Lumpy Skin Disease Virus Pathogenesis: Viral Protein Functions and Comparative Insights from Vaccinia Virus
Simple Summary
Abstract
1. Introduction
2. Methods
3. Lumpy Skin Disease Virus
4. LSDV Virion Architecture and Key Structural Proteins
4.1. Lateral Body Proteins
4.2. Membrane Proteins (IMV and EEV)
4.3. Core Proteins
5. Molecular Mechanisms of LSDV Proteins in the Viral Life Cycle
5.1. Proteins Orchestrating Viral Entry
5.2. Proteins Driving Viral Replication
5.3. DNA Integrity and Genome Maintenance
5.4. Viral Transcription Machinery
5.5. Viral Proteins Involved in Translation
5.6. Proteins Involved in Virion Assembly, Maturation, and Egress
6. LSDV Immunomodulatory Strategies
6.1. Interference with Host Innate Immune Signaling Pathways
6.2. LSDV-Encoded Proteins That Subvert Host Adaptive Immunity
6.3. Modulation of Host Cell Apoptosis and Autophagy
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Kutish, G.F.; Rock, D.L. Genome of lumpy skin disease virus. J. Virol. 2001, 75, 7122–7130. [Google Scholar] [CrossRef] [PubMed]
- Abdalhamed, A.M.; Naser, S.M.; Mohamed, A.H.; Zeedan, G.S.G. Development of gold nanoparticles-lateral flow test as a novel field diagnostic assay for detecting foot-and-mouth disease and lumpy skin disease viruses. Iran. J. Microbiol. 2022, 14, 574–586. [Google Scholar] [CrossRef]
- Smaraki, N.; Jogi, H.R.; Kamothi, D.J.; Savsani, H.H. An insight into emergence of lumpy skin disease virus: A threat to Indian cattle. Arch. Microbiol. 2024, 206, 210. [Google Scholar] [CrossRef]
- Modethed, W.; Kreausukon, K.; Singhla, T.; Boonsri, K.; Pringproa, K.; Sthitmatee, N.; Vinitchaikul, P.; Srisawang, S.; Salvador, R.; Gubbins, S.; et al. An evaluation of financial losses due to lumpy skin disease outbreaks in dairy farms of northern Thailand. Front. Vet. Sci. 2024, 11, 1501460. [Google Scholar] [CrossRef]
- Tuppurainen, E.; Dietze, K.; Wolff, J.; Bergmann, H.; Beltran-Alcrudo, D.; Fahrion, A.; Lamien, C.E.; Busch, F.; Sauter-Louis, C.; Conraths, F.J.; et al. Review: Vaccines and Vaccination against Lumpy Skin Disease. Vaccines 2021, 9, 1136. [Google Scholar] [CrossRef]
- Hamdi, J.; Munyanduki, H.; Omari Tadlaoui, K.; El Harrak, M.; Fassi Fihri, O. Capripoxvirus Infections in Ruminants: A Review. Microorganisms 2021, 9, 902. [Google Scholar] [CrossRef]
- Whittle, L.; Chapman, R.; Williamson, A.L. Lumpy Skin Disease-An Emerging Cattle Disease in Europe and Asia. Vaccines 2023, 11, 578. [Google Scholar] [CrossRef]
- Ul-Rahman, A.; Shabbir, M.Z.; Raza, M.A.; Rossiter, P. The expanding host range of lumpy skin disease virus in wild and domestic animals. Trop. Anim. Health Prod. 2024, 56, 269. [Google Scholar] [CrossRef]
- Sprygin, A.; Van Schalkwyk, A.; Shumilova, I.; Nesterov, A.; Kononova, S.; Prutnikov, P.; Byadovskaya, O.; Kononov, A. Full-length genome characterization of a novel recombinant vaccine-like lumpy skin disease virus strain detected during the climatic winter in Russia, 2019. Arch. Virol. 2020, 165, 2675–2677. [Google Scholar] [CrossRef] [PubMed]
- Schlosser-Perrin, L.; Holzmuller, P.; Fernandez, B.; Miotello, G.; Dahmani, N.; Neyret, A.; Bertagnoli, S.; Armengaud, J.; Caufour, P. Constitutive proteins of lumpy skin disease virion assessed by next-generation proteomics. J. Virol. 2023, 97, e0072323. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Venkatesan, G.; Kushwaha, A.; Poulinlu, G.; Saha, T.; Ramakrishnan, M.A.; Dhar, P.; Singh, R.K. Genomic characterization of Lumpy Skin Disease virus (LSDV) from India: Circulation of Kenyan-like LSDV strains with unique kelch-like proteins. Acta. Trop. 2023, 241, 106838. [Google Scholar] [CrossRef]
- Zhang, M.; Shi, Y.; Lu, X.; Zhang, Q.; Zhao, Y.; Li, S.; Wen, Z.; Ge, J.; Wang, X.; Li, J.; et al. Lumpy skin disease virus 001/156 protein is a virulence factor that suppresses interferon production through impairing IRF3 dimerization. PLoS Pathog. 2025, 21, e1013362. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Sharma, S.; Tripathi, B.N. Pathogenicity and virulence of lumpy skin disease virus: A comprehensive update. Virulence 2025, 16, 2495108. [Google Scholar] [CrossRef]
- Biswas, S.; Noyce, R.S.; Babiuk, L.A.; Lung, O.; Bulach, D.M.; Bowden, T.R.; Boyle, D.B.; Babiuk, S.; Evans, D.H. Extended sequencing of vaccine and wild-type capripoxvirus isolates provides insights into genes modulating virulence and host range. Transbound. Emerg. Dis. 2020, 67, 80–97. [Google Scholar] [CrossRef] [PubMed]
- Sprygin, A.; Pestova, Y.; Bjadovskaya, O.; Prutnikov, P.; Zinyakov, N.; Kononova, S.; Ruchnova, O.; Lozovoy, D.; Chvala, I.; Kononov, A. Evidence of recombination of vaccine strains of lumpy skin disease virus with field strains, causing disease. PLoS ONE 2020, 15, e0232584. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, F.; Mathijs, E.; Philips, W.; Saduakassova, M.; De Leeuw, I.; Sultanov, A.; Haegeman, A.; De Clercq, K. Recombinant LSDV Strains in Asia: Vaccine Spillover or Natural Emergence? Viruses 2022, 14, 1429. [Google Scholar] [CrossRef]
- Sendow, I.; Meki, I.K.; Dharmayanti, N.L.P.I.; Hoerudin, H.; Ratnawati, A.; Settypalli, T.B.K.; Ahmed, H.O.; Nuradji, H.; Saepulloh, M.; Adji, R.S.; et al. Molecular characterization of recombinant LSDV isolates from 2022 outbreak in Indonesia through phylogenetic networks and whole-genome SNP-based analysis. BMC Genom. 2024, 25, 240. [Google Scholar] [CrossRef]
- Nesterov, A.; Mazloum, A.; Byadovskaya, O.; Shumilova, I.; Van Schalkwyk, A.; Krotova, A.; Kirpichenko, V.; Donnik, I.; Chvala, I.; Sprygin, A. Experimentally controlled study indicates that the naturally occurring recombinant vaccine-like lumpy skin disease strain Udmurtiya/2019, detected during freezing winter in northern latitudes, is transmitted via indirect contact. Front. Vet. Sci. 2022, 9, 1001426. [Google Scholar] [CrossRef]
- Kononova, S.; Kononov, A.; Shumilova, I.; Byadovskaya, O.; Nesterov, A.; Prutnikov, P.; Babiuk, S.; Sprygin, A. A lumpy skin disease virus which underwent a recombination event demonstrates more aggressive growth in primary cells and cattle than the classical field isolate. Transbound. Emerg. Dis. 2021, 68, 1377–1383. [Google Scholar] [CrossRef]
- Hollinshead, M.; Vanderplasschen, A.; Smith, G.L.; Vaux, D.J. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 1999, 73, 1503–1517. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.L.; Vanderplasschen, A.; Law, M. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 2002, 83, 2915–2931. [Google Scholar] [CrossRef] [PubMed]
- Meade, N.; King, M.; Munger, J.; Walsh, D. mTOR Dysregulation by Vaccinia Virus F17 Controls Multiple Processes with Varying Roles in Infection. J. Virol. 2019, 93, e00784-19. [Google Scholar] [CrossRef] [PubMed]
- Meade, N.; Toreev, H.K.; Chakrabarty, R.P.; Hesser, C.R.; Park, C.; Chandel, N.S.; Walsh, D. The poxvirus F17 protein counteracts mitochondrially orchestrated antiviral responses. Nat. Commun. 2023, 14, 7889. [Google Scholar] [CrossRef]
- Schmidt, F.I.; Bleck, C.K.; Reh, L.; Novy, K.; Wollscheid, B.; Helenius, A.; Stahlberg, H.; Mercer, J. Vaccinia virus entry is followed by core activation and proteasome-mediated release of the immunomodulatory effector VH1 from lateral bodies. Cell Rep. 2013, 4, 464–476. [Google Scholar] [CrossRef]
- Bidgood, S.R.; Samolej, J.; Novy, K.; Collopy, A.; Albrecht, D.; Krause, M.; Burden, J.J.; Wollscheid, B.; Mercer, J. Poxviruses package viral redox proteins in lateral bodies and modulate the host oxidative response. PLoS Pathog. 2022, 18, e1010614. [Google Scholar] [CrossRef] [PubMed]
- Chervyakova, O.V.; Zaitsev, V.L.; Iskakov, B.K.; Tailakova, E.T.; Strochkov, V.M.; Sultankulova, K.T.; Sandybayev, N.T.; Stanbekova, G.E.; Beisenov, D.K.; Abduraimov, Y.O.; et al. Recombinant Sheep Pox Virus Proteins Elicit Neutralizing Antibodies. Viruses 2016, 8, 159. [Google Scholar] [CrossRef]
- Kutumbetov, L.; Ragatova, A.; Azanbekova, M.; Myrzakhmetova, B.; Aldayarov, N.; Zhugunissov, K.; Abduraimov, Y.; Nissanova, R.; Sarzhigitova, A.; Kemalova, N.; et al. Investigation of the Pathogenesis of Lumpy Skin Disease Virus in Indigenous Cattle in Kazakhstan. Pathogens 2025, 14, 577. [Google Scholar] [CrossRef]
- Ho, Y.; Hsiao, J.C.; Yang, M.H.; Chung, C.S.; Peng, Y.C.; Lin, T.H.; Chang, W.; Tzou, D.L. The oligomeric structure of vaccinia viral envelope protein A27L is essential for binding to heparin and heparan sulfates on cell surfaces: A structural and functional approach using site-specific mutagenesis. J. Mol. Biol. 2005, 349, 1060–1071. [Google Scholar] [CrossRef]
- Singh, K.; Gittis, A.G.; Gitti, R.K.; Ostazeski, S.A.; Su, H.P.; Garboczi, D.N. The Vaccinia Virus H3 Envelope Protein, a Major Target of Neutralizing Antibodies, Exhibits a Glycosyltransferase Fold and Binds UDP-Glucose. J. Virol. 2016, 90, 5020–5030. [Google Scholar] [CrossRef]
- Erlandson, K.J.; Bisht, H.; Weisberg, A.S.; Hyun, S.I.; Hansen, B.T.; Fischer, E.R.; Hinshaw, J.E.; Moss, B. Poxviruses Encode a Reticulon-Like Protein that Promotes Membrane Curvature. Cell Rep. 2016, 14, 2084–2091. [Google Scholar] [CrossRef]
- Wagenaar, T.R.; Ojeda, S.; Moss, B. Vaccinia virus A56/K2 fusion regulatory protein interacts with the A16 and G9 subunits of the entry fusion complex. J. Virol. 2008, 82, 5153–5160. [Google Scholar] [CrossRef]
- Bryk, P.; Brewer, M.G.; Ward, B.M. Vaccinia Virus Phospholipase Protein F13 Promotes Rapid Entry of Extracellular Virions into Cells. J. Virol. 2018, 92, e02154-17. [Google Scholar] [CrossRef] [PubMed]
- Monticelli, S.R.; Earley, A.K.; Tate, J.; Ward, B.M. The Ectodomain of the Vaccinia Virus Glycoprotein A34 Is Required for Cell Binding by Extracellular Virions and Contains a Large Region Capable of Interaction with Glycoprotein B5. J. Virol. 2019, 93, e01343-18. [Google Scholar] [CrossRef]
- Monticelli, S.R.; Earley, A.K.; Stone, R.; Norbury, C.C.; Ward, B.M. Vaccinia Virus Glycoproteins A33, A34, and B5 Form a Complex for Efficient Endoplasmic Reticulum to trans-Golgi Network Transport. J. Virol. 2020, 94, e02155-19. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.M.; Moss, B. Vaccinia virus A36R membrane protein provides a direct link between intracellular enveloped virions and the microtubule motor kinesin. J. Virol. 2004, 78, 2486–2493. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, D.C.J.; Van Loggerenberg, A.; Dieckmann, N.M.G.; Smith, G.L. Vaccinia virus egress mediated by virus protein A36 is reliant on the F12 protein. J. Gen. Virol. 2017, 98, 1500–1514. [Google Scholar] [CrossRef]
- Teffera, M.; Boshra, H.; Bowden, T.R.; Babiuk, S. Which Proteins? The Challenge of Identifying the Protective Antigens for Next-Generation Capripoxvirus Vaccines. Vaccines 2025, 13, 219. [Google Scholar] [CrossRef]
- Jesus, D.M.; Moussatche, N.; McFadden, B.B.; Nielsen, C.P.; D’Costa, S.M.; Condit, R.C. Vaccinia virus protein A3 is required for the production of normal immature virions and for the encapsidation of the nucleocapsid protein L4. Virology 2015, 481, 1–12. [Google Scholar] [CrossRef]
- Liu, J.; Corroyer-Dulmont, S.; Pražák, V.; Khusainov, I.; Bahrami, K.; Welsch, S.; Vasishtan, D.; Obarska-Kosińska, A.; Thorkelsson, S.R.; Grünewald, K.; et al. The palisade layer of the poxvirus core is composed of flexible A10 trimers. Nat. Struct. Mol. Biol. 2024, 31, 1105–1113. [Google Scholar] [CrossRef]
- Jesus, D.M.; Moussatche, N.; Condit, R.C. Vaccinia virus mutations in the L4R gene encoding a virion structural protein produce abnormal mature particles lacking a nucleocapsid. J. Virol. 2014, 88, 14017–14029. [Google Scholar] [CrossRef]
- Wilcock, D.; Smith, G.L. Vaccinia virions lacking core protein VP8 are deficient in early transcription. J. Virol. 1996, 70, 934–943. [Google Scholar] [CrossRef]
- Williams, O.; Wolffe, E.J.; Weisberg, A.S.; Merchlinsky, M. Vaccinia virus WR gene A5L is required for morphogenesis of mature virions. J. Virol. 1999, 73, 4590–4599. [Google Scholar] [CrossRef]
- Risco, C.; Rodríguez, J.R.; Demkowicz, W.; Heljasvaara, R.; Carrascosa, J.L.; Esteban, M.; Rodríguez, D. The vaccinia virus 39-kDa protein forms a stable complex with the p4a/4a major core protein early in morphogenesis. Virology 1999, 265, 375–386. [Google Scholar] [CrossRef]
- Wang, S.; Cheng, P.; Guo, K.; Ren, S.; Tadele, B.A.; Liang, Z.; Sun, Y.; Yin, X.; Wang, X. Lumpy skin disease virus enters into host cells via dynamin-mediated endocytosis and macropinocytosis. Vet. Microbiol. 2024, 298, 110254. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, C.L.; Ojeda, S.; Moss, B. Transcriptional repression and RNA silencing act synergistically to demonstrate the function of the eleventh component of the vaccinia virus entry-fusion complex. J. Virol. 2012, 86, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.; Kao, C.F.; Chang, W.; Ku, C. Widespread Distribution and Evolution of Poxviral Entry-Fusion Complex Proteins in Giant Viruses. Microbiol. Spectr. 2023, 11, e0494422. [Google Scholar] [CrossRef] [PubMed]
- Tak, A.I.; Americo, J.L.; Diesterbeck, U.S.; Moss, B. Loss of the vaccinia virus 35-amino acid hydrophobic O3 protein is partially compensated by mutations in the transmembrane domains of other entry proteins. J. Virol. 2021, 95, e02228-20. [Google Scholar] [CrossRef]
- Bisht, H.; Weisberg, A.S.; Moss, B. Vaccinia virus L1 protein is required for cell entry and membrane fusion. J. Virol. 2008, 82, 8687–8694. [Google Scholar] [CrossRef]
- Priyamvada, L.; Kallemeijn, W.W.; Faronato, M.; Wilkins, K.; Goldsmith, C.S.; Cotter, C.A.; Ojeda, S.; Solari, R.; Moss, B.; Tate, E.W.; et al. Inhibition of vaccinia virus L1 N-myristoylation by the host N-myristoyltransferase inhibitor IMP-1088 generates non-infectious virions defective in cell entry. PLoS Pathog. 2022, 18, e1010662. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Moss, B. Vaccinia virus H2 protein is an essential component of a complex involved in virus entry and cell-cell fusion. J. Virol. 2005, 79, 4744–4754. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Ward, B.M.; Moss, B. Vaccinia virus entry into cells is dependent on a virion surface protein encoded by the A28L gene. J. Virol. 2004, 78, 2357–2366. [Google Scholar] [CrossRef]
- Townsley, A.C.; Senkevich, T.G.; Moss, B. Vaccinia virus A21 virion membrane protein is required for cell entry and fusion. J. Virol. 2005, 79, 9458–9469. [Google Scholar] [CrossRef]
- Czarnecki, M.W.; Traktman, P. The vaccinia virus DNA polymerase and its processivity factor. Virus Res. 2017, 234, 193–206. [Google Scholar] [CrossRef]
- Boutin, L.; Mosca, E.; Iseni, F. Efficient Method for Generating Point Mutations in the Vaccinia Virus Genome Using CRISPR/Cas9. Viruses 2022, 14, 1559. [Google Scholar] [CrossRef]
- Boyle, K.A.; Arps, L.; Traktman, P. Biochemical and genetic analysis of the vaccinia virus D5 protein: Multimerization-dependent ATPase activity is required to support viral DNA replication. J. Virol. 2007, 81, 844–859. [Google Scholar] [CrossRef] [PubMed]
- Gammon, D.B.; Gowrishankar, B.; Duraffour, S.; Andrei, G.; Upton, C.; Evans, D.H. Vaccinia virus-encoded ribonucleotide reductase subunits are differentially required for replication and pathogenesis. PLoS Pathog. 2010, 6, e1000984. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.; Shen, L.; Tcherepanov, V.; Watson, C.; Upton, C. Predicted function of the vaccinia virus G5R protein. Bioinformatics 2006, 22, 2846–2850. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Koonin, E.V.; Moss, B. Predicted poxvirus FEN1-like nuclease required for homologous recombination, double-strand break repair and full-size genome formation. Proc. Natl. Acad. Sci. USA 2009, 106, 17921–17926. [Google Scholar] [CrossRef]
- De Silva, F.S.; Moss, B. Effects of vaccinia virus uracil DNA glycosylase catalytic site and deoxyuridine triphosphatase deletion mutations individually and together on replication in active and quiescent cells and pathogenesis in mice. Virol. J. 2008, 5, 145. [Google Scholar] [CrossRef] [PubMed]
- Zia, S.; Sumon, M.M.; Ashik, M.A.; Basar, A.; Lim, S.; Oh, Y.; Park, Y.; Rahman, M.M. Potential Inhibitors of Lumpy Skin Disease’s Viral Protein (DNA Polymerase): A Combination of Bioinformatics Approaches. Animals 2024, 14, 1283. [Google Scholar] [CrossRef]
- Grimm, C.; Hillen, H.S.; Bedenk, K.; Bartuli, J.; Neyer, S.; Zhang, Q.; Hüttenhofer, A.; Erlacher, M.; Dienemann, C.; Schlosser, A.; et al. Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes. Cell 2019, 179, 1537–1550.e1519. [Google Scholar] [CrossRef]
- Sanz, P.; Moss, B. Identification of a transcription factor, encoded by two vaccinia virus early genes, that regulates the intermediate stage of viral gene expression. Proc. Natl. Acad. Sci. USA 1999, 96, 2692–2697. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.A.; Greseth, M.D.; Traktman, P. Genetic Confirmation that the H5 Protein Is Required for Vaccinia Virus DNA Replication. J. Virol. 2015, 89, 6312–6327. [Google Scholar] [CrossRef]
- Ly, M.; Burgess, H.M.; Shah, S.B.; Mohr, I.; Glaunsinger, B.A. Vaccinia virus D10 has broad decapping activity that is regulated by mRNA splicing. PLoS Pathog. 2022, 18, e1010099. [Google Scholar] [CrossRef]
- Burgess, H.M.; Mohr, I. Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 2015, 17, 332–344. [Google Scholar] [CrossRef]
- Jha, S.; Rollins, M.G.; Fuchs, G.; Procter, D.J.; Hall, E.A.; Cozzolino, K.; Sarnow, P.; Savas, J.N.; Walsh, D. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 2017, 546, 651–655. [Google Scholar] [CrossRef]
- Liu, Y.; Chin, J.M.; Choo, E.L.; Phua, K.K.L. Messenger RNA translation enhancement by immune evasion proteins: A comparative study between EKB (vaccinia virus) and NS1 (influenza A virus). Sci. Rep. 2019, 9, 11972. [Google Scholar] [CrossRef] [PubMed]
- Carten, J.D.; Greseth, M.; Traktman, P. Structure-Function Analysis of Two Interacting Vaccinia Proteins That Are Critical for Viral Morphogenesis: L2 and A30.5. J. Virol. 2022, 96, e0157721. [Google Scholar] [CrossRef]
- Meng, X.; Embry, A.; Rose, L.; Yan, B.; Xu, C.; Xiang, Y. Vaccinia virus A6 is essential for virion membrane biogenesis and localization of virion membrane proteins to sites of virion assembly. J. Virol. 2012, 86, 5603–5613. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Rose, L.; Han, Y.; Deng, J.; Xiang, Y. Vaccinia Virus A6 Is a Two-Domain Protein Requiring a Cognate N-Terminal Domain for Full Viral Membrane Assembly Activity. J. Virol. 2017, 91, e02405-16. [Google Scholar] [CrossRef]
- Resch, W.; Weisberg, A.S.; Moss, B. Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane. J. Virol. 2005, 79, 6598–6609. [Google Scholar] [CrossRef]
- Satheshkumar, P.S.; Weisberg, A.; Moss, B. Vaccinia virus H7 protein contributes to the formation of crescent membrane precursors of immature virions. J. Virol. 2009, 83, 8439–8450. [Google Scholar] [CrossRef] [PubMed]
- Maruri-Avidal, L.; Weisberg, A.S.; Moss, B. Vaccinia virus L2 protein associates with the endoplasmic reticulum near the growing edge of crescent precursors of immature virions and stabilizes a subset of viral membrane proteins. J. Virol. 2011, 85, 12431–12441. [Google Scholar] [CrossRef] [PubMed]
- Szajner, P.; Jaffe, H.; Weisberg, A.S.; Moss, B. A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions. Virology 2004, 330, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Heljasvaara, R.; Rodríguez, D.; Risco, C.; Carrascosa, J.L.; Esteban, M.; Rodríguez, J.R. The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleoprotein complex to form immature viral particles. J. Virol. 2001, 75, 5778–5795. [Google Scholar] [CrossRef]
- Ansarah-Sobrinho, C.; Moss, B. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J. Virol. 2004, 78, 6335–6343. [Google Scholar] [CrossRef]
- Eldi, P.; Cooper, T.H.; Liu, L.; Prow, N.A.; Diener, K.R.; Howley, P.M.; Suhrbier, A.; Hayball, J.D. Production of a Chikungunya Vaccine Using a CHO Cell and Attenuated Viral-Based Platform Technology. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 2332–2344. [Google Scholar] [CrossRef]
- Liu, L.; Cooper, T.; Howley, P.M.; Hayball, J.D. From Crescent to Mature Virion: Vaccinia Virus Assembly and Maturation. Viruses 2014, 6, 3787–3808. [Google Scholar] [CrossRef]
- Mercer, J.; Traktman, P. Genetic and cell biological characterization of the vaccinia virus A30 and G7 phosphoproteins. J. Virol. 2005, 79, 7146–7161. [Google Scholar] [CrossRef]
- Satheshkumar, P.S.; Weisberg, A.S.; Moss, B. Vaccinia virus A19 protein participates in the transformation of spherical immature particles to barrel-shaped infectious virions. J. Virol. 2013, 87, 10700–10709. [Google Scholar] [CrossRef]
- White, C.L.; Weisberg, A.S.; Moss, B. A glutaredoxin, encoded by the G4L gene of vaccinia virus, is essential for virion morphogenesis. J. Virol. 2000, 74, 9175–9183. [Google Scholar] [CrossRef]
- White, C.L.; Senkevich, T.G.; Moss, B. Vaccinia virus G4L glutaredoxin is an essential intermediate of a cytoplasmic disulfide bond pathway required for virion assembly. J. Virol. 2002, 76, 467–472. [Google Scholar] [CrossRef]
- Lynn, H.; Howell, L.M.; Diefenbach, R.J.; Newsome, T.P. Phototracking Vaccinia Virus Transport Reveals Dynamics of Cytoplasmic Dispersal and a Requirement for A36R and F12L for Exit from the Site of Wrapping. Viruses 2018, 10, 390. [Google Scholar] [CrossRef]
- Morgan, G.W.; Hollinshead, M.; Ferguson, B.J.; Murphy, B.J.; Carpentier, D.C.; Smith, G.L. Vaccinia protein F12 has structural similarity to kinesin light chain and contains a motor binding motif required for virion export. PLoS Pathog. 2010, 6, e1000785. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, D.C.J.; Gao, W.N.D.; Ewles, H.; Morgan, G.W.; Smith, G.L. Vaccinia Virus Protein Complex F12/E2 Interacts with Kinesin Light Chain Isoform 2 to Engage the Kinesin-1 Motor Complex. PLoS Pathog. 2015, 11, e1004723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Y.; Pallett, M.A.; French, J.; Ren, H.; Smith, G.L. Vaccinia virus BTB-Kelch proteins C2 and F3 inhibit NF-κB activation. J. Gen. Virol. 2022, 103, 001786. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Varga, J.; Deschambault, Y. Poxvirus encoded eIF2α homolog, K3 family proteins, is a key determinant of poxvirus host species specificity. Virology 2020, 541, 101–112. [Google Scholar] [CrossRef]
- Park, C.; Peng, C.; Rahman, M.J.; Haller, S.L.; Tazi, L.; Brennan, G.; Rothenburg, S. Orthopoxvirus K3 orthologs show virus- and host-specific inhibition of the antiviral protein kinase PKR. PLoS Pathog. 2021, 17, e1009183. [Google Scholar] [CrossRef]
- Parrish, S.; Moss, B. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J. Virol. 2007, 81, 12973–12978. [Google Scholar] [CrossRef]
- Liu, S.W.; Wyatt, L.S.; Orandle, M.S.; Minai, M.; Moss, B. The D10 Decapping Enzyme of Vaccinia Virus Contributes to Decay of Cellular and Viral mRNAs and to Virulence in Mice. J. Virol. 2014, 88, 202–211. [Google Scholar] [CrossRef]
- Conrad, S.J.; Raza, T.; Peterson, E.A.; Liem, J.; Connor, R.; Nounamo, B.; Cannon, M.; Liu, J. Myxoma virus lacking the host range determinant M062 stimulates cGAS-dependent type 1 interferon response and unique transcriptomic changes in human monocytes/macrophages. PLoS Pathog. 2022, 18, e1010316. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wennier, S.; Zhang, L.; McFadden, G. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J. Virol. 2011, 85, 3270–3282. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Wang, Y.; Dai, P.; Li, T.; Zierhut, C.; Tan, A.; Zhang, T.; Xiang, J.Z.; Ordureau, A.; Funabiki, H.; et al. Vaccinia E5 is a major inhibitor of the DNA sensor cGAS. Nat. Commun. 2023, 14, 2898. [Google Scholar] [CrossRef]
- Liang, Z.; Wang, S.; Yao, K.; Ren, S.; Cheng, P.; Qu, M.; Ma, X.; Gao, X.; Yin, X.; Wang, X.; et al. Lumpy skin disease virus ORF127 protein suppresses type I interferon responses by inhibiting K63-linked ubiquitination of tank binding kinase 1. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2024, 38, e23467. [Google Scholar] [CrossRef]
- Pallett, M.A.; Ren, H.; Zhang, R.Y.; Scutts, S.R.; Gonzalez, L.; Zhu, Z.; Maluquer de Motes, C.; Smith, G.L. Vaccinia Virus BBK E3 Ligase Adaptor A55 Targets Importin-Dependent NF-κB Activation and Inhibits CD8(+) T-Cell Memory. J. Virol. 2019, 93, e00051-19. [Google Scholar] [CrossRef]
- Bowie, A.; Kiss-Toth, E.; Symons, J.A.; Smith, G.L.; Dower, S.K.; O’Neill, L.A. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 2000, 97, 10162–10167. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, J.; Lu, B.; Li, H.; Liu, C.; Zeng, H.; Chen, J.; Liu, S.; Jiang, Q.; Jia, K. Lumpy skin disease virus ORF142 protein inhibits type I interferon production by disrupting interactions of TBK1 and IRF3. BMC Vet. Res. 2025, 21, 257. [Google Scholar] [CrossRef]
- Novy, K.; Kilcher, S.; Omasits, U.; Bleck, C.K.E.; Beerli, C.; Vowinckel, J.; Martin, C.K.; Syedbasha, M.; Maiolica, A.; White, I.; et al. Proteotype profiling unmasks a viral signalling network essential for poxvirus assembly and transcriptional competence. Nat. Microbiol. 2018, 3, 588–599. [Google Scholar] [CrossRef]
- Mann, B.A.; Huang, J.H.; Li, P.; Chang, H.C.; Slee, R.B.; O’Sullivan, A.; Anita, M.; Yeh, N.; Klemsz, M.J.; Brutkiewicz, R.R.; et al. Vaccinia virus blocks Stat1-dependent and Stat1-independent gene expression induced by type I and type II interferons. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2008, 28, 367–380. [Google Scholar] [CrossRef]
- Xie, S.; Fang, Y.; Liao, Z.; Cui, L.; Niu, K.; Ren, S.; Zhu, J.; Wu, W.; Jing, Z.; Peng, C. A poxvirus ankyrin protein LSDV012 inhibits IFIT1 in a host-species-specific manner by compromising its RNA binding ability. PLoS Pathog. 2025, 21, e1012994. [Google Scholar] [CrossRef]
- Früh, K.; Bartee, E.; Gouveia, K.; Mansouri, M. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. 2002, 88, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, M.; Bartee, E.; Gouveia, K.; Hovey Nerenberg, B.T.; Barrett, J.; Thomas, L.; Thomas, G.; McFadden, G.; Früh, K. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 2003, 77, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- Guerin, J.L.; Gelfi, J.; Boullier, S.; Delverdier, M.; Bellanger, F.A.; Bertagnoli, S.; Drexler, I.; Sutter, G.; Messud-Petit, F. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J. Virol. 2002, 76, 2912–2923. [Google Scholar] [CrossRef] [PubMed]
- Rehm, K.E.; Connor, R.F.; Jones, G.J.; Yimbu, K.; Roper, R.L. Vaccinia virus A35R inhibits MHC class II antigen presentation. Virology 2010, 397, 176–186. [Google Scholar] [CrossRef]
- Shchelkunov, S.N.; Yakubitskiy, S.N.; Sergeev, A.A.; Starostina, E.V.; Titova, K.A.; Pyankov, S.A.; Shchelkunova, G.A.; Borgoyakova, M.B.; Zadorozhny, A.M.; Orlova, L.A.; et al. Enhancing the Immunogenicity of Vaccinia Virus. Viruses 2022, 14, 1453. [Google Scholar] [CrossRef]
- Barry, M.; Hnatiuk, S.; Mossman, K.; Lee, S.F.; Boshkov, L.; McFadden, G. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 1997, 239, 360–377. [Google Scholar] [CrossRef]
- Byun, M.; Wang, X.; Pak, M.; Hansen, T.H.; Yokoyama, W.M. Cowpox virus exploits the endoplasmic reticulum retention pathway to inhibit MHC class I transport to the cell surface. Cell Host Microbe 2007, 2, 306–315. [Google Scholar] [CrossRef]
- Alzhanova, D.; Hammarlund, E.; Reed, J.; Meermeier, E.; Rawlings, S.; Ray, C.A.; Edwards, D.M.; Bimber, B.; Legasse, A.; Planer, S.; et al. T cell inactivation by poxviral B22 family proteins increases viral virulence. PLoS Pathog. 2014, 10, e1004123. [Google Scholar] [CrossRef]
- Forsyth, K.S.; Roy, N.H.; Peauroi, E.; DeHaven, B.C.; Wold, E.D.; Hersperger, A.R.; Burkhardt, J.K.; Eisenlohr, L.C. Ectromelia-encoded virulence factor C15 specifically inhibits antigen presentation to CD4+ T cells post peptide loading. PLoS Pathog. 2020, 16, e1008685. [Google Scholar] [CrossRef]
- Peauroi, E.M.; Carro, S.D.; Pei, L.; Reynoso, G.V.; Hickman, H.D.; Eisenlohr, L.C. The ectromelia virus virulence factor C15 facilitates early viral spread by inhibiting NK cell contact. iScience 2022, 25, 105510. [Google Scholar] [CrossRef]
- Matozaki, T.; Murata, Y.; Okazawa, H.; Ohnishi, H. Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol. 2009, 19, 72–80. [Google Scholar] [CrossRef]
- Cameron, C.M.; Barrett, J.W.; Mann, M.; Lucas, A.; McFadden, G. Myxoma virus M128L is expressed as a cell surface CD47-like virulence factor that contributes to the downregulation of macrophage activation in vivo. Virology 2005, 337, 55–67. [Google Scholar] [CrossRef]
- Yadav, P.; Kumar, A.; Nath, S.S.; Devasurmutt, Y.; Shashidhar, G.; Joshi, M.; Puvar, A.; Sharma, S.; Raval, J.; Pandit, R.; et al. Unravelling the genomic origins of lumpy skin disease virus in recent outbreaks. BMC Genom. 2024, 25, 196. [Google Scholar] [CrossRef]
- Boshra, H.; Truong, T.; Nfon, C.; Bowden, T.R.; Gerdts, V.; Tikoo, S.; Babiuk, L.A.; Kara, P.; Mather, A.; Wallace, D.B.; et al. A lumpy skin disease virus deficient of an IL-10 gene homologue provides protective immunity against virulent capripoxvirus challenge in sheep and goats. Antivir. Res. 2015, 123, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Kara, P.D.; Mather, A.S.; Pretorius, A.; Chetty, T.; Babiuk, S.; Wallace, D.B. Characterisation of putative immunomodulatory gene knockouts of lumpy skin disease virus in cattle towards an improved vaccine. Vaccine 2018, 36, 4708–4715. [Google Scholar] [CrossRef]
- Chervyakova, O.; Issabek, A.; Sultankulova, K.; Bopi, A.; Kozhabergenov, N.; Omarova, Z.; Tulendibayev, A.; Aubakir, N.; Orynbayev, M. Lumpy Skin Disease Virus with Four Knocked Out Genes Was Attenuated In Vivo and Protects Cattle from Infection. Vaccines 2022, 10, 1705. [Google Scholar] [CrossRef]
- Maluquer de Motes, C.; Cooray, S.; Ren, H.; Almeida, G.M.; McGourty, K.; Bahar, M.W.; Stuart, D.I.; Grimes, J.M.; Graham, S.C.; Smith, G.L. Inhibition of apoptosis and NF-κB activation by vaccinia protein N1 occur via distinct binding surfaces and make different contributions to virulence. PLoS Pathog. 2011, 7, e1002430. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, M.F.; Waseem, Q.U.A.; Awan, M.F.; Ali, S.; Ahmad, A.; Malook, S.U.; Ali, Q. In-silico characterization of LSDV132 protein divulged its BCL-2-like nature. Heliyon 2024, 10, e27657. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Zhang, S.W.; Li, H.L.; Feng, Y.L.; Zhang, Y. orf132: A critical gene for LSDV replication and its role in Er stress-related apoptosis. Virol. J. 2025, 22, 172. [Google Scholar] [CrossRef]
- Hasib, F.M.Y.; Islam, M.S. Structural, Functional and Inhibitory Annotations of Lumpy Skin Disease Virus Hypothetical Protein LSDV004: An In-Silico Study. Vet Med. Sci. 2025, 11, e70437. [Google Scholar] [CrossRef]
- Su, J.; Wang, G.; Barrett, J.W.; Irvine, T.S.; Gao, X.; McFadden, G. Myxoma virus M11L blocks apoptosis through inhibition of conformational activation of Bax at the mitochondria. J. Virol. 2006, 80, 1140–1151. [Google Scholar] [CrossRef]
- Everett, H.; Barry, M.; Lee, S.F.; Sun, X.; Graham, K.; Stone, J.; Bleackley, R.C.; McFadden, G. M11L: A novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J. Exp. Med. 2000, 191, 1487–1498. [Google Scholar] [CrossRef]
- Teoh, M.L.; Turner, P.V.; Evans, D.H. Tumorigenic poxviruses up-regulate intracellular superoxide to inhibit apoptosis and promote cell proliferation. J. Virol. 2005, 79, 5799–5811. [Google Scholar] [CrossRef]
- Munyanduki, H.; Douglass, N.; Offerman, K.; Carulei, O.; Williamson, A.L. Influence of the lumpy skin disease virus (LSDV) superoxide dismutase homologue on host transcriptional activity, apoptosis and histopathology. J. Gen. Virol. 2020, 101, 645–650. [Google Scholar] [CrossRef]
- Bloomer, D.T.; Kitevska-Ilioski, T.; Pantaki-Eimany, D.; Ji, Y.H.; Miles, M.A.; Heras, B.; Hawkins, C.J. CrmA orthologs from diverse poxviruses potently inhibit caspases-1 and-8, yet cleavage site mutagenesis frequently produces caspase-1-specific variants. Biochem. J. 2019, 476, 1335–1357. [Google Scholar] [CrossRef] [PubMed]
- Veyer, D.L.; de Motes, C.M.; Sumner, R.P.; Ludwig, L.; Johnson, B.F.; Smith, G.L. Analysis of the anti-apoptotic activity of four vaccinia virus proteins demonstrates that B13 is the most potent inhibitor in isolation and during viral infection. J. Gen. Virol. 2014, 95, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Gao, X.; Li, Y.; Zhang, Z.; Xie, S.; Ren, S.; Li, Y.; Li, H.; Niu, K.; Fu, S.; et al. Human FAM111A inhibits vaccinia virus replication by degrading viral protein I3 and is antagonized by poxvirus host range factor SPI-1. Proc. Natl. Acad. Sci. USA 2023, 120, e2304242120. [Google Scholar] [CrossRef]
- Tuppurainen, E.S.; Pearson, C.R.; Bachanek-Bankowska, K.; Knowles, N.J.; Amareen, S.; Frost, L.; Henstock, M.R.; Lamien, C.E.; Diallo, A.; Mertens, P.P. Characterization of sheep pox virus vaccine for cattle against lumpy skin disease virus. Antivir. Res. 2014, 109, 1–6. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199. [Google Scholar] [CrossRef] [PubMed]
- Alcami, A. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 2003, 3, 36–50. [Google Scholar] [CrossRef]
- Wickramasekera, N.T.; Traktman, P. Structure/Function analysis of the vaccinia virus F18 phosphoprotein, an abundant core component required for virion maturation and infectivity. J. Virol. 2010, 84, 6846–6860. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Tong, J.; Zhang, X.; Tuohetiniyazi, M.; Zhang, Y.; Li, Y. Comparative analysis of ankyrin (ANK) genes of five capripoxviruses isolate strains from Xinjiang province in China. Virol. J. 2020, 17, 133. [Google Scholar] [CrossRef] [PubMed]
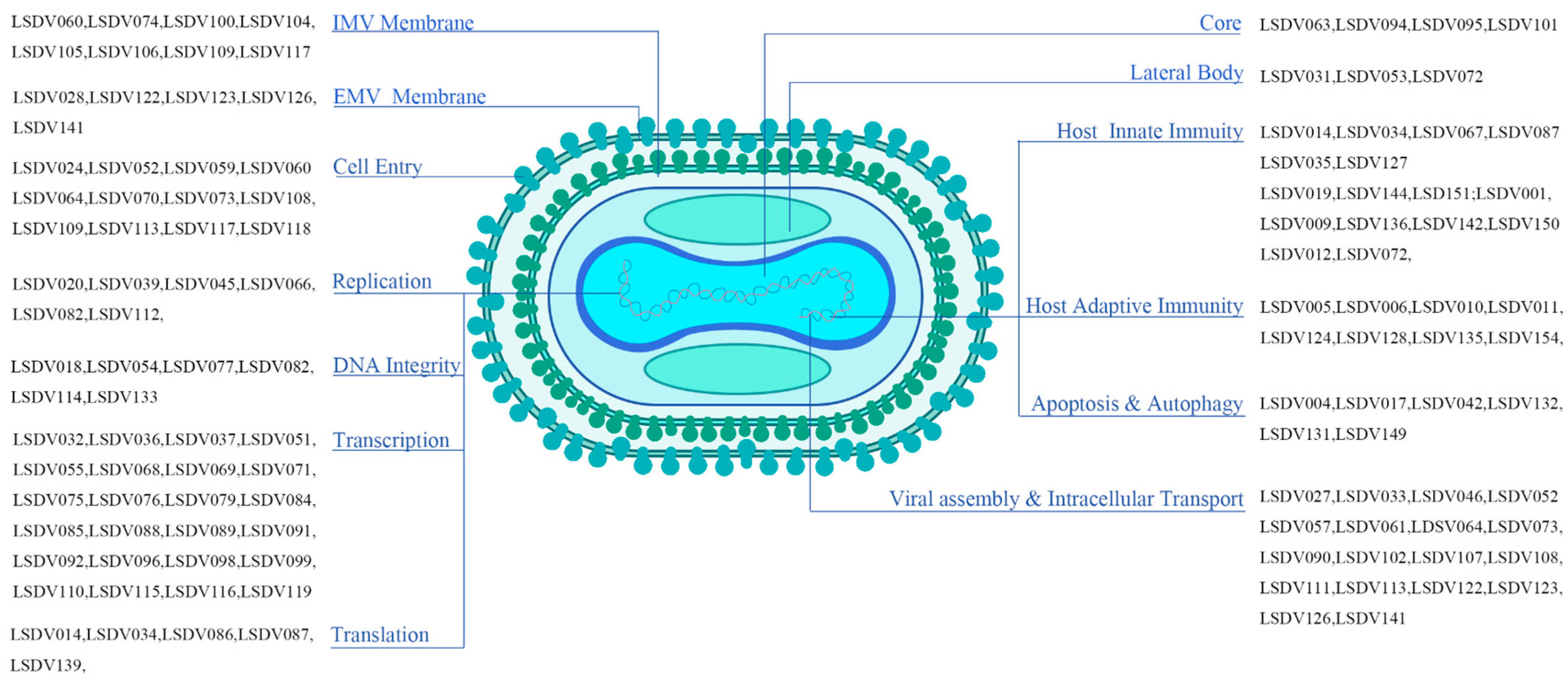
| LSDV ORF | Predicted Structure and/or Function | VACV ORF |
|---|---|---|
| LSDV028 | Palmitoylated EEV membrane protein | F13L |
| LSDV038 | Membrane protein | E8R |
| LSDV060 | IMV membrane protein | L1R |
| LSDV064 | Putative membrane protein, required for cell entry | L5R |
| LSDV070 | Late putative membrane protein | J5L |
| LSDV074 | IMV heparin binding surface protein | H3L |
| LSDV100 | IMV membrane protein | A9L |
| LSDV104 | IMV membrane protein | A13L |
| LSDV105 | Phosphorylated IMV membrane protein | A14L |
| LSDV106 | Nonessential hydrophobic IV and IMV membrane protein | A14.5 |
| LSDV109 | IMV membrane protein | A17L |
| LSDV117 | IMV surface protein | A27L |
| LSDV122 | EEV membrane phosphoglycoprotein | A33R |
| LSDV123 | EEV glycoprotein | A34R |
| LSDV126 | EEV glycoprotein, TM | A36R |
| LSDV141 | EEV host range protein | B5R |
| LSDV ORF | Predicted Structure and/or Function | VACV ORF |
|---|---|---|
| LSDV063 | DNA-binding virion core protein VP8 | L4R |
| LSDV094 | Virion core protein P4b | A3L |
| LSDV095 | Virion core protein, virion morphogenesis | A4L |
| LSDV101 | Virion core protein P4a | A10L |
| LSDV ORF | Predicted Structure and/or Function | VACV ORF |
|---|---|---|
| LSDV024 | S-S bond formation pathway protein | F9L |
| LSDV052 | Component of the entry–fusion complex | G3L |
| LSDV059 | Myristylated protein | G9R |
| LSDV060 | Myristylated IMV envelope protein | L1R |
| LSDV064 | Putative membrane protein, required for cell entry | L5R |
| LSDV070 | Late putative membrane protein | J5L |
| LSDV073 | Component of the entry–fusion complex | H2R |
| LSDV108 | Soluble myristylprotein, required for cell entry | A16L |
| LSDV109 | Phosphorylated IMV membrane protein | A17L |
| LSDV113 | Required for cell entry and fusion | A21L |
| LSDV117 | Fusion protein, virus assembly | A27L |
| LSDV118 | Required for cell entry and fusion | A28L |
| LSDV ORF | Predicted Structure and/or Function | VACV ORF |
|---|---|---|
| LSDV018 | dUTPase | F2L |
| LSDV020 | Ribonucleotide reductase, small subunit, | F4L |
| LSDV039 | DNA polymerase | E9L |
| LSDV043 | DNA-binding virion core protein | I1L |
| LSDV045 | DNA-binding phosphoprotein | I3L |
| LSDV054 | Fen1-like nuclease | G5R |
| LSDV066 | Thymidine kinase | J2R |
| LSDV077 | DNA topoisomerase | H6R |
| LSDV082 | uracil DNA glycosylase | D4R |
| LSDV083 | NTPase; DNA replication | D5R |
| LSDV112 | DNA polymerase processivity factor | A20R |
| LSDV114 | Holliday junction endonuclease | A22R |
| LSDV133 | DNA ligase | A50R |
| LSDV139 | Ser/Thr protein kinase, DNA replication | B1R |
| LSDV ORF | Predicted Structure and/or Function | VACV ORF |
|---|---|---|
| LSDV032 | Poly(A) polymerase PAPL | E1L |
| LSDV036 | RNA polymerase subunit RPO30 | E4L |
| LSDV037 | Unidentified role in viral transcription | E6R |
| LSDV051 | Putative transcriptional elongation factor | G2R |
| LSDV055 | RNA polymerase subunit RPO7 | G5.5R |
| LSDV068 | Poly(A) polymerase PAPs | J3R |
| LSDV069 | RNA polymerase subunit RPO22 | J4R |
| LSDV071 | RNA polymerase subunit RPO147 | J6R |
| LSDV075 | RNA polymerase-associated protein | H4L |
| LSDV076 | Late transcription factor VLTF-4 | H5R |
| LSDV079 | mRNA capping enzyme, large | D1R |
| LSDV084 | Early transcription factor VETFa1 | D6R |
| LSDV085 | RNA polymerase subunit RPO18 | D7R |
| LSDV088 | ATPase, nucleoside triphosphate phosphohydrolase-I, homolog of VAC NPH-I | D11L |
| LSDV089 | mRNA capping enzyme, small | D12L |
| LSDV091 | Late gene transcription factor, homolog of VAC VLTF-2 | A1L |
| LSDV092 | Putative late transcription factor (VAC VLTF-3) | A2L |
| LSDV096 | RNA polymerase subunit RPO19 | A5R |
| LSDV098 | A large subunit of early gene transcription factor, homolog of VAC VETF | A7L |
| LSDV099 | A small subunit of transcription factor, homolog of VAC VITF-3 | A8R |
| LSDV110 | DNA helicase; transcriptional elongation | A18R |
| LSDV115 | Large subunit of intermediate gene transcription factor, homolog of VAC VITF-3 | A23R |
| LSDV116 | RNA polymerase subunit RPO132 | A24R |
| LSDV119 | RNA polymerase subunit RPO35 | A29L |
| LSDV ORF | Predicted Structure and/or Function | VACV ORF |
|---|---|---|
| LSDV027 | EEV maturation | F12L |
| LSDV033 | Required for IEV transport, VAC F12/E2 complex associates with kinesin-1 | E2L |
| LSDV046 | IMV protein, homolog of VAC VP13 | I5L |
| LSDV052 | Required for entry as a component of the entry-Fusion complex | G3L |
| LSDV057 | Virion core protein | G7L |
| LSDV061 | Required for virion assembly, crescent formation | L2R |
| LSDV064 | Putative membrane protein, required for cell entry | L5R |
| LSDV073 | VAC H2R homolog, component of the entry fusion complex | H2R |
| LSDV090 | Rifampin resistance protein, IMV assembly | D13L |
| LSDV102 | Stabilizes membranes during virus assembly | A11R |
| LSDV107 | Absence produces empty immature virions | A15L |
| LSDV108 | Soluble myristylprotein, required for cell entry | A16L |
| LSDV111 | Required for maturation of virus particles | A19L |
| LSDV113 | Required for cell entry and fusion | A21L |
| LSDV122 | EEV membrane phosphoglycoprotein | A33R |
| LSDV123 | EEV glycoprotein | A34R |
| LSDV126 | EEV glycoprotein | A36R |
| LSDV141 | EEV host range protein | B5R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Zhai, R.; Cai, C.; Zhu, X.; Jung, Y.-S.; Qian, Y. Lumpy Skin Disease Virus Pathogenesis: Viral Protein Functions and Comparative Insights from Vaccinia Virus. Animals 2025, 15, 3176. https://doi.org/10.3390/ani15213176
Chen H, Zhai R, Cai C, Zhu X, Jung Y-S, Qian Y. Lumpy Skin Disease Virus Pathogenesis: Viral Protein Functions and Comparative Insights from Vaccinia Virus. Animals. 2025; 15(21):3176. https://doi.org/10.3390/ani15213176
Chicago/Turabian StyleChen, Huan, Ruiyu Zhai, Chang Cai, Xiaojie Zhu, Yong-Sam Jung, and Yingjuan Qian. 2025. "Lumpy Skin Disease Virus Pathogenesis: Viral Protein Functions and Comparative Insights from Vaccinia Virus" Animals 15, no. 21: 3176. https://doi.org/10.3390/ani15213176
APA StyleChen, H., Zhai, R., Cai, C., Zhu, X., Jung, Y.-S., & Qian, Y. (2025). Lumpy Skin Disease Virus Pathogenesis: Viral Protein Functions and Comparative Insights from Vaccinia Virus. Animals, 15(21), 3176. https://doi.org/10.3390/ani15213176





