Identification of the Bovine CSN3 Core Promoter Region and the Relationships Between CSN3 Promoter Polymorphisms and the CSN3 A and B Alleles
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
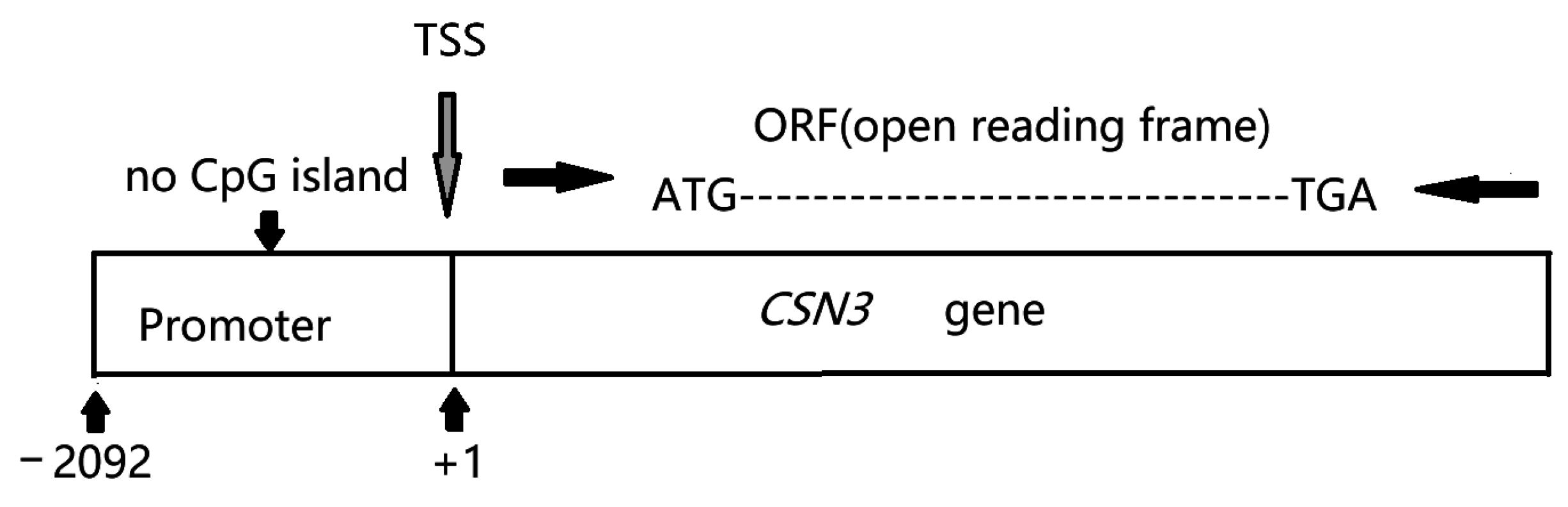
2.1. Animals, Samples, and Promoter Analysis
2.2. Construction of a Progressively Shortened pGL3-Basic Recombinant Vector
2.3. Cell Transfection and the Dual-Luciferase Reporter Assay
2.4. Sequencing the Core Promoter Region of Bovine CSN3
2.5. Promoter Mutations Outside the CSN3 Core Promoter Region
2.6. Identification of the CSN3 A and B Alleles and Their Relationships with CSN3 Promoter Polymorphisms
2.7. Construction of the Promoter Mutation Recombinant Vector and the Dual-Luciferase Reporter Assay
2.8. Statistical Analysis
3. Results
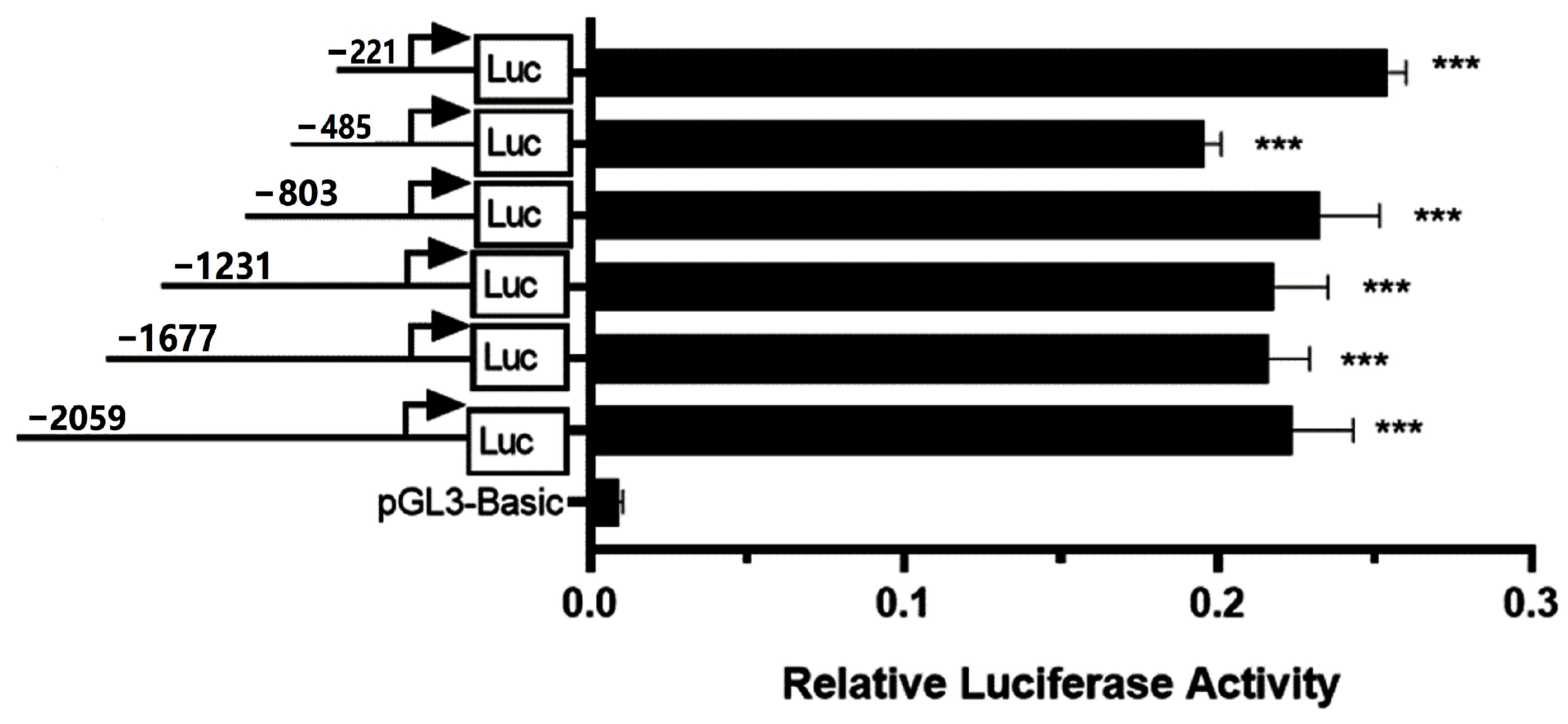
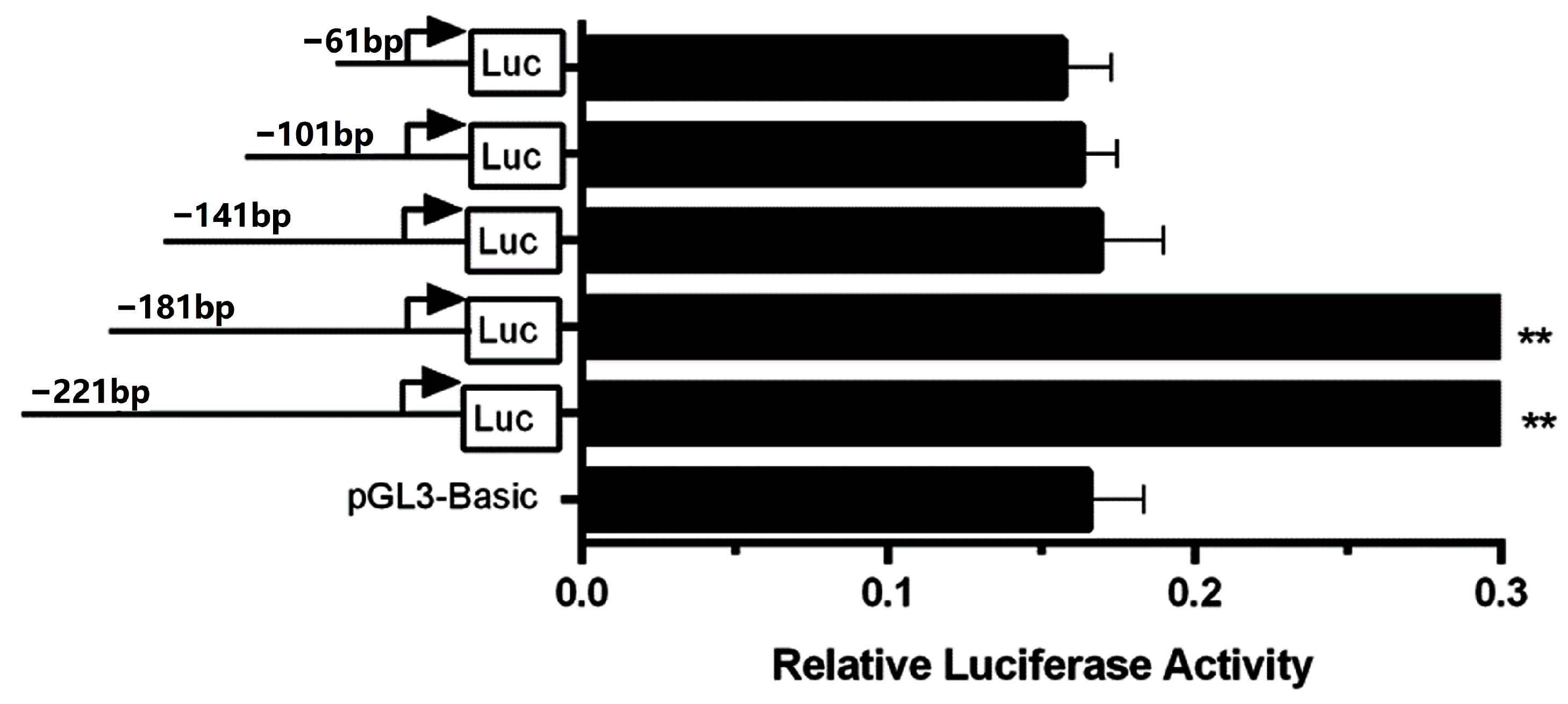
3.1. Identification of the Bovine CSN3 Core Promoter
3.2. Sequence Variations in the Bovine CSN3 Core Promoter Region
3.3. CSN3 Promoter Polymorphisms
3.4. Identification of CSN3*A and CSN3*B Variants and Their Relationships with CSN3 Promoter Polymorphisms
3.5. Differences in the Promoter Activities of CSN3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef]
- Sanchez, M.P.; Govignon-Gion, A.; Croiseau, P.; Fritz, S.; Hozé, C.; Miranda, G.; Martin, P.; Barbat-Leterrier, A.; Letaïef, R.; Rocha, D.; et al. Within-breed and multi-breed GWAS on imputed whole-genome sequence variants reveal candidate mutations affecting milk protein composition in dairy cattle. Genet. Sel. Evol. 2017, 49, 68. [Google Scholar] [CrossRef] [PubMed]
- Čítek, J.; Brzáková, M.; Hanusová, L.; Hanuš, O.; Večerek, L.; Samková, E.; Křížová, Z.; Hoštičková, I.; Kávová, T.; Straková, K.; et al. Gene polymorphisms influencing yield, composition and technological properties of milk from Czech Simmental and Holstein cows. Anim. Biosci. 2021, 34, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, P.; Rostamzadeh, J.; Rashidi, A.; Zergani, E.; Razmkabir, M. A meta-analysis on association between CSN3 gene variants and milk yield and composition in cattle. Anim. Genet. 2020, 51, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, C.; Arcangeli, C.; Ciullo, M.; Torricelli, M.; Cinti, G.; Fisichella, S.; Biagetti, M. Frequencies Evaluation of beta-Casein Gene Polymorphisms in Dairy Cows Reared in Central Italy. Animals 2020, 10, 252. [Google Scholar] [CrossRef]
- Caffaro, M.E.; Raschia, M.A.; Maizon, D.O.; Poli, M.A. Frequencies of β-casein variants and their influence on genetic merit for production traits in Holstein cows. Anim. Prod. Sci. 2024, 64, AN24146. [Google Scholar] [CrossRef]
- Poulsen, N.A.; Bertelsen, H.P.; Jensen, H.B.; Gustavsson, F.; Glantz, M.; Månsson, H.L.; Andrén, A.; Paulsson, M.; Bendixen, C.; Buitenhuis, A.J.; et al. The occurrence of noncoagulating milk and the association of bovine milk coagulation properties with genetic variants of the caseins in 3 Scandinavian dairy breeds. J. Dairy Sci. 2013, 96, 4830–4842. [Google Scholar] [CrossRef]
- Gai, N.; Uniacke-Lowe, T.; O’Regan, J.; Faulkner, H.; Kelly, A.L. Effect of Protein Genotypes on Physicochemical Properties and Protein Functionality of Bovine Milk: A Review. Foods 2021, 10, 2409. [Google Scholar] [CrossRef]
- Zhang, W.; Zheng, S.; Zhu, H.; Lu, J.; Zhang, Y.; Hettinga, K.; Pang, X.; Lyu, J.; Zhang, S. Effects of protein genetic variants on their phosphorylation levels, milk composition, milk proteome, and milk coagulation ability in Chinese Holstein bovine milk. Int. J. Biol. Macromol. 2024, 262 Pt 2, 129844. [Google Scholar] [CrossRef]
- Pinders, J.; Perryc, B.N.; Skidmoraend, J.; Sawa, D. Analysis of polymorphism in the bovine casein genes by use of the polymerase chain reaction. Anim Genet. 1991, 22, 11–20. [Google Scholar] [CrossRef]
- McLean, D.M.; Graham, E.R.B.; Ponzoni, R.W.; McKenzie, H.A. Effects of milk protein genetic variants on milk yield and composition. J. Dairy Res. 1984, 51, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Bobe, G.; Beitz, D.C.; Freeman, A.E.; Lindberg, G.L. Effect of milk protein genotypes on milk protein composition and its genetic parameter estimates. J. Dairy Sci. 1999, 82, 2797–2804. [Google Scholar] [CrossRef] [PubMed]
- Hallén, E.; Wedholm, A.; Andrén, A.; Lundén, A. Effect of beta-casein, kappa-casein and beta-lactoglobulin genotypes on concentration of milk protein variants. J. Anim. Breed. Genet. 2008, 125, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Amalfitano, N.; Macedo Mota, L.F.; Rosa, G.M.; Cecchinato, A.; Bittante, G. Role of CSN2, CSN3, and BLG genes and the polygenic background in the cattle milk protein profile. J. Dairy Sci. 2022, 105, 6001–6020. [Google Scholar] [CrossRef]
- Ehrmann, S.; Bartenschlager, H.; Geldermann, H. Polymorphism in the 5’ flanking region of the bovine-lactoglobulin-encoding gene and its association with β-lactoglobulin in the milk. J. Anim. Breed. Genet. 1997, 114, 49–53. [Google Scholar] [CrossRef]
- Robitaille, G.; Britten, M.; Morisset, J.; Petitclerc, D. Polymorphism in the bovine κ-casein (CSN3) gene and the 5’-flanking region: Sequence analysis of CSN3 A and B alleles. Anim. Genet. 2005, 36, 184–185. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, M.; Rong, Y.; Liu, Z.; Yang, S.; Zhang, W.; Li, J.; Cai, Y. A Novel SNPs in Alpha-Lactalbumin Gene Effects on Lactation Traits in Chinese Holstein Dairy Cows. Animals 2019, 10, 60. [Google Scholar] [CrossRef]
- Hobor, S.; Kunej, T.; Dovc, P. Polymorphisms in the kappa casein (CSN3) gene in horse and comparative analysis of its promoter and coding region. Anim. Genet. 2008, 39, 520–530. [Google Scholar] [CrossRef]
- Vanvanhossou, S.F.U.; Giambra, I.J.; Yin, T.; Brügemann, K.; Dossa, L.H.; König, S. First DNA Sequencing in Beninese Indigenous Cattle Breeds Captures New Milk Protein Variants. Genes 2021, 12, 1702. [Google Scholar] [CrossRef]
- Damiani, G.; Caroli, A.; Leone, P.; Budelli, E.; Florio, S. Effect of Bov-A2 Sine elements on quantitative traits. In Proceedings of the ASPA Congress-Recent Progress in Animal Production Science, Florence, Italy, 12–15 June 2021; University of Florence Press: Florence, Italy, 2001; pp. 49–51. [Google Scholar]
- Lundén, A.; Nilsson, M.; Janson, L. Marked Effect of β-Lactoglobulin Polymorphism on the Ratio of Casein to Total Protein in Milk. J. Dairy Sci. 1997, 80, 2996–3005. [Google Scholar] [CrossRef]
- Martin, P.; Szymanowska, M.; Zwierzchowski, L.; Leroux, C. The Impact of Genetic Polymorphisms on the Protein Composition of Ruminant Milks. Reprod. Nutr. Dev. 2002, 42, 433–459. [Google Scholar] [CrossRef] [PubMed]
- Farrell, H.M., Jr.; Jimenez-Flores, R.; Bleck, G.T.; Brown, E.M.; Butler, J.E.; Creamer, L.K.; Hicks, C.L.; Hollar, C.M.; Ng-Kwai-Hang, K.F.; Swaisgood, H.E. Nomenclature of the proteins of cows’ milk—Sixth revision. J. Dairy Sci. 2004, 87, 1641–1674. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.L.; Singer, D.S. Core promoters in transcription: Old problem, new insights. Trends Biochem. Sci. 2015, 40, 165–171. [Google Scholar] [CrossRef]
- Decker, K.B.; Hinton, D.M. Transcription regulation at the core: Similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu. Rev. Microbiol. 2013, 67, 113–139. [Google Scholar] [CrossRef]



| Primer Name | Primer Sequences (5′-3′) | Size (bp) | Positions Relative to the TSS |
|---|---|---|---|
| CSN3-P1F | atctgcgatctaagtaagcttACCAGAGCAACAATTTTTGCCT | 2059 | −2000/+59 |
| CSN3-P2F | atctgcgatctaagtaagcttCTCCTGCCCCACTGTAGAA | 1677 | −1618/+59 |
| CSN3-P3F | atctgcgatctaagtaagcttCATGACCATCACTCTGAACATTC | 1231 | −1172/+59 |
| CSN3-P4F | atctgcgatctaagtaagcttTGTGAAGAAAGGGGAATCCTC | 803 | −744/+59 |
| CSN3-P5F | atctgcgatctaagtaagcttCAACCACAGCCCATAATATATGTAG | 485 | −426/+59 |
| CSN3-P6F | atctgcgatctaagtaagcttCTGCATTCCATTAACCGAGAC | 221 | −162/+59 |
| CSN3-P-R | cagtaccggaatgccaagcttTTCCTTGTGACCGTCAGCT |
| Primer Name | Primer Sequences (5′-3′) | Size (bp) | Positions Relative to the TSS |
|---|---|---|---|
| CSN3-P6F | atctgcgatctaagtaagcttCTGCATTCCATTAACCGAGACTG | 221 | −162/+9 |
| CSN3-P7F | atctgcgatctaagtaagcttCTATCGTCAGATCTTTCCTTTCTGTC | 181 | −122/+59 |
| CSN3-P8F | atctgcgatctaagtaagcttGCAATGTAAAAGGAAGATAAATCTCATG | 141 | −82/+59 |
| CSN3-P9F | atctgcgatctaagtaagcttAACACCCTTTAATTAGTCTCTGGTTATT | 101 | −42/+59 |
| CSN3-P10F | atctgcgatctaagtaagcttTCCTTACAGTGGAAAGGCCAAC | 61 | −2/+59 |
| CSN3-P-R | cagtaccggaatgccaagcttTTCCTTGTGACCGTCAGCTCTT |
| Allele Position Before The TSS and SNP ID | −66 A>G No SNP ID | −394 T>A No SNP ID | −641 A>− rs134762502(AT>−) | −982 A>T No SNP ID | −1002 T>− rs136772334 | −1269 T>C No SNP ID | −1654 T>A rs110303451 | −1867 T>A No SNP ID | −2039 T>G rs109870556 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of Mutant Bases (Sample Number) and Mutation Frequencies | ||||||||||
| No base mutation (Nos. 2, 3, 9, 11, 12, 16, 20, 25, 26, 27, 28, 31, 33, 34, and 39) | ||||||||||
| One base mutation (no. 5) | C | |||||||||
| One base mutation (no. 10) | A | |||||||||
| One base mutation (no. 19) | G | |||||||||
| One base mutation (no. 37) | — | |||||||||
| Two base mutations (no. 15) | T | — | ||||||||
| Two base mutations (no. 18) | T | A | ||||||||
| Two base mutations (no. 35) | A | G | ||||||||
| Two base mutations (no. 40) | — | G | ||||||||
| Three base mutations (nos. 1, 4, 6, 7, 8, 17, 22, 23, 24, 29, 30, 32, 36, and 38) | — | A | G | |||||||
| Three base mutations (no. 14) | G | A | A | |||||||
| Four base mutations (no. 13) | G | — | A | G | ||||||
| Five base mutations (no. 21) | — | — | C | A | G | |||||
| Mutation frequencies | 5% | 5% | 5% | 5% | 45% | 5% | 42.5% | 5% | 47.5% | |
| Allele | CSN3*A | CSN3*B |
|---|---|---|
| Sample numbers | 4, 5, 7, 8, 10, 13, 14, 17, 20, 21, 22, 24, 27, 28, 29, 30, 34, 37, 40 | 1, 2, 3, 6, 9, 11, 12, 15, 16, 18, 19, 23, 25, 26, 31, 32, 33, 35, 36, 38, 39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Wang, X.; Xu, X.; Wu, P.; Fu, T.; Zhang, L.; Gao, T. Identification of the Bovine CSN3 Core Promoter Region and the Relationships Between CSN3 Promoter Polymorphisms and the CSN3 A and B Alleles. Animals 2025, 15, 134. https://doi.org/10.3390/ani15020134
Li W, Wang X, Xu X, Wu P, Fu T, Zhang L, Gao T. Identification of the Bovine CSN3 Core Promoter Region and the Relationships Between CSN3 Promoter Polymorphisms and the CSN3 A and B Alleles. Animals. 2025; 15(2):134. https://doi.org/10.3390/ani15020134
Chicago/Turabian StyleLi, Wenqing, Xiaoyang Wang, Xiuyang Xu, Pinhui Wu, Tong Fu, Liyang Zhang, and Tengyun Gao. 2025. "Identification of the Bovine CSN3 Core Promoter Region and the Relationships Between CSN3 Promoter Polymorphisms and the CSN3 A and B Alleles" Animals 15, no. 2: 134. https://doi.org/10.3390/ani15020134
APA StyleLi, W., Wang, X., Xu, X., Wu, P., Fu, T., Zhang, L., & Gao, T. (2025). Identification of the Bovine CSN3 Core Promoter Region and the Relationships Between CSN3 Promoter Polymorphisms and the CSN3 A and B Alleles. Animals, 15(2), 134. https://doi.org/10.3390/ani15020134






