Integrative Analysis of Gene Networks Associated with Adipose and Muscle Traits in Hanwoo Steers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and RNA-Seq Data Generation
2.2. Quality Control and Mapping of RNA-Seq Data
2.3. Construction of Gene Co-Expression Network of Fat and Muscle Tissues
2.4. Identification of Co-Expressed Gene Modules in Fat and Muscle Tissues via WGCNA
2.5. Functional Enrichment Analysis of Tissue-Specific Gene Co-Expression Modules
2.6. Identification of Module-Specific Hub Genes via PPI and Functional Network Analysis
3. Results
3.1. QC and Mapping of RNA-Seq Data
3.2. Construction of Gene Co-Expression Network of Fat and Muscle Tissues
3.3. Identification of Hub Genes in Fat and Muscle Tissues via WGCNA
3.4. Functional Enrichment Analysis in Each Module Using Gene Ontology (GO) and KEGG Pathway
3.5. Identification of the Trait-Associated Hub Genes from Gene Modules
3.6. Functional Interpretation of Tissue-Specific Gene Modules Using ClueGO and CluePedia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declaration of Generative AI
References
- Hocquette, J.-F.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef]
- Kwon, E.G.; Park, B.K.; Kim, H.C.; Cho, Y.M.; Kim, T.I.; Chang, S.S.; Oh, Y.K.; Kim, N.K.; Kim, J.H.; Kim, Y.J. Effects of fattening period on growth performance, carcass characteristics and lipogenic gene expression in Hanwoo steers. Asian-Australas. J. Anim. Sci. 2009, 22, 1654–1660. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Yu, H.; Yang, Z.; Wang, J.; Li, H.; Li, X.; Liang, E.; Mei, C.; Zan, L. Identification of key genes and metabolites involved in meat quality performance in Qinchuan cattle by WGCNA. J. Integr. Agric. 2024, 23, 3923–3937. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Chen, H.; Yang, J.; Wu, W. Seven key hub genes identified by gene co-expression network in cutaneous squamous cell carcinoma. BMC Cancer 2021, 21, 852. [Google Scholar] [CrossRef]
- Salcedo-Tacuma, D.; Parales-Giron, J.; Prom, C.; Chirivi, M.; Laguna, J.; Lock, A.L.; Contreras, G.A. Transcriptomic profiling of adipose tissue inflammation, remodeling, and lipid metabolism in periparturient dairy cows (Bos taurus). BMC Genom. 2020, 21, 824. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Du, L.; Zhang, Y.; Liu, G. NEFAs influence the inflammatory and insulin signaling pathways through TLR4 in primary calf hepatocytes in vitro. Front. Vet. Sci. 2021, 8, 755505. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, N.; Fernández-Veledo, S.; Punzón, C.; Moreno, C.; Barrocal, B.; Sreeramkumar, V.; Desco, M.; Fresno, M. Opposing actions of TLR2 and TLR4 in adipocyte differentiation and mature-onset obesity. Int. J. Mol. Sci. 2022, 23, 15682. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Petkova, A.P.; Granneman, J.G. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metab. 2013, 18, 355–367. [Google Scholar] [CrossRef]
- Li, G.; Fang, X.; Liu, Y.; Lu, X.; Liu, Y.; Li, Y.; Zhao, Z.; Liu, B.; Yang, R. Lipid Regulatory Element Interact with CD44 on Mitochondrial Bioenergetics in Bovine Adipocyte Differentiation and Lipometabolism. J. Agric. Food Chem. 2024, 72, 17481–17498. [Google Scholar] [CrossRef]
- Kodama, K.; Toda, K.; Morinaga, S.; Yamada, S.; Butte, A.J. Anti-CD44 antibody treatment lowers hyperglycemia and improves insulin resistance, adipose inflammation, and hepatic steatosis in diet-induced obese mice. Diabetes 2015, 64, 867–875. [Google Scholar] [CrossRef]
- Chirivi, M.; Abou-Rjeileh, U.; Myers, M.; Parales-Giron, J.; Worden, L.; Lock, A.L.; Contreras, G.A. TLR4 and prostaglandin pathways at the crossroads of endotoxemia-induced lipolysis. Front. Immunol. 2025, 16, 1591210. [Google Scholar] [CrossRef]
- Sadhu, L.; Tsopoulidis, N.; Hasanuzzaman, M.; Laketa, V.; Way, M.; Fackler, O.T. ARPC5 isoforms and their regulation by calcium-calmodulin-N-WASP drive distinct Arp2/3-dependent actin remodeling events in CD4 T cells. eLife 2023, 12, e82450. [Google Scholar] [CrossRef] [PubMed]
- Garcia, I.S.; Silva-Vignato, B.; Cesar, A.S.M.; Petrini, J.; da Silva, V.H.; Morosini, N.S.; Goes, C.P.; Afonso, J.; da Silva, T.R.; Lima, B.D.; et al. Novel putative causal mutations associated with fat traits in Nellore cattle uncovered by eQTLs located in open chromatin regions. Sci. Rep. 2024, 14, 10094. [Google Scholar] [CrossRef] [PubMed]
- Olaso, E.; Labrador, J.-P.; Wang, L.; Ikeda, K.; Eng, F.J.; Klein, R.; Lovett, D.H.; Lin, H.C.; Friedman, S.L. Discoidin domain receptor 2 regulates fibroblast proliferation and migration through the extracellular matrix in association with transcriptional activation of matrix metalloproteinase-2. J. Biol. Chem. 2002, 277, 3606–3613. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Choi, Y.M. Correlation of marbling characteristics with meat quality and histochemical characteristics in longissimus thoracis muscle from Hanwoo steers. Food Sci. Anim. Resour. 2019, 39, 151. [Google Scholar] [CrossRef]
- Gajaweera, C.; Kang, D.H.; Lee, D.H.; Kim, Y.-K.; Park, B.H.; Chang, S.S.; Kim, U.H.; Lee, S.H.; Chung, K.Y. Development of nutrigenomic based precision management model for Hanwoo steers. J. Anim. Sci. Technol. 2023, 65, 596. [Google Scholar] [CrossRef]
- Chen, C.; Ren, H.; Li, H.; Deng, Y.; Cui, Q.; Zhu, J.; Zhang, S.; Yu, J.; Wang, H.; Yu, X. Identification of crucial modules and genes associated with backfat tissue development by WGCNA in Ningxiang pigs. Front. Genet. 2023, 14, 1234757. [Google Scholar] [CrossRef]
- Prasad, S.S.; Garg, A.; Agarwal, A.K. Enzymatic activities of the human AGPAT isoform 3 and isoform 5: Localization of AGPAT5 to mitochondria [S]. J. Lipid Res. 2011, 52, 451–462. [Google Scholar] [CrossRef]
- Tan, Z.; Jiang, H. Molecular and cellular mechanisms of intramuscular fat development and growth in cattle. Int. J. Mol. Sci. 2024, 25, 2520. [Google Scholar] [CrossRef]
- Li, X.; Wang, M.; Li, S.; Chen, Y.; Wang, M.; Wu, Z.; Sun, X.; Yao, L.; Dong, H.; Song, Y. HIF-1-induced mitochondrial ribosome protein L52: A mechanism for breast cancer cellular adaptation and metastatic initiation in response to hypoxia. Theranostics 2021, 11, 7337. [Google Scholar] [CrossRef]
- Dorji, J.; MacLeod, I.M.; Chamberlain, A.J.; Vander Jagt, C.J.; Ho, P.N.; Khansefid, M.; Mason, B.A.; Prowse-Wilkins, C.P.; Marett, L.C.; Wales, W.J. Mitochondrial protein gene expression and the oxidative phosphorylation pathway associated with feed efficiency and energy balance in dairy cattle. J. Dairy Sci. 2021, 104, 575–587. [Google Scholar] [CrossRef]
- McKenna, C.; Porter, R.; Fitzsimons, C.; Waters, S.; McGee, M.; Kenny, D. Mitochondrial abundance and function in skeletal muscle and liver from Simmental beef cattle divergent for residual feed intake. Animal 2020, 14, 1710–1717. [Google Scholar] [CrossRef]
- Tegeler, A.P.; Ford, H.R.; Fiallo-Diez, J.F.; Michelotti, T.C.; Johnson, B.J.; Benitez, O.J.; Woerner, D.R.; Strieder-Barboza, C. Transcriptome and Cellular Evidence of Depot-Specific Function in Beef Cattle Intramuscular, Subcutaneous, and Visceral Adipose Tissues. Biology 2025, 14, 848. [Google Scholar] [CrossRef]
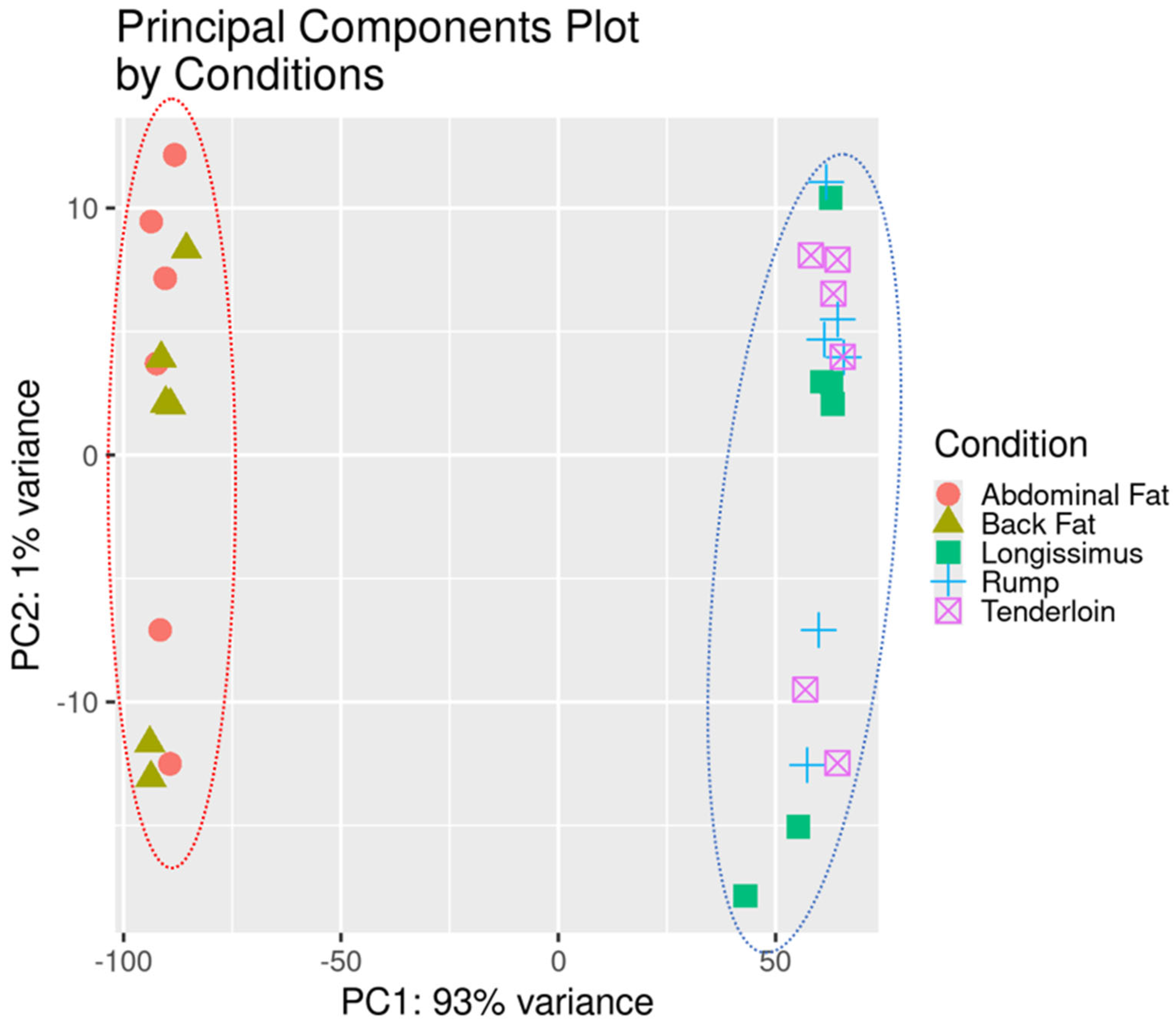

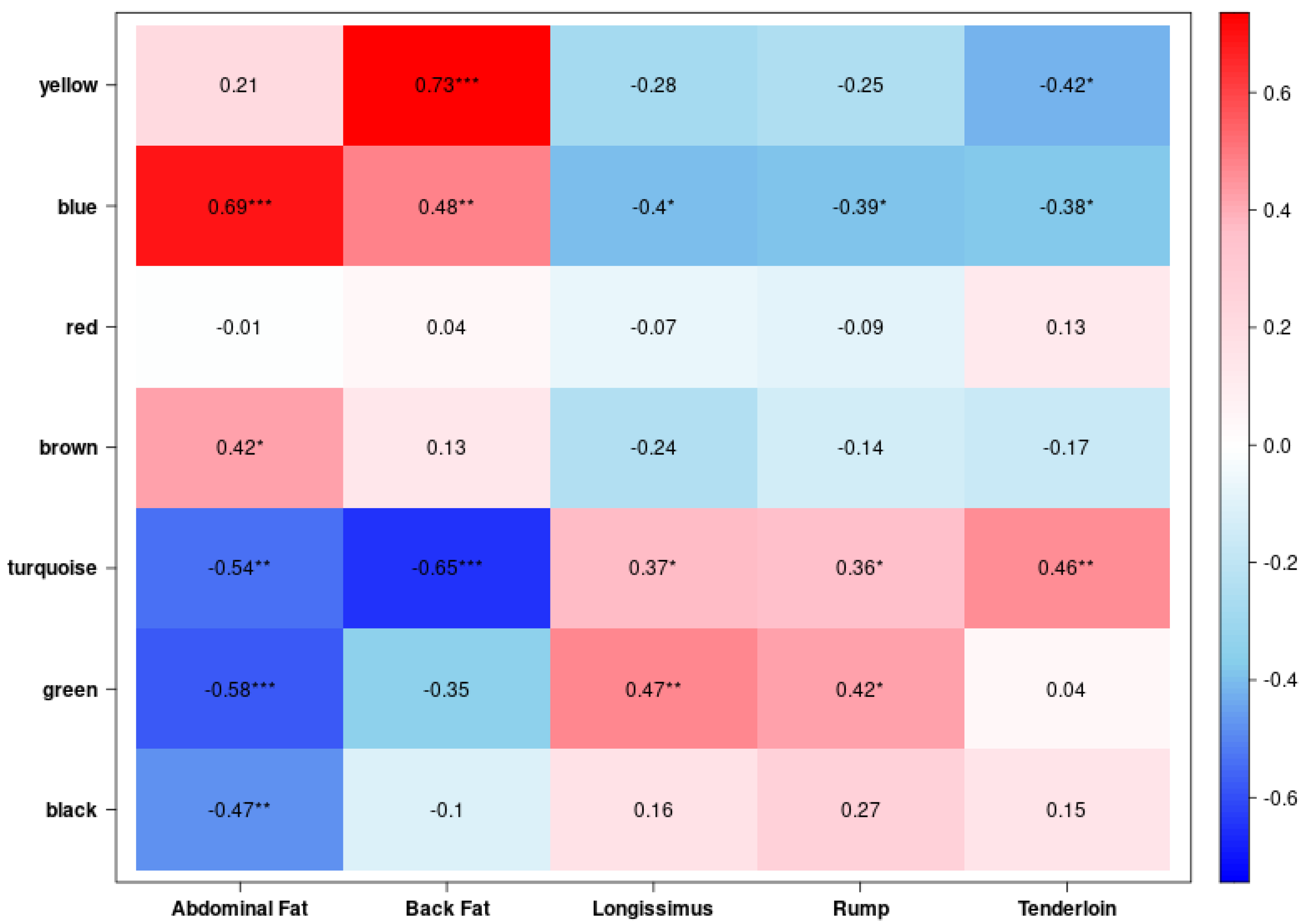
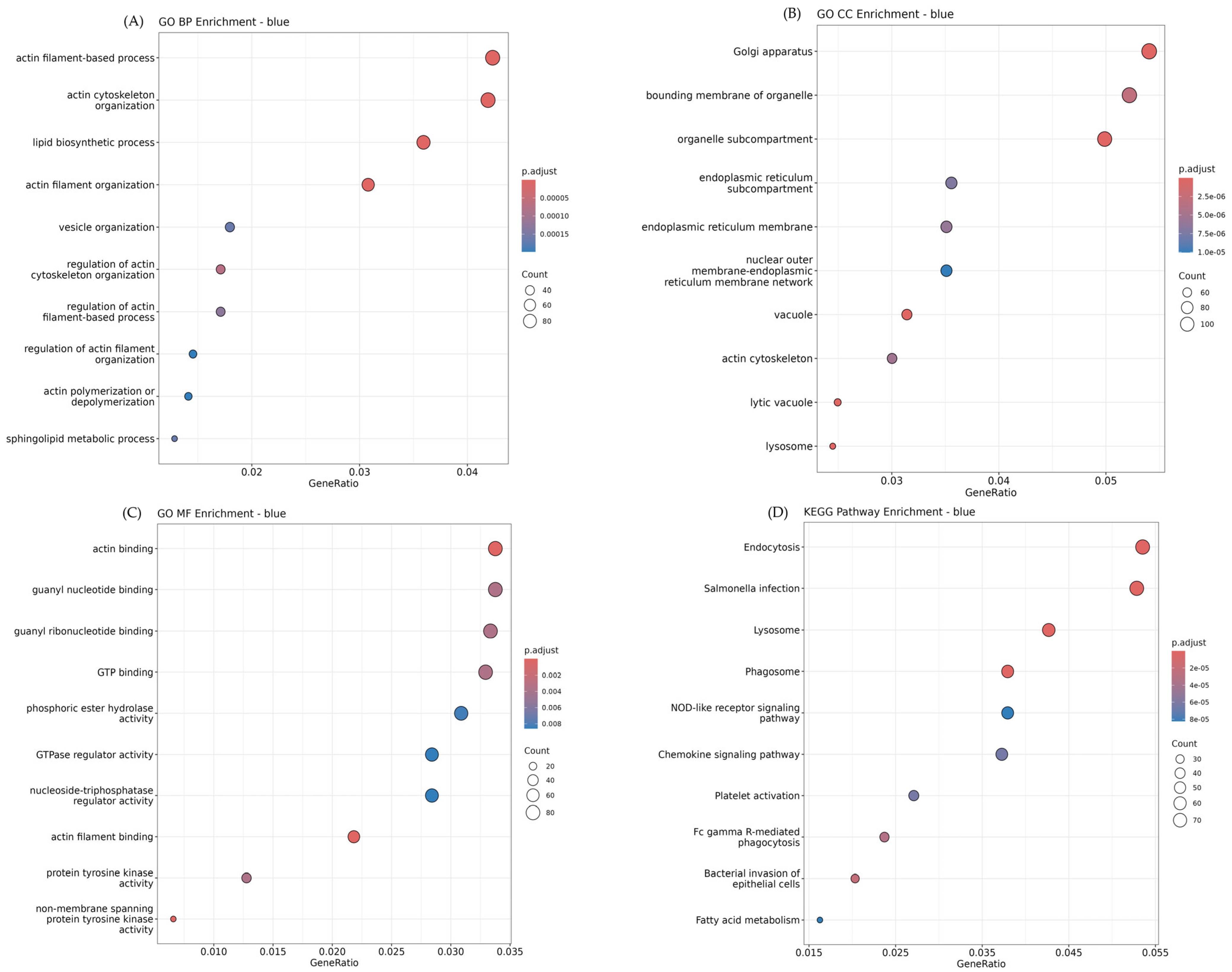

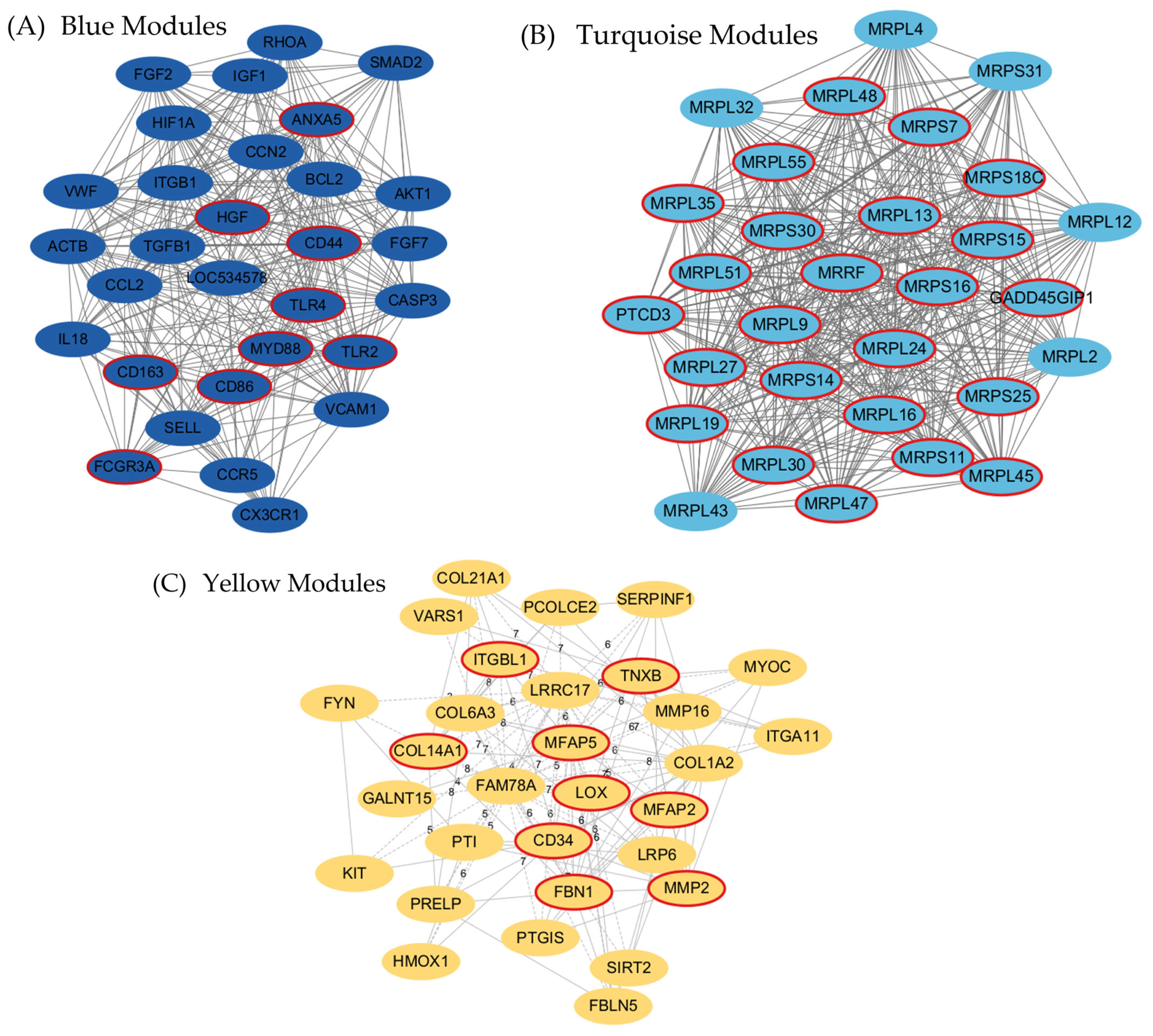

| Module | Gene Count in Module | Hub Gene |
|---|---|---|
| Yellow | 170 | DDR2 |
| Turquoise | 5987 | AGPAT5 |
| Brown | 280 | RREB1 |
| Red | 79 | RPL34 |
| Blue | 3501 | ARPC5 |
| Black | 55 | LOC112442783 |
| Green | 145 | LOC112443631 |
| Module | Total Genes in Module | Genes MM > 0.9 | PPI Top 30 | True Hub Genes |
|---|---|---|---|---|
| Blue | 3501 | 568 | 30 | 9 |
| Turquoise | 5987 | 487 | 30 | 24 |
| Yellow | 170 | 21 | 30 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, S.; Jeong, T.; Lee, J.; Park, W.; Jang, S.; Lim, D. Integrative Analysis of Gene Networks Associated with Adipose and Muscle Traits in Hanwoo Steers. Animals 2025, 15, 3201. https://doi.org/10.3390/ani15213201
Hwang S, Jeong T, Lee J, Park W, Jang S, Lim D. Integrative Analysis of Gene Networks Associated with Adipose and Muscle Traits in Hanwoo Steers. Animals. 2025; 15(21):3201. https://doi.org/10.3390/ani15213201
Chicago/Turabian StyleHwang, Suk, Taejoon Jeong, Junyoung Lee, Woncheoul Park, Sunsik Jang, and Dajeong Lim. 2025. "Integrative Analysis of Gene Networks Associated with Adipose and Muscle Traits in Hanwoo Steers" Animals 15, no. 21: 3201. https://doi.org/10.3390/ani15213201
APA StyleHwang, S., Jeong, T., Lee, J., Park, W., Jang, S., & Lim, D. (2025). Integrative Analysis of Gene Networks Associated with Adipose and Muscle Traits in Hanwoo Steers. Animals, 15(21), 3201. https://doi.org/10.3390/ani15213201





