Metabolomic Analysis of Cauda Epididymal Fluid in Yaks and Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Metabolite Extraction and Quality Control
2.3. The UHPLC-MS/MS Analysis
2.4. Data Processing and Metabolite Identification
2.5. Data Analysis
3. Result
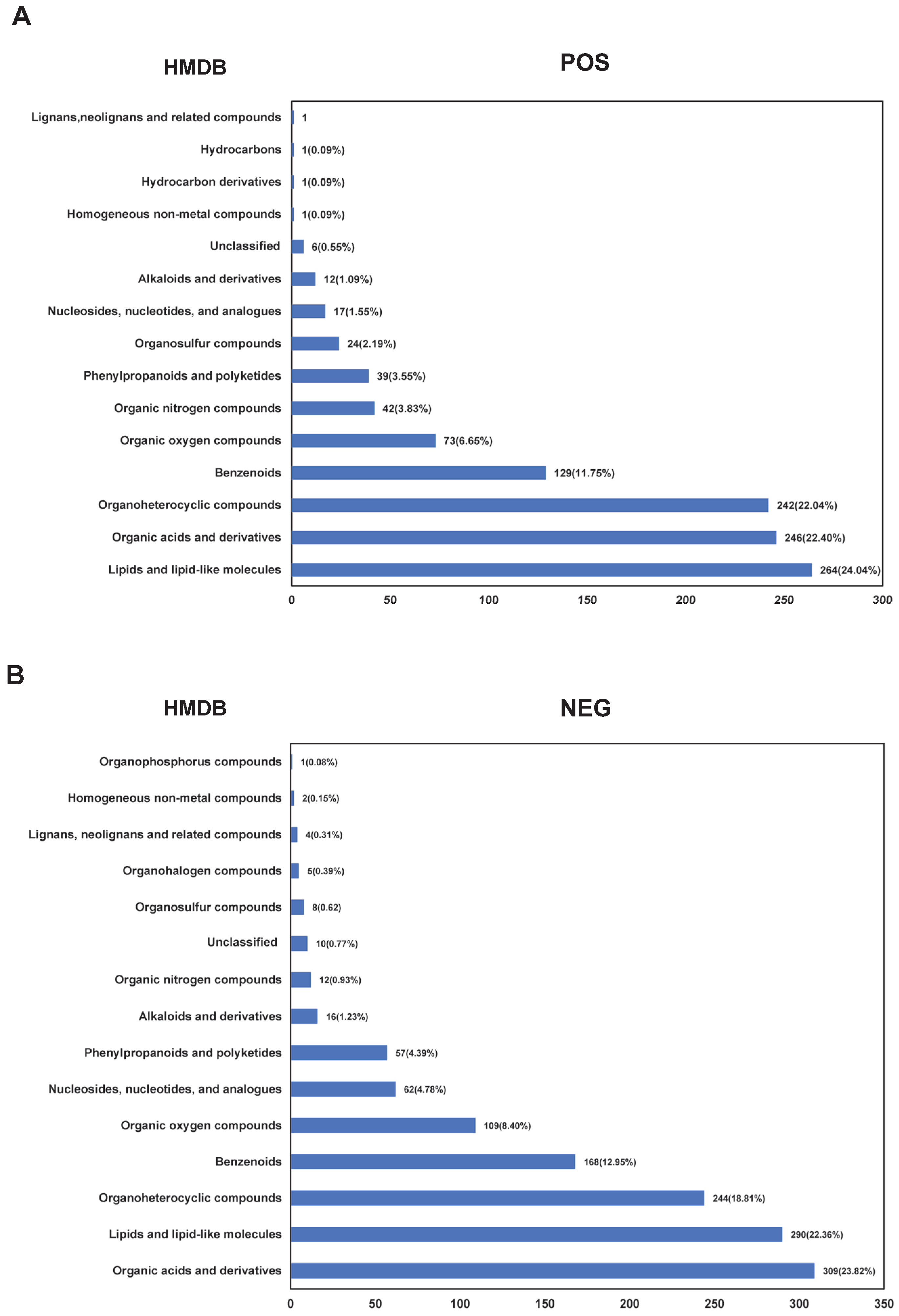
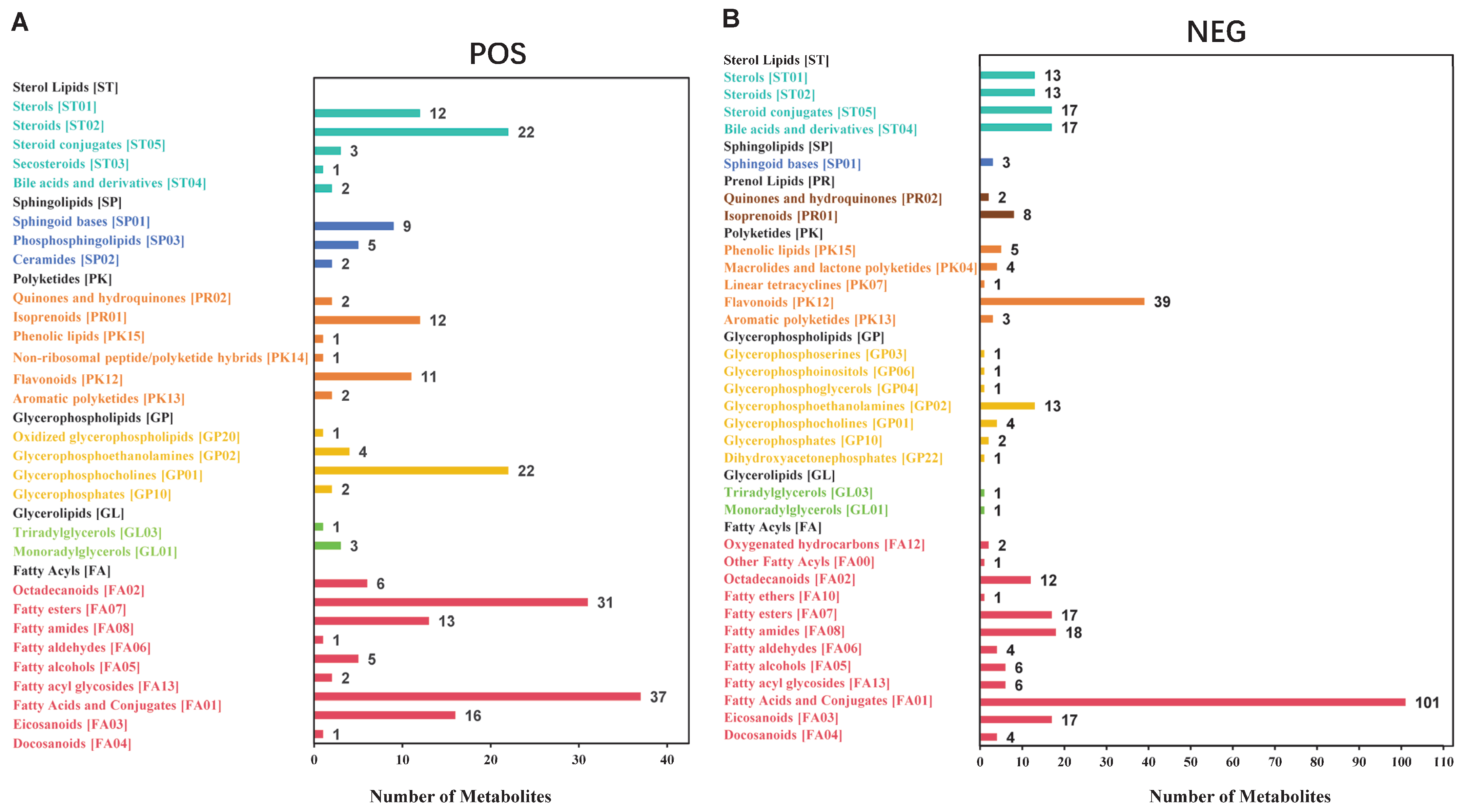
3.1. Data Quality Control and Metabolite Identification in Cauda Epididymal Fluid from Yak and Cattle
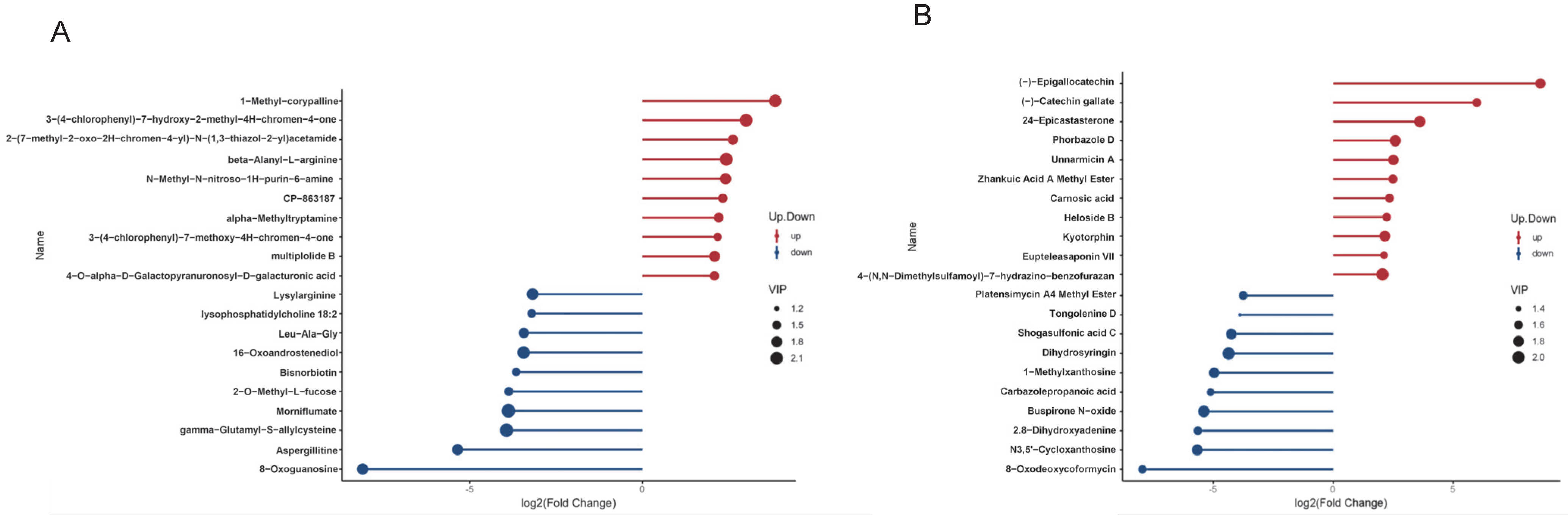
3.2. Identification of Differential Metabolites in Cauda Epididymal Fluid Between Yak and Cattle
3.3. Functional Enrichment Analyses of Differential Metabolites in Cauda Epididymal Fluid Between Yak and Cattle
3.4. Identification of Differential Metabolites Related to Sperm Quality, Function or Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HMBD | Human Metabolome Database |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PBS | Phosphate Buffered Saline |
| QC | Quality Control |
| CV | Coefficient of variation |
| PLS-DA | Partial least squares-discriminant analysis |
| VIP | Variable Importance in the Projection |
| FC | Fold Change |
| GSEA | Gene Set Enrichment Analysis |
| ES | Enrichment Score |
| NES | Normalized Enrichment Score |
| FDR | False Discovery Rate |
| PLS-DA | Partial Least Squares Discrimination Analysis |
| R2 | Goodness-of-fit |
| Q2 | Predictive ability |
| ncRNA | non-coding RNA |
| XCMS | Xtracted Chromatograms and Mass Spectra |
References
- Lee, V.; Hinton, B.T.; Hirashima, T. Collective cell dynamics and luminal fluid flow in the epididymis: A mechanobiological perspective. Andrology 2024, 12, 939–948. [Google Scholar] [CrossRef]
- Sosnicki, D.M.; Cohen, R.; Asano, A.; Nelson, J.L.; Mukai, C.; Comizzoli, P.; Travis, A.J. Segmental differentiation of the murine epididymis: Identification of segment-specific, GM1-enriched vesicles and regulation by luminal fluid factors. Biol. Reprod. 2023, 109, 864–877. [Google Scholar] [CrossRef]
- Cichowska, A.W.; Wisniewski, J.; Bromke, M.A.; Olejnik, B.; Mogielnicka-Brzozowska, M. Proteome Profiling of Canine Epididymal Fluid: In Search of Protein Markers of Epididymal Sperm Motility. Int. J. Mol. Sci. 2023, 24, 14790. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Argenti, L.E.; de Souza, A.P.B.; Santi, L.; Beys-da-Silva, W.O.; Yates, J.R., 3rd; Bustamante-Filho, I.C. Ready for the journey: A comparative proteome profiling of porcine cauda epididymal fluid and spermatozoa. Cell Tissue Res. 2020, 379, 389–405. [Google Scholar] [CrossRef]
- Xue, X.R.; Ma, Y.M.; Hu, N.X.; Zhao, C.C.; Jin, S.H. Experimental Comparison of Post-thaw Motility and Survival Time of Semen from Different Breeds of Bulls in Plateau Area. China Dairy Cattle 2017, 17–20. [Google Scholar] [CrossRef]
- Rosenfeld, C.S.; Javurek, A.B.; Johnson, S.A.; Lei, Z.; Sumner, L.W.; Hess, R.A. Seminal fluid metabolome and epididymal changes after antibiotic treatment in mice. Reproduction 2018, 156, 1–10. [Google Scholar] [CrossRef] [PubMed]
- James, E.R.; Carrell, D.T.; Aston, K.I.; Jenkins, T.G.; Yeste, M.; Salas-Huetos, A. The Role of the Epididymis and the Contribution of Epididymosomes to Mammalian Reproduction. Int. J. Mol. Sci. 2020, 21, 5377. [Google Scholar] [CrossRef]
- Sullivan, R.; Saez, F.; Girouard, J.; Frenette, G. Role of exosomes in sperm maturation during the transit along the male reproductive tract. Blood Cells Mol. Dis. 2005, 35, 1–10. [Google Scholar] [CrossRef]
- Wang, G.; Li, Y.; Yang, Q.; Xu, S.; Ma, S.; Yan, R.; Zhang, R.; Jia, G.; Ai, D.; Yang, Q. Gene expression dynamics during the gonocyte to spermatogonia transition and spermatogenesis in the domestic yak. J. Anim. Sci. Biotechnol. 2019, 10, 64. [Google Scholar] [CrossRef]
- Dacheux, J.L.; Belleannée, C.; Jones, R.; Labas, V.; Belghazi, M.; Guyonnet, B.; Druart, X.; Gatti, J.L.; Dacheux, F. Mammalian epididymal proteome. Mol. Cell. Endocrinol. 2009, 306, 45–50. [Google Scholar] [CrossRef][Green Version]
- Gatti, J.L.; Castella, S.; Dacheux, F.; Ecroyd, H.; Métayer, S.; Thimon, V.; Dacheux, J.L. Post-testicular sperm environment and fertility. Anim. Reprod. Sci. 2004, 82–83, 321–339. [Google Scholar] [CrossRef]
- Belleannée, C.; Labas, V.; Teixeira-Gomes, A.P.; Gatti, J.L.; Dacheux, J.L.; Dacheux, F. Identification of luminal and secreted proteins in bull epididymis. J. Proteom. 2011, 74, 59–78. [Google Scholar] [CrossRef]
- Jones, R.C.; Dacheux, J.L.; Nixon, B.; Ecroyd, H.W. Role of the epididymis in sperm competition. Asian J. Androl. 2007, 9, 493–499. [Google Scholar] [CrossRef]
- Chioccarelli, T.; Manfrevola, F.; Porreca, V.; Fasano, S.; Altucci, L.; Pierantoni, R.; Cobellis, G. The Cannabinoid Receptor CB1 Stabilizes Sperm Chromatin Condensation Status During Epididymal Transit by Promoting Disulphide Bond Formation. Int. J. Mol. Sci. 2020, 21, 3117. [Google Scholar] [CrossRef]
- Shan, S.; Xu, F.; Bleyer, M.; Becker, S.; Melbaum, T.; Wemheuer, W.; Hirschfeld, M.; Wacker, C.; Zhao, S.; Schütz, E.; et al. Association of α/β-Hydrolase D16B with Bovine Conception Rate and Sperm Plasma Membrane Lipid Composition. Int. J. Mol. Sci. 2020, 21, 627. [Google Scholar] [CrossRef]
- Sellem, E.; Marthey, S.; Rau, A.; Jouneau, L.; Bonnet, A.; Le Danvic, C.; Guyonnet, B.; Kiefer, H.; Jammes, H.; Schibler, L. Dynamics of cattle sperm sncRNAs during maturation, from testis to ejaculated sperm. Epigenetics Chromatin 2021, 14, 24. [Google Scholar] [CrossRef]
- Zhou, W.; De Iuliis, G.N.; Dun, M.D.; Nixon, B. Characteristics of the Epididymal Luminal Environment Responsible for Sperm Maturation and Storage. Front. Endocrinol. 2018, 9, 59. [Google Scholar] [CrossRef]
- Koziorowska-Gilun, M.; Koziorowski, M.; Fraser, L.; Strzeżek, J. Antioxidant defence system of boar cauda epididymidal spermatozoa and reproductive tract fluids. Reprod. Domest. Anim. = Zuchthyg. 2011, 46, 527–533. [Google Scholar] [CrossRef]
- Murdoch, R.N.; Jones, R.C.; Wade, M.; Lin, M. The ultrastructure and metabolism of ejaculated tammar wallaby sperm are impaired by swim-up procedures when compared with sperm from the cauda epididymidis. Reprod. Fertil. Dev. 1999, 11, 263–271. [Google Scholar] [CrossRef]
- Mayorga, L.S.; Bertini, F. The origin of some acid hydrolases of the fluid of the rat cauda epididymidis. J. Androl. 1985, 6, 243–245. [Google Scholar] [CrossRef]
- Caflisch, C.R.; DuBose, T.D., Jr. Cadmium-induced changes in luminal fluid pH in testis and epididymis of the rat in vivo. J. Toxicol. Environ. Health 1991, 32, 49–57. [Google Scholar] [CrossRef]
- Nagdas, S.K.; Britney, T.; Simpson, D.; Salters, T.; Raychoudhury, S.S. Fibrinogen-Related Protein, Fgl2, of Hamster Cauda Epididymal Fluid: Enzymatic Characterization, and Identification of Fgl2-Binding Proteins and Ligand of Defective Hamster Sperm Organelles. Int. J. Biochem. Physiol. 2023, 8, 000228. [Google Scholar] [CrossRef]
- Erikson, D.W.; Way, A.L.; Chapman, D.A.; Killian, G.J. Detection of osteopontin on Holstein bull spermatozoa, in cauda epididymal fluid and testis homogenates, and its potential role in bovine fertilization. Reproduction 2007, 133, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.Y.; Zhang, B.L.; Gao, D.Y.; Li, Q.; Xu, X.Y.; Shum, W. Epididymal epithelial degeneration and lipid metabolism impairment account for male infertility in occludin knockout mice. Front. Endocrinol. 2022, 13, 1069319. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.P.B.; Lopes, T.N.; da Silva, A.F.T.; Santi, L.; Beys-da-Silva, W.O.; Yates, J.R., 3rd; Bustamante-Filho, I.C. Changes in porcine cauda epididymal fluid proteome by disrupting the HPT axis: Unveiling potential mechanisms of male infertility. Mol. Reprod. Dev. 2020, 87, 952–965. [Google Scholar] [CrossRef]
- Khaw, S.C.; Wong, Z.Z.; Anderson, R.; Martins da Silva, S. L-carnitine and L-acetylcarnitine supplementation for idiopathic male infertility. Reprod. Fertil. 2020, 1, 67–81. [Google Scholar] [CrossRef]
- Xu, C.; Yuan, Y.; Zhang, C.; Zhou, Y.; Yang, J.; Yi, H.; Gyawali, I.; Lu, J.; Guo, S.; Ji, Y.; et al. Smooth muscle AKG/OXGR1 signaling regulates epididymal fluid acid-base balance and sperm maturation. Life Metab. 2022, 1, 67–80. [Google Scholar] [CrossRef]
- Zhao, X.; Nie, J.; Zhou, W.; Zeng, X.; Sun, X. The metabolomics changes in epididymal lumen fluid of CABS1 deficient male mice potentially contribute to sperm deformity. Front. Endocrinol. 2024, 15, 1432612. [Google Scholar] [CrossRef]
- Serri, O.; Boguenet, M.; Chao de la Barca, J.M.; Bouet, P.E.; El Hachem, H.; Blanchet, O.; Reynier, P.; May-Panloup, P. A Metabolomic Profile of Seminal Fluid in Extremely Severe Oligozoopermia Suggesting an Epididymal Involvement. Metabolites 2022, 12, 1266. [Google Scholar] [CrossRef]
- Zhao, S.; Chen, T.; Luo, X.; Chen, S.; Wang, J.; Lai, S.; Jia, X. Identification of Novel lncRNA and Differentially Expressed Genes (DEGs) of Testicular Tissues among Cattle, Yak, and Cattle-Yak Associated with Male Infertility. Animals 2021, 11, 2420. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Wang, X.; Guo, S.; Kang, Y.; Pei, J.; Guo, X. F1 Male Sterility in Cattle-Yak Examined through Changes in Testis Tissue and Transcriptome Profiles. Animals 2022, 12, 2711. [Google Scholar] [CrossRef]
- Zhu, Y.; Yu, J.; Yang, Q.; Xie, Y.; Li, X.; Chen, Z.; Xiong, Y.; Fu, W.; He, H.; Yin, S.; et al. Mitochondria-targeted antioxidant MitoQ improves the quality of low temperature-preserved yak semen via alleviating oxidative stress. Anim. Reprod. Sci. 2025, 273, 107680. [Google Scholar] [CrossRef]
- Want, E.J.; O’Maille, G.; Smith, C.A.; Brandon, T.R.; Uritboonthai, W.; Qin, C.; Trauger, S.A.; Siuzdak, G. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal. Chem. 2006, 78, 743–752. [Google Scholar] [CrossRef]
- Barri, T.; Dragsted, L.O. UPLC-ESI-QTOF/MS and multivariate data analysis for blood plasma and serum metabolomics: Effect of experimental artefacts and anticoagulant. Anal. Chim. Acta 2013, 768, 118–128. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, D.A.; Piccolo, B.D.; Vaziri, N.D.; Liu, S.; Lau, W.L.; Khazaeli, M.; Nazertehrani, S.; Moore, M.E.; Marco, M.L.; Martin, R.J.; et al. Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats. Am. J. physiology. Ren. Physiol. 2016, 310, F857–F871. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.; Sui, J.; Zhang, J. Metabolomics reveals significant variations in metabolites and correlations regarding the maturation of walnuts (Juglans regia L.). Biol. Open 2016, 5, 829–836. [Google Scholar] [CrossRef]
- Heischmann, S.; Quinn, K.; Cruickshank-Quinn, C.; Liang, L.P.; Reisdorph, R.; Reisdorph, N.; Patel, M. Exploratory Metabolomics Profiling in the Kainic Acid Rat Model Reveals Depletion of 25-Hydroxyvitamin D3 during Epileptogenesis. Sci. Rep. 2016, 6, 31424. [Google Scholar] [CrossRef]
- Haspel, J.A.; Chettimada, S.; Shaik, R.S.; Chu, J.H.; Raby, B.A.; Cernadas, M.; Carey, V.; Process, V.; Hunninghake, G.M.; Ifedigbo, E.; et al. Circadian rhythm reprogramming during lung inflammation. Nat. Commun. 2014, 5, 4753. [Google Scholar] [CrossRef] [PubMed]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.D.; Eisman, A.S.; Tahir, U.A.; Katz, D.H.; Deng, S.; Ngo, D.; Robbins, J.M.; Hofmann, A.; Shi, X.; Zheng, S.; et al. Protein-metabolite association studies identify novel proteomic determinants of metabolite levels in human plasma. Cell Metab. 2023, 35, 1646–1660.e3. [Google Scholar] [CrossRef] [PubMed]
- Boulesteix, A.L.; Strimmer, K. Partial least squares: A versatile tool for the analysis of high-dimensional genomic data. Brief. Bioinform. 2007, 8, 32–44. [Google Scholar] [CrossRef]
- Wang, J.B.; Pu, S.B.; Sun, Y.; Li, Z.F.; Niu, M.; Yan, X.Z.; Zhao, Y.L.; Wang, L.F.; Qin, X.M.; Ma, Z.J.; et al. Metabolomic Profiling of Autoimmune Hepatitis: The Diagnostic Utility of Nuclear Magnetic Resonance Spectroscopy. J. Proteome Res. 2014, 13, 3792–3801. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Bellezza, I.; Minelli, A. Adenosine in sperm physiology. Mol. Asp. Med. 2017, 55, 102–109. [Google Scholar] [CrossRef]
- Espinosa, F.; de la Vega-Beltrán, J.L.; López-González, I.; Delgado, R.; Labarca, P.; Darszon, A. Mouse sperm patch-clamp recordings reveal single Cl- channels sensitive to niflumic acid, a blocker of the sperm acrosome reaction. FEBS Lett. 1998, 426, 47–51. [Google Scholar] [CrossRef]
- Guerrero, A.; Espinal, J.; Wood, C.D.; Rendón, J.M.; Carneiro, J.; Martínez-Mekler, G.; Darszon, A. Niflumic acid disrupts marine spermatozoan chemotaxis without impairing the spatiotemporal detection of chemoattractant gradients. J. Cell Sci. 2013, 126, 1477–1487. [Google Scholar] [CrossRef]
- Partyka, A.; Kostrzewa Susłow, E.; Dymarska, M.; Ligocka, Z.; Smalec, B.; Kalinin, J.; Meco, M.; Niżański, W. Flavone and 3-hydroxyflavone supplementation in cryopreservation medium protects canine sperm against apoptosis and lipid peroxidation. Theriogenology 2024, 226, 319–327. [Google Scholar] [CrossRef]
- Akarsu, S.A.; Güngör, İ.H.; Cihangiroğlu, A.; Acısu, T.C.; Koca, R.H.; Türk, G.; Sönmez, M.; Gür, S. Effect of sulforaphane on long-term storage of rabbit semen. Anim. Reprod. 2023, 20, e20230001. [Google Scholar] [CrossRef]
- Mu, Y.; Yin, T.L.; Huang, X.X.; Hu, X.; Yin, L.; Yang, J. Sulforaphane ameliorates high-fat diet-induced spermatogenic deficiency in mice. Biol. Reprod. 2019, 101, 223–234. [Google Scholar] [CrossRef]
- Ahmed, H.; Ijaz, M.U.; Riaz, M.; Jahan, S. Sulforaphane inclusion in a freezing medium augments post-thaw motility, functional and biochemical features, and fertility potential of buffalo (Bubalus bubalis) spermatozoa. Res. Vet. Sci. 2023, 158, 196–202. [Google Scholar] [CrossRef]
- Jiang, X.P.; Tang, J.Y.; Xu, Z.; Han, P.; Qin, Z.Q.; Yang, C.D.; Wang, S.Q.; Tang, M.; Wang, W.; Qin, C.; et al. Sulforaphane attenuates di-N-butylphthalate-induced reproductive damage in pubertal mice: Involvement of the Nrf2-antioxidant system. Environ. Toxicol. 2017, 32, 1908–1917. [Google Scholar] [CrossRef]
- Anand, L.; Choudhury, A.; Sarin, S.K. Reply. Hepatology 2019, 70, 755–756. [Google Scholar] [CrossRef] [PubMed]
- Oyovwi, M.O.; Nwangwa, E.K.; Ben-Azu, B.; Rotue, R.A.; Edesiri, T.P.; Emojevwe, V.; Igweh, J.C.; Uruaka, C.I. Prevention and reversal of chlorpromazine induced testicular dysfunction in rats by synergistic testicle-active flavonoids, taurine and coenzyme-10. Reprod. Toxicol. 2021, 101, 50–62. [Google Scholar] [CrossRef]
- Ma, Q.; Gui, Y.; Ma, X.; Zhang, B.; Xiong, W.; Yang, S.; Cao, C.; Mo, S.; Shu, G.; Ye, J.; et al. N6-methyladenosine writer METTL16-mediated alternative splicing and translation control are essential for murine spermatogenesis. Genome Biol. 2024, 25, 193. [Google Scholar] [CrossRef]
- Irvine, D.S. Glutathione as a treatment for male infertility. Rev. Reprod. 1996, 1, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, Y.; Nishimura, M.; Matsumoto, K.; Miyashita, M.; Takeo, T.; Nakagata, N.; Hosoi, Y.; Anzai, M. The influence of reduced glutathione in fertilization medium on the fertility of in vitro-matured C57BL/6 mouse oocytes. Theriogenology 2013, 80, 421–426. [Google Scholar] [CrossRef]
- Itahashi, T.; Oikawa, T.; Numabe, T. Effects of glutathione treatments during sperm washing and in vitro fertilization on the in vitro early development of embryos of Japanese Black cattle. J. Reprod. Dev. 2022, 68, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, D.; He, Y.; Zhang, W.; He, H.; Du, R.; Pang, W.; Yang, G.; Yu, T. Supplementation of salvianic acid A to boar semen extender to improve seminal quality and antioxidant capacity. Anim. Sci. J. = Nihon Chikusan Gakkaiho 2019, 90, 1142–1148. [Google Scholar] [CrossRef]
- Hibi, H.; Kato, K.; Mitsui, K.; Taki, T.; Yamada, Y.; Honda, N.; Fukatsu, H.; Yamamoto, M. The treatment with tranilast, a mast cell blocker, for idiopathic oligozoospermia. Arch. Androl. 2001, 47, 107–111. [Google Scholar] [CrossRef][Green Version]
- Hibi, H.; Kato, K.; Mitsui, K.; Taki, T.; Yamada, Y.; Honda, N.; Fukatsu, H.; Yamamoto, M. Treatment of oligoasthenozoospermia with tranilast, a mast cell blocker, after long-term administration. Arch. Androl. 2002, 48, 451–459. [Google Scholar] [CrossRef]
- Dwivedi, C.; Long, N.J. Effect of cholinergic agents on human spermatozoa motility. Biochem. Med. Metab. Biol. 1989, 42, 66–70. [Google Scholar] [CrossRef]
- Piroozmanesh, H.; Jannatifar, R.; Ebrahimi, S.M.; Fazaeli, H.; Naserpoor, L.; Dimitriadis, E.; Nejatbakhsh, R. Cyclic adenosine monophosphate (cAMP) analog and phosphodiesterase inhibitor (IBMX) ameliorate human sperm capacitation and motility. Rev. Int. Androl. 2022, 20 (Suppl. 1), S24–S30. [Google Scholar] [CrossRef] [PubMed]
- Lone, F.A.; Naikoo, M.; Khatun, A.; Shah, R.A.; Pampori, Z.A.; Khan, H.M.; Ahanger, A.A. Idebenone improves quality of ram sperm by mitigating oxidative stress during cryopreservation. Cryobiology 2019, 90, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Eslami, M.; Jahan-Roshan, N.; Farrokhi-Ardabili, F. Influence of idebenone on ram semen quality stored at 4°C. Reprod. Domest. Anim. = Zuchthyg. 2019, 54, 486–497. [Google Scholar] [CrossRef]
- Nikitaras, V.; Zander-Fox, D.; McPherson, N.O. Improving Sperm Oxidative Stress and Embryo Quality in Advanced Paternal Age Using Idebenone In Vitro-A Proof-of-Concept Study. Antioxidants 2021, 10, 1079. [Google Scholar] [CrossRef]
- Li, Y.; Peng, Q.; Shang, J.; Dong, W.; Wu, S.; Guo, X.; Xie, Z.; Chen, C. The role of taurine in male reproduction: Physiology, pathology and toxicology. Front. Endocrinol. 2023, 14, 1017886. [Google Scholar] [CrossRef]
- Elkina, Y.L.; Atroshchenko, M.M.; Bragina, E.E.; Muronetz, V.I.; Schmalhausen, E.V. Oxidation of glyceraldehyde-3-phosphate dehydrogenase decreases sperm motility. Biochem. Biokhimiia 2011, 76, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.A.; Isa, A.M.; El-Kholy, W.M.; Nour, S.E. Testicular disorders induced by plant growth regulators: Cellular protection with proanthocyanidins grape seeds extract. Cytotechnology 2013, 65, 851–862. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bae, J.W.; Hwang, J.M.; Yoon, M.; Kwon, W.S. Bifenthrin Diminishes Male Fertility Potential by Inducing Protein Defects in Mouse Sperm. Toxics 2024, 12, 53. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.W.; Kwon, W.S. The deleterious toxic effects of bifenthrin on male fertility. Reprod. Toxicol. 2021, 101, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Gungor, S.; Inanc, M.E.; Ozturk, C.; Korkmaz, F.; Bastan, I.; Cil, B.; Kastelic, J.P. Gallic and carnosic acids improve quality of frozen-thawed ram spermatozoa. Andrologia 2019, 51, e13393. [Google Scholar] [CrossRef]
- Shum, W.W.; Da Silva, N.; Brown, D.; Breton, S. Regulation of luminal acidification in the male reproductive tract via cell-cell crosstalk. J. Exp. Biol. 2009, 212, 1753–1761. [Google Scholar] [CrossRef]
- Shum, W.W.; Ruan, Y.C.; Da Silva, N.; Breton, S. Establishment of cell-cell cross talk in the epididymis: Control of luminal acidification. J. Androl. 2011, 32, 576–586. [Google Scholar] [CrossRef]
- Jakop, U.; Svetlichnyy, V.; Schiller, J.; Schulze, M.; Schroeter, F.; Mueller, K. In vitro supplementation with unsaturated fatty acids improves boar sperm viability after storage at 6 °C. Anim. Reprod. Sci. 2019, 206, 60–68. [Google Scholar] [CrossRef]
- Teveroni, E.; Di Nicuolo, F.; Vergani, E.; Bruno, C.; Maulucci, G.; Bianchetti, G.; Astorri, A.L.; Grande, G.; Gervasoni, J.; Santucci, L.; et al. Short-Chain Fatty Acids Modulate Sperm Migration through Olfactory Receptor 51E2 Activity. Int. J. Mol. Sci. 2022, 23, 12726. [Google Scholar] [CrossRef]
- Islam, M.M.; Umehara, T.; Tsujita, N.; Shimada, M. Saturated fatty acids accelerate linear motility through mitochondrial ATP production in bull sperm. Reprod. Med. Biol. 2021, 20, 289–298. [Google Scholar] [CrossRef]
- Cooper, T.G. Role of the epididymis in mediating changes in the male gamete during maturation. Adv. Exp. Med. Biol. 1995, 377, 87–101. [Google Scholar] [CrossRef]
- Batini, C.; Buisseret-Delmas, C.; Conrath-Verrier, M. Olivo-cerebellar activity during harmaline-induced tremor. A 2-[14C] deoxyglucose study. Neurosci. Lett. 1979, 12, 241–246. [Google Scholar] [CrossRef]
- Gyamera-Acheampong, C.; Tantibhedhyangkul, J.; Weerachatyanukul, W.; Tadros, H.; Xu, H.; van de Loo, J.W.; Pelletier, R.M.; Tanphaichitr, N.; Mbikay, M. Sperm from mice genetically deficient for the PCSK4 proteinase exhibit accelerated capacitation, precocious acrosome reaction, reduced binding to egg zona pellucida, and impaired fertilizing ability. Biol. Reprod. 2006, 74, 666–673. [Google Scholar] [CrossRef][Green Version]
- Breitbart, H.; Grinshtein, E. Mechanisms That Protect Mammalian Sperm from the Spontaneous Acrosome Reaction. Int. J. Mol. Sci. 2023, 24, 17005. [Google Scholar] [CrossRef]
- Karanwal, S.; Pal, A.; Josan, F.; Patel, A.; Chera, J.S.; Yadav, S.; Gaur, V.; Verma, P.; Badrhan, S.; Chauhan, V.; et al. Higher abundance of DLD protein in buffalo bull spermatozoa causes elevated ROS production leading to early sperm capacitation and reduction in fertilizing ability. J. Anim. Sci. Biotechnol. 2024, 15, 126. [Google Scholar] [CrossRef] [PubMed]
- Dahan, T.; Breitbart, H. Involvement of metabolic pathway in the sperm spontaneous acrosome reaction. Theriogenology 2022, 192, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wang, X.; Guo, S.; Kang, Y.; Cao, M.; Hu, L.; Zhang, B.; Xiong, L.; Pei, J.; Yang, T.; et al. Characteristic analysis of N(6)-methyladenine in different parts of yak epididymis. BMC Genom. 2025, 26, 500. [Google Scholar] [CrossRef]
- Paulusma, C.C.; Lamers, W.H.; Broer, S.; van de Graaf, S.F.J. Amino acid metabolism, transport and signalling in the liver revisited. Biochem. Pharmacol. 2022, 201, 115074. [Google Scholar] [CrossRef]
- Katayama, S.; Mine, Y. Antioxidative activity of amino acids on tissue oxidative stress in human intestinal epithelial cell model. J. Agric. Food Chem. 2007, 55, 8458–8464. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Carvalho, B.; Carrageta, D.F.; Maurício, T.; Pereira, S.C.; Barros, A.; Carvalho, R.A.; Alves, M.G.; Domingues, P.; Oliveira, P.F. Metabolomics analysis of human spermatozoa reveals impaired metabolic pathways in asthenozoospermia. Eur. J. Clin. Investig. 2024, 54, e14289. [Google Scholar] [CrossRef]



| Name | Formula | Classification | Log2FC | p-Value | Function Related to Sperm |
|---|---|---|---|---|---|
| Adenosine | C10H13N5O4 | Nucleosides, nucleotides, and analogues | −2.8923 | 0.0145 | Inhibition of spontaneous acrosome reaction [47] |
| Niflumic acid | C13H9F3N2O2 | Benzenoids | −2.3234 | 0.0470 | Blocking sperm acrosome reaction and chemotaxis [48,49] |
| FLAVONE | C15H10O2 | Phenylpropanoids and polyketides | −1.4766 | 0.0042 | Protecting against acrosome damage, capacitation, apoptosis, and prevents lipid peroxidation during sperm cryopreservation [50] |
| Sulforaphane | C6H11NOS2 | Organosulfur compounds | −1.1168 | 0.0246 | Reducing oxidative stress and endoplasmic reticulum stress in sperm, thus improving sperm motility and fertilization rates in frozen semen [51,52,53,54] |
| Acetylcarnitine | C9H17NO4 | Lipids and lipid-like molecules | 0.6234 | 0.0375 | Enhancing sperm energy metabolism and motility while reducing apoptosis [26,55]. |
| Chlorpromazine | C17H19ClN2S | Organoheterocyclic compounds | 1.1107 | 0.0110 | Inhibition of epididymal sperm capacitation and acrosomal reaction [56] |
| Name | Formula | Classification | Log2FC | p-Value | Function Related to Sperm |
|---|---|---|---|---|---|
| N6-Methyladenosine | C11H15N5O4 | Nucleosides, nucleotides, and analogues | −3.3979 | 0.0177 | Increased N6-Methyladeno-sine content is a risk factor for asthenozoospermia and affects sperm motility [57]. |
| Glutathione | C10H17N3O6S | Organic acids and derivatives | −3.1306 | 0.0152 | Increasing sperm count, motility and fertilization rate by reducing ROS levels [58,59,60]. |
| Danshensu | C9H10O5 | Phenylpropanoids and polyketides | −1.3530 | 0.0481 | Improving seminal quality and antioxidant capacity [61]. |
| Tranilast | C18H17NO5 | Phenylpropanoids and polyketides | −1.3146 | 0.0024 | Increasing sperm count [62,63]. |
| Succinylproline | C9H13NO5 | Organic acids and derivatives | −1.0481 | 0.0248 | Inhibiting spermatozoa motility [64]. |
| IBMX | C10H14N4O2 | Organoheterocyclic compounds | −1.0398 | 0.0028 | Ameliorate sperm capacitation and motility and mobility [65]. |
| Idebenone | C19H30O5 | Lipids and lipid-like molecules | −0.7619 | 0.0246 | Improving post thaw sperm quality by mitigating oxidative stress [66,67,68]. |
| Taurine | C2H7NO3S | Organic acids and derivatives | 0.7165 | 0.0287 | Enhancing sperm mitochondrial energy metabolism [69]. |
| Glyceraldehyde 3-phosphate | C3H7O6P | Organic oxygen compounds | 0.7332 | 0.0473 | Facilitating sperm flagellum motility [70]. |
| Indolelactic acid | C11H11NO3 | Organoheterocyclic compounds | 1.0075 | 0.0377 | Increasing ROS level, decreasing antioxidant activity and reducing sperm number [71]. |
| BIFENTHRIN | C23H22ClF3O2 | Benzenoids | 1.1856 | 0.0299 | Diminishing motility, spontaneous acrosome reaction, and capacitation in sperm [72,73]. |
| Carnosic acid | C20H28O4 | Lipids and lipid-like molecules | 2.3463 | 0.0296 | Improving the quality and mitochondrial function of frozen-thawed sperm [74]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Yang, X.; Liu, D.; Zhou, W.; Zhou, A.; Xiong, Y.; Xiong, X.; Fu, W.; Li, J.; Lan, D.; et al. Metabolomic Analysis of Cauda Epididymal Fluid in Yaks and Cattle. Animals 2025, 15, 2861. https://doi.org/10.3390/ani15192861
Yang G, Yang X, Liu D, Zhou W, Zhou A, Xiong Y, Xiong X, Fu W, Li J, Lan D, et al. Metabolomic Analysis of Cauda Epididymal Fluid in Yaks and Cattle. Animals. 2025; 15(19):2861. https://doi.org/10.3390/ani15192861
Chicago/Turabian StyleYang, Gan, Xiaolong Yang, Dongju Liu, Wending Zhou, Anjun Zhou, Yan Xiong, Xianrong Xiong, Wei Fu, Jian Li, Daoliang Lan, and et al. 2025. "Metabolomic Analysis of Cauda Epididymal Fluid in Yaks and Cattle" Animals 15, no. 19: 2861. https://doi.org/10.3390/ani15192861
APA StyleYang, G., Yang, X., Liu, D., Zhou, W., Zhou, A., Xiong, Y., Xiong, X., Fu, W., Li, J., Lan, D., & Yin, S. (2025). Metabolomic Analysis of Cauda Epididymal Fluid in Yaks and Cattle. Animals, 15(19), 2861. https://doi.org/10.3390/ani15192861







