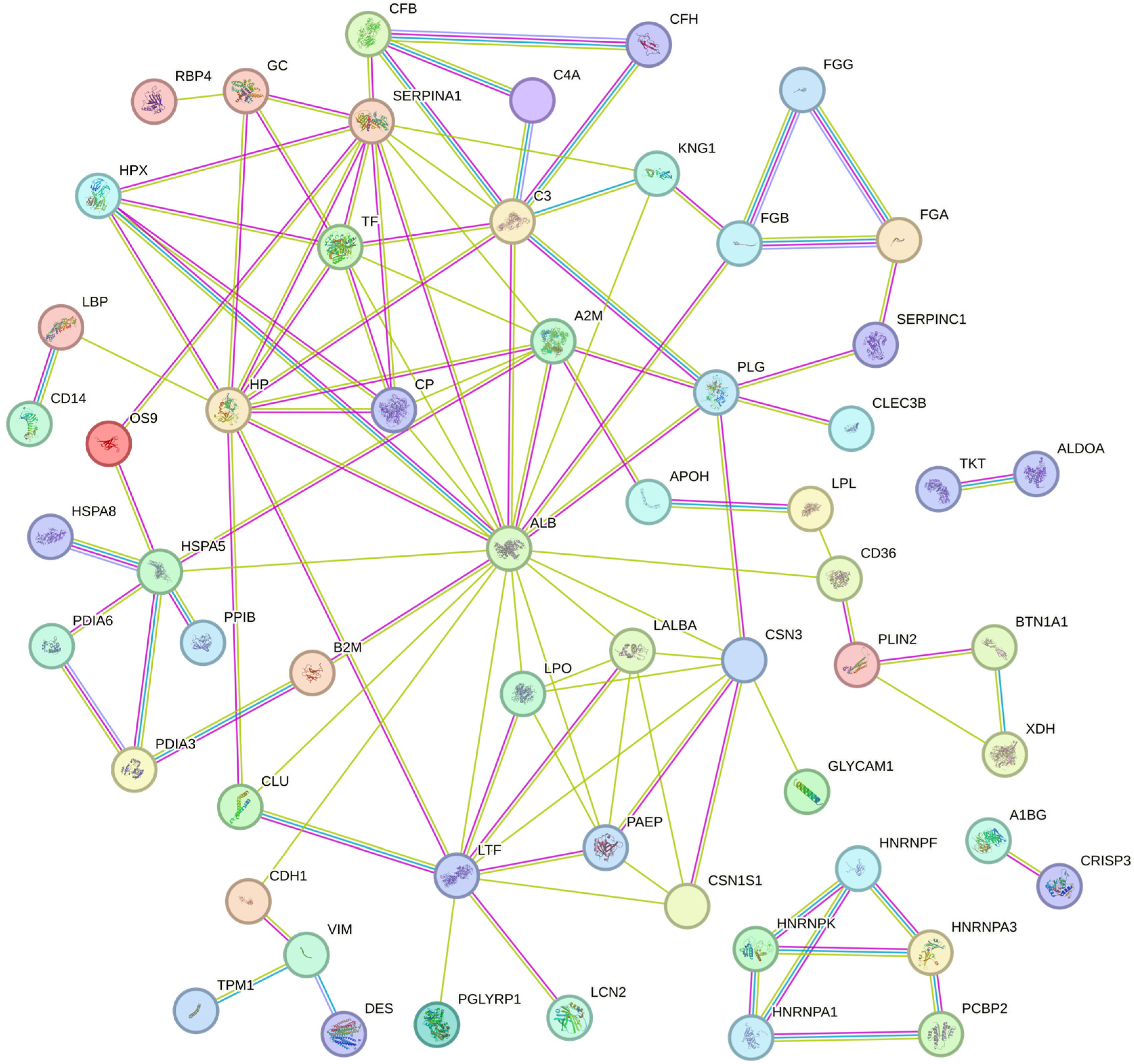
3.3. Clustering and Principal Component Analysis of Differentially Abundant Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schukken, Y.H.; Wilson, D.J.; Welcome, F.; Garrison-Tikofsky, L.; Gonzalez, R.N. Monitoring udder health and milk quality using somatic cell counts. Vet. Res. 2003, 34, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Dufour, S.; Dohoo, I.R. Monitoring herd incidence of intramammary infection in lactating cows using repeated longitudinal somatic cell count measurements. J. Dairy Sci. 2013, 96, 1568–1580. [Google Scholar] [CrossRef]
- Frossling, J.; Ohlson, A.; Hallen-Sandgren, C. Incidence and duration of increased somatic cell count in Swedish dairy cows and associations with milking system type. J. Dairy Sci. 2017, 100, 7368–7378. [Google Scholar] [CrossRef] [PubMed]
- Forsback, L.; Lindmark-Mansson, H.; Svennersten-Sjaunja, K.; Bach Larsen, L.; Andren, A. Effect of storage and separation of milk at udder quarter level on milk composition, proteolysis, and coagulation properties in relation to somatic cell count. J. Dairy Sci. 2011, 94, 5341–5349. [Google Scholar] [CrossRef] [PubMed]
- Hadrich, J.C.; Wolf, C.A.; Lombard, J.; Dolak, T.M. Estimating milk yield and value losses from increased somatic cell count on US dairy farms. J. Dairy Sci. 2018, 101, 3588–3596. [Google Scholar] [CrossRef] [PubMed]
- Testa, F.; Marano, G.; Ambrogi, F.; Boracchi, P.; Casula, A.; Biganzoli, E.; Moroni, P. Study of the association of atmospheric temperature and relative humidity with bulk tank milk somatic cell count in dairy herds using Generalized additive mixed models. Res. Vet. Sci. 2017, 114, 511–517. [Google Scholar] [CrossRef]
- Fauteux, V.; Bouchard, E.; Haine, D.; Scholl, D.T.; Roy, J.P. Prediction of bulk tank somatic cell count violations based on monthly individual cow somatic cell count data. J. Dairy Sci. 2015, 98, 2312–2321. [Google Scholar] [CrossRef] [PubMed]
- van Schaik, G.; Lotem, M.; Schukken, Y.H. Trends in somatic cell counts, bacterial counts, and antibiotic residue violations in New York State during 1999–2000. J. Dairy Sci. 2002, 85, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.F.; Hegazy, Y.M.; Ibrahem, S.A. Interrelationship of milk acute-phase proteins and casein percentage in cows and buffaloes subclinical mastitis. Proc. Vet. Res. Forum 2021, 12, 409–414. [Google Scholar]
- Bisutti, V.; Vanzin, A.; Toscano, A.; Pegolo, S.; Giannuzzi, D.; Tagliapietra, F.; Schiavon, S.; Gallo, L.; Trevisi, E.; Negrini, R. Impact of somatic cell count combined with differential somatic cell count on milk protein fractions in Holstein cattle. J. Dairy Sci. 2022, 105, 6447–6459. [Google Scholar] [CrossRef] [PubMed]
- Pegolo, S.; Giannuzzi, D.; Bisutti, V.; Tessari, R.; Gelain, M.; Gallo, L.; Schiavon, S.; Tagliapietra, F.; Trevisi, E.; Marsan, P.A. Associations between differential somatic cell count and milk yield, quality, and technological characteristics in Holstein cows. J. Dairy Sci. 2021, 104, 4822–4836. [Google Scholar] [CrossRef]
- Sah, B.N.P.; Vasiljevic, T.; McKechnie, S.; Donkor, O.N. Antioxidative and antibacterial peptides derived from bovine milk proteins. Crit. Rev. Food Sci. Nutr. 2018, 58, 726–740. [Google Scholar] [CrossRef] [PubMed]
- Giagu, A.; Penati, M.; Traini, S.; Dore, S.; Addis, M.F. Milk proteins as mastitis markers in dairy ruminants-a systematic review. Vet. Res. Commun. 2022, 46, 329–351. [Google Scholar] [CrossRef] [PubMed]
- Caggiano, N.; Lorenzo Smirnoff, A.; Bottini, J.M.; De Simone, E.A. Protease activity and protein profile in milk from healthy dairy cows and cows with different types of mastitis. Int. Dairy J. 2019, 89, 1–5. [Google Scholar] [CrossRef]
- Maity, S.; Das, D.; Ambatipudi, K. Quantitative alterations in bovine milk proteome from healthy, subclinical and clinical mastitis during S. aureus infection. J. Proteom. 2020, 223, 103815. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.-h.; Zhang, Y.; Shen, Y.; Su, Z.-t.; Yu, S.-m.; Cao, S.-z.; Zong, X.-l. Effect of anemoside B4 on milk whey in clinical mastitis-affected cows elucidated using tandem mass tag (TMT)-based quantitative proteomics. Sci. Rep. 2022, 12, 18829. [Google Scholar] [CrossRef]
- Winther, A.R.; da Silva Duarte, V.; Porcellato, D. Metataxonomic analysis and host proteome response in dairy cows with high and low somatic cell count: A quarter level investigation. Vet. Res. 2023, 54, 32. [Google Scholar] [CrossRef]
- Vanzin, A.; Franchin, C.; Arrigoni, G.; Battisti, I.; Masi, A.; Squartini, A.; Bisutti, V.; Giannuzzi, D.; Gallo, L.; Cecchinato, A. Subclinical mastitis from Streptococcus agalactiae and Prototheca spp. induces changes in milk peptidome in Holstein cattle. J. Agric. Food Chem. 2023, 71, 16827–16839. [Google Scholar] [CrossRef]
- She, Y.; Liu, J.; Su, M.; Li, Y.; Guo, Y.; Liu, G.; Deng, M.; Qin, H.; Sun, B.; Guo, J. A Study on Differential Biomarkers in the Milk of Holstein Cows with Different Somatic Cells Count Levels. Animals 2023, 13, 2446. [Google Scholar] [CrossRef]
- Huppertz, T.; Fox, P.F.; Kelly, A.L. High pressure treatment of bovine milk: Effects on casein micelles and whey proteins. J. Dairy Res. 2004, 71, 97–106. [Google Scholar] [CrossRef]
- Addis, M.F.; Tedde, V.; Puggioni, G.M.G.; Pisanu, S.; Casula, A.; Locatelli, C.; Rota, N.; Bronzo, V.; Moroni, P.; Uzzau, S. Evaluation of milk cathelicidin for detection of bovine mastitis. J. Dairy Sci. 2016, 99, 8250–8258. [Google Scholar] [CrossRef] [PubMed]
- Bourganou, M.V.; Kontopodis, E.; Tsangaris, G.T.; Pierros, V.; Vasileiou, N.G.; Mavrogianni, V.S.; Fthenakis, G.C.; Katsafadou, A.I. Unique Peptides of Cathelicidin-1 in the Early Detection of Mastitis—In Silico Analysis. Int. J. Mol. Sci. 2023, 24, 10160. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 2004, 75, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Damm, M.; Holm, C.; Blaabjerg, M.; Bro, M.N.; Schwarz, D. Differential somatic cell count-A novel method for routine mastitis screening in the frame of Dairy Herd Improvement testing programs. J. Dairy Sci. 2017, 100, 4926–4940. [Google Scholar] [CrossRef] [PubMed]
- Rambault, M.; Gilbert, F.B.; Roussel, P.; Tessier, A.; David, V.; Germon, P.; Winter, N.; Remot, A. Neutrophils expressing major histocompatibility complex class II molecules circulate in blood and milk during mastitis and show high microbicidal activity. J. Dairy Sci. 2023, 106, 4245–4256. [Google Scholar] [CrossRef]
- Tapper, H.; Karlsson, A.; Morgelin, M.; Flodgaard, H.; Herwald, H. Secretion of heparin-binding protein from human neutrophils is determined by its localization in azurophilic granules and secretory vesicles. Blood 2002, 99, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.P.; Vanet, A.; Witko-Sarsat, V.; Melchior, M.; McCabe, D.; Gabay, J.E. Azurocidin, a natural antibiotic from human neutrophils: Expression, antimicrobial activity, and secretion. Protein Expr. Purif. 1996, 7, 355–366. [Google Scholar] [CrossRef]
- Prame Kumar, K.; Nicholls, A.J.; Wong, C.H.Y. Partners in crime: Neutrophils and monocytes/macrophages in inflammation and disease. Cell Tissue Res. 2018, 371, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Roosen, S.; Exner, K.; Paul, S.; Schroder, J.M.; Kalm, E.; Looft, C. Bovine beta-defensins: Identification and characterization of novel bovine beta-defensin genes and their expression in mammary gland tissue. Mamm. Genome 2004, 15, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Kosciuczuk, E.M.; Lisowski, P.; Jarczak, J.; Krzyzewski, J.; Zwierzchowski, L.; Bagnicka, E. Expression patterns of beta-defensin and cathelicidin genes in parenchyma of bovine mammary gland infected with coagulase-positive or coagulase-negative Staphylococci. BMC Vet. Res. 2014, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Siegert, S.; Fischer, A. β-defensin-4 as an endogenous biomarker in cows with mastitis. Front. Vet. Sci. 2023, 10, 1154386. [Google Scholar] [CrossRef] [PubMed]
- Bastos, P.A.; Wheeler, R.; Boneca, I.G. Uptake, recognition and responses to peptidoglycan in the mammalian host. FEMS Microbiol. Rev. 2021, 45, fuaa044. [Google Scholar] [CrossRef]
- Zabolewicz, T.; Puckowska, P.; Brym, P.; Oleński, K.; Kamiński, S. Relationship between polymorphism within peptidoglycan recognition protein 1 gene (PGLYRP1) and somatic cell counts in milk of Holstein cows. Ann. Anim. Sci. 2022, 22, 593–599. [Google Scholar] [CrossRef]
- Mudaliar, M.; Tassi, R.; Thomas, F.C.; McNeilly, T.N.; Weidt, S.K.; McLaughlin, M.; Wilson, D.; Burchmore, R.; Herzyk, P.; Eckersall, P.D.; et al. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 2. Label-free relative quantitative proteomics. Mol. Biosyst. 2016, 12, 2748–2761. [Google Scholar] [CrossRef]
- Wang, C.; Liu, J.; Liu, H. Proteomic analysis of the effects of lutein on mammary gland metabolism in dairy cows. J. Dairy Res. 2018, 85, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Mahadev, K.; Raval, G.; Bharadwaj, S.; Willingham, M.C.; Lange, E.M.; Vonderhaar, B.; Salomon, D.; Prasad, G.L. Suppression of the transformed phenotype of breast cancer by tropomyosin-1. Exp. Cell Res. 2002, 279, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Lacolley, P.; Challande, P.; Boumaza, S.; Cohuet, G.; Laurent, S.; Boutouyrie, P.; Grimaud, J.A.; Paulin, D.; Lamaziere, J.M.; Li, Z. Mechanical properties and structure of carotid arteries in mice lacking desmin. Cardiovasc. Res. 2001, 51, 178–187. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Andrei, S.; Culea, M.; Matei, S.; Pintea, A.; Groza, I.S. Amino acid concentration in normal and subclinical mastitis milk. Phys. Fluids 2011, 23, 15. [Google Scholar]
- Yoshida, M.; Shinohara, H.; Sugiyama, T.; Kumagai, M.; Muto, H.; Kodama, H. Taste of milk from inflamed breasts of breastfeeding mothers with mastitis evaluated using a taste sensor. Breastfeed Med. 2014, 9, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Mitschke, L.; Parthier, C.; Schroder-Tittmann, K.; Coy, J.; Ludtke, S.; Tittmann, K. The crystal structure of human transketolase and new insights into its mode of action. J. Biol. Chem. 2010, 285, 31559–31570. [Google Scholar] [CrossRef]
- Burrai, G.P.; Tanca, A.; Cubeddu, T.; Abbondio, M.; Polinas, M.; Addis, M.F.; Antuofermo, E. A first immunohistochemistry study of transketolase and transketolase-like 1 expression in canine hyperplastic and neoplastic mammary lesions. BMC Vet. Res. 2017, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Fhaikrue, I.; Srisawat, W.; Nambooppha, B.; Pringproa, K.; Thongtharb, A.; Prachasilchai, W.; Sthitmatee, N. Identification of potential canine mammary tumour cell biomarkers using proteomic approach: Differences in protein profiles among tumour and normal mammary epithelial cells by two-dimensional electrophoresis-based mass spectrometry. Vet. Comp. Oncol. 2020, 18, 787–795. [Google Scholar] [CrossRef]
- Mendes, S.R.; Gomis-Rüth, F.X.; Goulas, T. Frozen fresh blood plasma preserves the functionality of native human α2-macroglobulin. Sci. Rep. 2023, 13, 4579. [Google Scholar] [CrossRef]
- Kusebauch, U.; Hernandez-Castellano, L.E.; Bislev, S.L.; Moritz, R.L.; Rontved, C.M.; Bendixen, E. Selected reaction monitoring mass spectrometry of mastitis milk reveals pathogen-specific regulation of bovine host response proteins. J. Dairy Sci. 2018, 101, 6532–6541. [Google Scholar] [CrossRef] [PubMed]
- Rohde, H.; Burdelski, C.; Bartscht, K.; Hussain, M.; Buck, F.; Horstkotte, M.A.; Knobloch, J.K.; Heilmann, C.; Herrmann, M.; Mack, D. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 2005, 55, 1883–1895. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.E.; Lazarus, L.H.; Tomer, K.B. Bradykinin and kininogens in bovine milk. J. Biol. Chem. 1989, 264, 17777–17783. [Google Scholar] [CrossRef] [PubMed]
- Mulakala, B.; Smith, K.; Snider, M.; Ayers, A.; Honan, M.; Greenwood, S. Use of milk proteins as biomarkers of changes in the rumen metaproteome of Holstein cows fed low fiber, high starch diets. J. Dairy Sci. 2023, 106, 9630–9643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Boeren, S.; van Hooijdonk, A.C.; Vervoort, J.M.; Hettinga, K.A. A proteomic perspective on the changes in milk proteins due to high somatic cell count. J. Dairy Sci. 2015, 98, 5339–5351. [Google Scholar] [CrossRef]
- Alonso-Fauste, I.; Andres, M.; Iturralde, M.; Lampreave, F.; Gallart, J.; Alava, M.A. Proteomic characterization by 2-DE in bovine serum and whey from healthy and mastitis affected farm animals. J. Proteom. 2012, 75, 3015–3030. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, H.; Heikaus, L.; Long, A.T.; Naudin, C.; Schluter, H.; Renne, T. The plasma contact system, a protease cascade at the nexus of inflammation, coagulation and immunity. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2118–2127. [Google Scholar] [CrossRef] [PubMed]

| Accession No. | Protein Name | Selected Peptide Sequence | S1 Group 1 | S2 Group 2 | S3 Group 2 | S4 Group 2 | S5 Group 2 | S1/S2 3 | S1/S3 3 | S1/S4 3 | S1/S5 3 | −Log p Value 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G3MYN2 | Interleukin 18 binding protein | LWEGSTR | 7.75 × 105 | 1.37 × 106 | 1.71 × 106 | 2.38 × 106 | 3.23 × 106 | 0.57 | 0.45 | 0.33 | 0.24 | 5.41 |
| G3N0Q8 | Azurocidin 1 | ARPQELPFLASIQNQGR; GTDFFAR | 4.79 × 105 | 2.60 × 106 | 5.92 × 106 | 8.51 × 106 | 2.13 × 107 | 0.19 | 0.08 | 0.06 | 0.02 | 6.79 |
| O62654 | Desmin | VELQELNDR | 8.12 × 106 | 1.15 × 106 | 3.87 × 105 | 4.20 × 105 | 3.76 × 105 | 7.07 | 20.95 | 19.34 | 21.57 | 1.33 |
| P22226 | Cathelicidin-1 | AVDQLNEQSSEPNIYR | 3.86 × 106 | 1.79 × 107 | 2.59 × 107 | 4.58 × 107 | 8.47 × 107 | 0.22 | 0.15 | 0.08 | 0.05 | 8.44 |
| P33046 | Cathelicidin-4 | AVDQLNELSSEANLYR; TIQQPAEQCDFK | 1.73 × 106 | 6.56 × 106 | 1.55 × 107 | 2.55 × 107 | 6.87 × 107 | 0.26 | 0.11 | 0.07 | 0.02 | 6.94 |
| P54228 | Cathelicidin-6 | TSQQPAEQCDFK | 5.54 × 105 | 2.44 × 106 | 4.82 × 106 | 8.01 × 106 | 1.90 × 107 | 0.23 | 0.12 | 0.07 | 0.03 | 8.06 |
| P54229 | Cathelicidin-5 | TSQQSPEQCDFK; YGPIIVPIIR | 3.93 × 105 | 1.90 × 106 | 3.16 × 106 | 6.52 × 106 | 1.13 × 107 | 0.21 | 0.13 | 0.07 | 0.04 | 5.44 |
| Q0VCW4 | Serine dehydratase/ threonine deaminase | LVTLPCITSVAK | 2.89 × 105 | 4.36 × 105 | 3.59 × 105 | 4.94 × 105 | 1.73 × 106 | 0.66 | 0.80 | 0.58 | 0.17 | 2.55 |
| Q1JPB0 | Leukocyte elastase inhibitor | VLELPYEGK; IEQQLTLEK | 5.61 × 105 | 1.52 × 106 | 2.16 × 106 | 3.02 × 106 | 5.20 × 106 | 0.34 | 0.23 | 0.15 | 0.08 | 5.48 |
| Q3ZC00 | Lymphocyte cytosolic protein 1 | AYYHLLEQVAPK | 0.00 | 0.00 | 0.00 | 1.52 × 105 | 4.78 × 105 | 0.00 | 0.00 | 0.00 | 0.00 | 2.97 |
| Q8SPP7 | Peptidoglycan recognition protein 1 | QAQNVQYYHVR; DVQQTLSPGDELYK | 6.37 × 106 | 2.33 × 107 | 4.31 × 107 | 6.07 × 107 | 1.39E+08 | 0.28 | 0.16 | 0.11 | 0.05 | 6.82 |
| Pathway Name | Counts 1 | Percent (%) 2 | p Value 3 | Fold Enrichment 4 |
|---|---|---|---|---|
| Complement and coagulation cascades | 13 | 9.85 | 1.22E-12 | 19.51 |
| Staphylococcus aureus infection | 7 | 5.30 | 1.21E-05 | 13.17 |
| Salivary secretion | 7 | 5.30 | 8.59E-05 | 9.36 |
| Antigen processing and presentation | 6 | 4.55 | 5.05E-04 | 8.88 |
| Protein processing in endoplasmic reticulum | 7 | 5.30 | 0.004 | 4.60 |
| Spliceosome | 6 | 4.55 | 0.006 | 5.16 |
| Biosynthesis of amino acids | 4 | 3.03 | 0.025 | 6.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Chen, K.; Zang, C.; Zhao, X.; Cheng, Z.; Li, X.; Wang, C.; Chen, Y.; Yang, K. Alterations in Whey Protein Abundance Correlated with the Somatic Cell Count Identified via Label-Free and Selected Reaction Monitoring Proteomic Approaches. Animals 2025, 15, 675. https://doi.org/10.3390/ani15050675
Li J, Chen K, Zang C, Zhao X, Cheng Z, Li X, Wang C, Chen Y, Yang K. Alterations in Whey Protein Abundance Correlated with the Somatic Cell Count Identified via Label-Free and Selected Reaction Monitoring Proteomic Approaches. Animals. 2025; 15(5):675. https://doi.org/10.3390/ani15050675
Chicago/Turabian StyleLi, Jing, Kaixu Chen, Changjiang Zang, Xiaowei Zhao, Zhiqiang Cheng, Xiaobin Li, Caidie Wang, Yong Chen, and Kailun Yang. 2025. "Alterations in Whey Protein Abundance Correlated with the Somatic Cell Count Identified via Label-Free and Selected Reaction Monitoring Proteomic Approaches" Animals 15, no. 5: 675. https://doi.org/10.3390/ani15050675
APA StyleLi, J., Chen, K., Zang, C., Zhao, X., Cheng, Z., Li, X., Wang, C., Chen, Y., & Yang, K. (2025). Alterations in Whey Protein Abundance Correlated with the Somatic Cell Count Identified via Label-Free and Selected Reaction Monitoring Proteomic Approaches. Animals, 15(5), 675. https://doi.org/10.3390/ani15050675





