Polymorphism of the BMPR1B Variants for Prolific Traits in the Indonesian Local Ettawah Goat
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Isolation of DNA
2.3. PCR Amplification
2.4. DNA Sequencing
2.5. Polymorphism
2.6. Statistical Analysis
3. Results
3.1. PCR Results
3.2. Haplotype and Sequencing
3.3. BMPR1B Polymorphism of Does with Documented Prolificacy Records
3.4. Polymorphism Results Without Recorded Prolificacy Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMPR1B | Bone morphogenetic protein receptor type 1B |
| GDF9 | Growth Differentiation Factor 9 |
| ILEGs | Indonesian Local Ettawah Goats |
| INHA, INHB | Inhibin Subunit Alpha, Beta |
| KISS1 | Kisspeptins |
| PCR-RFLP | Polymerase Chain Reaction–Restriction Fragment Length Polymorphism |
References
- Icoutchika, K.L.M.; Ahozonlin, M.C.; Mitchikpe, C.E.S.; Bouraima, O.; Aboh, A.B.; Dossa, L.H. Socio-economic determinants of goat milk consumption by rural households in the Niger Valley of Benin and implications for the development of a smallholder dairy goat program. Front. Sustain. Food Syst. 2022, 6, 901293. [Google Scholar] [CrossRef]
- Skapetas, B.; Bampidis, V. Goat production in the world: Present situation and trends. Livest. Res. Rural. Dev. 2016, 28, 200. [Google Scholar]
- Ng’ambi, J.; John, A.O.; Norris, D. Role of goats in food security, poverty alleviation and prosperity with special reference to Sub-Saharan Africa: A Review. Indian J. Anim. Res. 2013, 47, 1–9. [Google Scholar]
- Al-Barakeh, F.; Khashroum, A.O.; Tarawneh, R.A.; Al-Lataifeh, F.A.; Al-Yacoub, A.N.; Dayoub, M.; Al-Najjar, K. Sustainable Sheep and Goat Farming in Arid Regions of Jordan. Ruminants 2024, 4, 241–255. [Google Scholar] [CrossRef]
- Idamokoro, E.M. The relevance of livestock husbandry in the context of food security: A bibliometric outlook of research studies from 1938 to 2020. Front. Sustain. Food Syst. 2023, 7, 1204221. [Google Scholar] [CrossRef]
- Utaaker, K.S.; Chaudhary, S.; Kifleyohannes, T.; Robertson, L.J. Global goat! Is the expanding goat population an important reservoir of cryptosporidium? Front. Vet. Sci. 2021, 8, 64850. [Google Scholar] [CrossRef] [PubMed]
- Mudawamah, M.; Ratnaningtyas, I.D.; Fadli, M.Z.; Ciptadi, G. Individual mutations in Indonesian local ettawah goats based on the GDF9 gene. J. Phys. Conf. Ser. 2019, 1146, 012023. [Google Scholar] [CrossRef]
- Mudawamah, M.; Ciptadi, G.; Retnaningtyas, I.D. The Prolific Variation, Body Morphometrics, and Breeding Value of Indonesian Local Etawah Goat Based in East Java. Anim. Prod. 2021, 23, 54–61. [Google Scholar] [CrossRef]
- Han, Y.; Cao, G.; Chen, W.; Wang, C.; Khan, M.Z. The Role of TGF-β Signaling Pathway in Determining Small Ruminant Litter Size. Biology 2025, 14, 786. [Google Scholar] [CrossRef]
- Margawati, E.T.; Putra, W.P.B.; Rizki, M.; Soetrisno, E.; Raadsma, H.W. Detection of carrier Booroola (Fec B) allele in BMPR1B gene of MEGA (Merino × Garut) sheep and its association with growth traits. J. Genet. Eng. Biotechnol. 2023, 21, 19. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; He, X.; Liu, Q.; Di, R.; Hu, W.; Cao, X.; Zhang, X.; Zhang, J.; Chu, M. Effects of FecB Mutation on Estrus, Ovulation, and Endocrine Characteristics in Small Tail Han Sheep. Front. Vet. Sci. 2021, 8, 709737. [Google Scholar] [CrossRef]
- de Lima, L.G.; de Souza, N.O.B.; Rios, R.R.; de Melo, B.A.; dos Santos, L.T.A.; Silva, K.d.M.; Murphy, T.W.; Fraga, A.B. Advance in molecular genetic techniques applied to selection for litter size in goats (Capra hircus): A Review. J. Appl. Anim. Res. 2020, 48, 38–44. [Google Scholar] [CrossRef]
- Sae-Foo, P.; Triwutanon, S.; Rukkwamsuk, T. Detection of Booroola Polymorphism of Bone Morphogenetic Protein Receptor 1b and Embrapa Polymorphism of Growth Differentiation Factor 9 in Sheep in Thailand. Animals 2024, 14, 809. [Google Scholar] [CrossRef] [PubMed]
- Acı, R.; Duman, E.; Kul, S.; Yiğit, S. Effect of Bone Morphogenetic Protein Receptor-1B (BMPR-1B) gene variant on litter size in Akkaraman Sheep Breed. J. Med. Vet. 2024, 7, 235–243. [Google Scholar] [CrossRef]
- Wen, Y.L.; Guo, X.F.; Ma, L.; Zhang, X.S.; Zhang, J.L.; Zhao, S.G.; Chu, M.X. The expression and mutation of BMPR1B and its association with litter size in small-tail Han sheep (Ovis aries). Arch. Anim. Breed. 2021, 64, 211–221. [Google Scholar] [CrossRef]
- Souza, C.J.; MacDougall, C.; Campbell, B.K.; McNeilly, A.S.; Baird, D.T. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene. J. Endocrinol. 2001, 169, R1–R6. [Google Scholar] [CrossRef]
- Hanrahan, P.J.; Gregan, S.M.; Mulsant, P.; Mullen, M.; Davis, G.H.; Powell, R.; Galloway, S.M. Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in cambridge and belclare sheep (Ovisaries). Biol. Reprod. 2004, 70, 900–909. [Google Scholar] [CrossRef]
- Galloway, S.M.; McNatty, K.P.; Cambridge, L.M.; Laitinen, M.P.; Jennifer, J.L.; Jokiranta, S.; McLaren, R.J.; Luiro, K.; Dodds, K.G.; Montgomery, G.W.; et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat. Genet. 2000, 25, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Ahlawati, S.; Sharma, R.; Maitra, A.; Tantia, A.; Roy, M.; Mandakmale, S. New genetic polymorphism in Indian goat BMPR1B gene. Indian J. Anim. Sci. 2014, 84, 39–44. [Google Scholar] [CrossRef]
- Polley, S.; De, S.; Batabyal, S.; Kaushika, R.; Yadava, P.; Aroraa, J.S.; Chattopadhyay, S.; Panc, S.; Brahmad, B.; Dattaa, T.K.; et al. Polymorphism of fecundity genes (BMPR1B, BMP15 and GDF9) in the Indian prolific Black Bengal goat. Small Rumin. Res. 2009, 85, 122–129. [Google Scholar] [CrossRef]
- Islam, M.; Basheer, A.; Javed, K.; Anjum, A.A.; Zahoor, I. PCR-RFLP-based identification of polymorphisms in BMPR1B, GDF9 and BMP15 genes associated with litter size in Beetal and Teddy goats. S. Afr. J. Anim. Sci. 2019, 49, 697–708. [Google Scholar] [CrossRef]
- Sasi, R.; Kanakkaparambil, R.; Thazhathuveettil, A. Polymorphism of fecundity genes, BMPR1B, BMP15, GDF9, in tropical goat breeds of Kerala. Gene Rep. 2020, 21, 100944. [Google Scholar] [CrossRef]
- Song, T.; Liu, Y.; Cuomu, R.; Tan, Y.; Wang, C.A.; De, J.; Cao, X.; Zeng, X. Polymorphisms Analysis of BMP15, GDF9 and BMPR1B in Tibetan Cashmere Goat (Capra hircus). Genes 2023, 14, 1102. [Google Scholar] [CrossRef]
- Rahawy, M.A.; AL-Mutar, H.A.K. Association of the KiSS1 gene with litter size in Cyprus and Iraqi black goats. Vet. World. 2021, 14, 1995–2001. [Google Scholar] [CrossRef]
- Mulyono, R.H.; Sumantri, C.; Noor, R.R.; Jakaria; Astuti, D.A. Association of BMP15, BMPR1B, and KISS1 Genes with Fecundity Traits on Etawah-Grade does. J. Ilmu Pertan. Indones. (JIPI) 2019, 24, 83–92. [Google Scholar] [CrossRef]
- Abuzahra, M.; Abu Eid, L.; Effendi, M.H.; Mustofa, I.; Lamid, M.; Rehman, S. Polymorphism studies and candidate genes associated with litter size traits in Indonesian goats. F1000Research 2023, 12, 61. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Isolation and Quantification of DNA. Cold Spring Harb. Protoc. 2018, 6, 11–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, J.; Bai, Y.; Wang, Q.; Wang, K.; Zhu, H.; Qu, L.; Guo, Z.; Pan, C.; Lan, X. A functional SNP of the core promoter region within goat CDC25A gene affects litter size. Front. Vet. Sci. 2024, 11, 1471123. [Google Scholar] [CrossRef]
- Liu, P.; Cao, C.; Wang, X.; Li, K.; Areb, E.; Li, R.; Zhang, Q.; Pan, C.; Lan, X. SNP variation landscape of BMP15 gene from 75 global sheep breeds and their genetic effect on lambing traits. Front. Vet. Sci. 2025, 12, 1612263. [Google Scholar] [CrossRef]
- Dutta, R.; Laskar, S.; Borah, P.; Kalita, D.; Das, B.; Zaman, G.; Barman, N.N.; Saika, D.P. Polymorphism and nucleotide sequencing of BMPR1B gene in prolific Assam hill goat. Mol. Biol. Rep. 2014, 41, 3677–3681. [Google Scholar] [CrossRef]
- Zamani, P.; Rabiei, F.; Hadiei, E.; Abdoli, R.; Ahmadi, A.; Rabiei, S. Screening for causative mutations in ovine BMPR1B and BMP15 genes and their homologous fragments in human. J. Assist. Reprod. Genet. 2023, 40, 1973–1982. [Google Scholar] [CrossRef]
- Abdelhamed, W.A.; El-Danasoury, M.M.; El-Hamamsy, S.M.; El-Sayed, M.A. Amino acids profile of three different Egyptian goat breeds. Egypt. J. Appl. Sci. 2020, 35, 261–271. [Google Scholar] [CrossRef]
- Guang-Xin, E.; Zhao, Y.J.; Chen, L.P.; Ma, Y.H.; Chu, M.X.; Li, X.L.; Hong, Q.H.; Li, L.H.; Guo, J.J.; Zhu, L.; et al. Genetic diversity of the Chinese goat in the littoral zone of the Yangtze River as assessed by microsatellite and mtDNA. Ecol. Evoution 2018, 8, 5111–5123. [Google Scholar] [CrossRef]
- Deniskova, T.; Bakoev, N.; Dotsev, N.; Selionova, N.; Zinovieva, N. Maternal Origins and Haplotype Diversity of Seven Russian Goat Populations based on the D-loop Sequens variability. Animals 2020, 10, 1603. [Google Scholar] [CrossRef]
- Wang, Y.L.; Chen, X.L.; Wang, X.L.; Yang, Z.X. The genetic diversity of seven indigenous Chinese goat breeds Chinese goat breeds. Small Rumin. Res. 2008, 74, 231–237. [Google Scholar] [CrossRef]
- Peng, W.; Zhang, Y.; Gao, L.; Feng, C.; Yang, Y.; Li, B.; Wu, L.; Wu, A.; Wang, S.; Ren, X. Analysis of World-Scale Mitochondrial DNA Reveals the Origin and Migration Route of East Asia Goats. Front. Genet. 2022, 13, 796979. [Google Scholar] [CrossRef] [PubMed]
- Basrin, R.; Khanom, R.; Hossain, M.M.; Islam, M.S.; Bhuiyan, A.K.F.H.; Bhuiyan, M.S.A. Polymorphisms in BMPR1B and INHβa genes are associated with littersize in indigenous sheep of Bangladesh. J. Anim. Plant Sci. 2023, 33, 483–489. [Google Scholar] [CrossRef]
- Dangar, N.S.; Pandya, G.M.; Ramani, U.V.; Kharadi, V.B.; Brahmkshtri, B.P. Association Study of Fecundity Gene BMP 15 with Prolificacy in Surti Goats under Farm and Field Condition of South Gujarat Region. Indian J. Vet. Sci. Biotechnol. 2022, 18, 100–104. [Google Scholar]
- Kazanski, C.E.; Balehegn, M.; Jones, K.; Bartlett, H.; Calle, A.; Garcia, E.; Hawkins, H.J.; Mayberry, D.; McDonald-Madden, E.; Odadi, W.O.; et al. Context is key to understand and improve livestock production systems. Glob. Food Secur. 2025, 45, 100840. [Google Scholar] [CrossRef]
- Brito, L.F.; Bedere, N.; Douhard, F.; Oliveira, H.R.; Arnal, M.; Peñagaricano, F.; Schinckel, A.P.; Baes, C.F.; Miglior, F. Review: Genetic selection of high-yielding dairy cattle toward sustainable farming systems in a rapidly changing world. Animals 2021, 15 (Suppl. S1), 100292. [Google Scholar] [CrossRef]
- Zhao, Y.; Han, Y.; Yang, Y.; Yuan, C.; Long, Y.; Wal, X. Genetic characterization and selection of litter size traits of Guizhou Black goat and Meigu goat. PLoS ONE 2024, 19, e0313297. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ma, S.; Zhao, B.; Qin, C.; Wu, Y.; Di, J.; Suo, L.; Fu, X. Drivers of plateau adaptability in cashmere goats revealed by genomic and transcriptomic analyses. BMC Genom. 2023, 24, 428. [Google Scholar] [CrossRef] [PubMed]
- Senczuk, G.; Macrì, M.; Civita, M.D.; Mastrangelo, S.; Fresno, M.d.L.; Capote, J.; Pilla, F.; Delgado, J.V.; Amills, M.; Martínez, A. The demographic history and adaptation of Canarian goat breeds to environmental conditions through the use of genome-wide SNP data. Genet. Sel. Evol. 2024, 56, 2. [Google Scholar] [CrossRef] [PubMed]

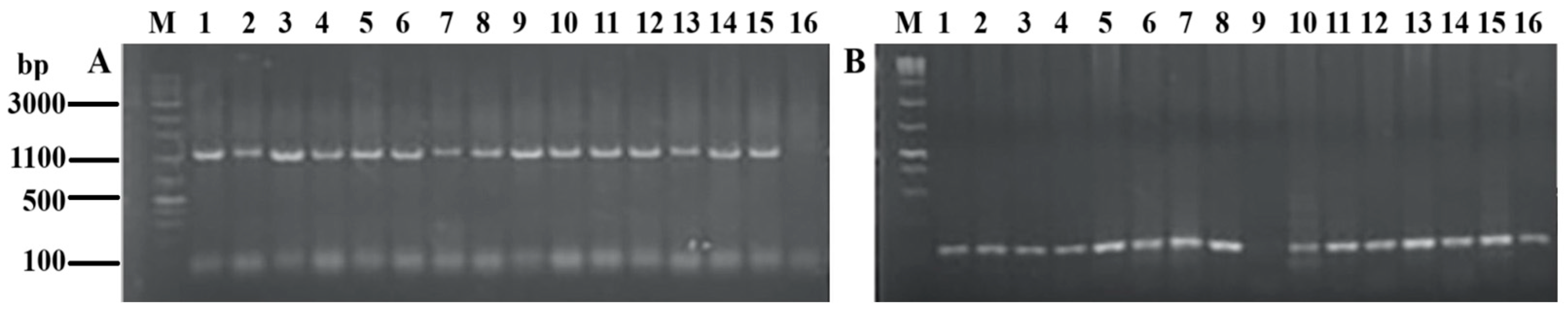
| Type | Nucleotide Sequence | Symbol |
|---|---|---|
| BMPR1B Allele A | R 5′-gctggttccgagagacagaaatatatca-3′ | A |
| F 5′-ccccgtccctttgatatctgcagcaatg-3′ | ||
| BMPR1B Allele G | R 5′-gtcgctatggggaagtttggatgggaa-3′ | G |
| F 5′-atgttttcatgcctcatcaacaccgtcc-3′ |
| Number of Nucleotide Sequences | n | Nucleotide | Frequency (%) | Nucleotide | Frequency (%) |
|---|---|---|---|---|---|
| 133 | 19 | A | 63 | C | 37 |
| 135 | 19 | G | 95 | T | 5 |
| 136 | 19 | A | 95 | G | 5 |
| 137 | 19 | G | 95 | A | 5 |
| 141 | 19 | A | 68 | C | 32 |
| 143 | 19 | G | 63 | A | 37 |
| 148 | 19 | T | 63 | G | 37 |
| Average | 77.43 | 22.57 | |||
| CDS Position(s) | REF→ALT (nuc) | Codon Change | HGVS.p (AA Change) | Effect (SnpEff) | Impact |
|---|---|---|---|---|---|
| 133 and 135 | A → C; G → T | AUG → CUU | p.Met45Leu | missense_variant | Moderate |
| 136 and 137 | A → G; G → A | AGC → GAC | p.Ser46Asp | missense_variant | Moderate |
| 141 | A → C | CCA → CCC | p.Pro47 = | synonymous_variant | Low |
| 143 | G → A | AGA → AAA | p.Arg48Lys | missense_variant | Moderate |
| 148 | T → G | TTA → GTA | p.Leu50Val | missense_variant | Moderate |
| No | Haplotype Position of ILEG |
|---|---|
| 1 | Hap_1: 1 [GR10] |
| 2 | Hap_2: 1 [AGI6] |
| 3 | Hap_3: 5 [AGI10 BW147; AGI4 AGI22 GR8] |
| 4 | Hap_4: 1 [AGI5] |
| 5 | Hap_5: 1 [BWB12] |
| 6 | Hap_6: 1 [AGI24] |
| 7 | Hap_7: 1 [LW2] |
| 8 | Hap_8: 1 [BW10] |
| 9 | Hap_9: 1 [GR2] |
| 10 | Hap_10: 1 [BW14] |
| 11 | Hap_11: 1 [AGI11] |
| 12 | Hap_12: 1 [AGI23] |
| 13 | Hap_13: 1 [BWC17] |
| 14 | Hap_14: 1 [AGI18] |
| 15 | Hap_15: 1 [AGI17] |
| Gene | n | Frequency of Genetic Group | Genetic Group | No. of Does | Average Prolificacy | ||
|---|---|---|---|---|---|---|---|
| AA | GA | GG | GG | 11 | 1.89 | ||
| BMPRIB | 73 | 0.15 | 0.70 | 0.15 | GA | 51 | 1.60 |
| (11) | (51) | (11) | AA | 11 | 1.49 | ||
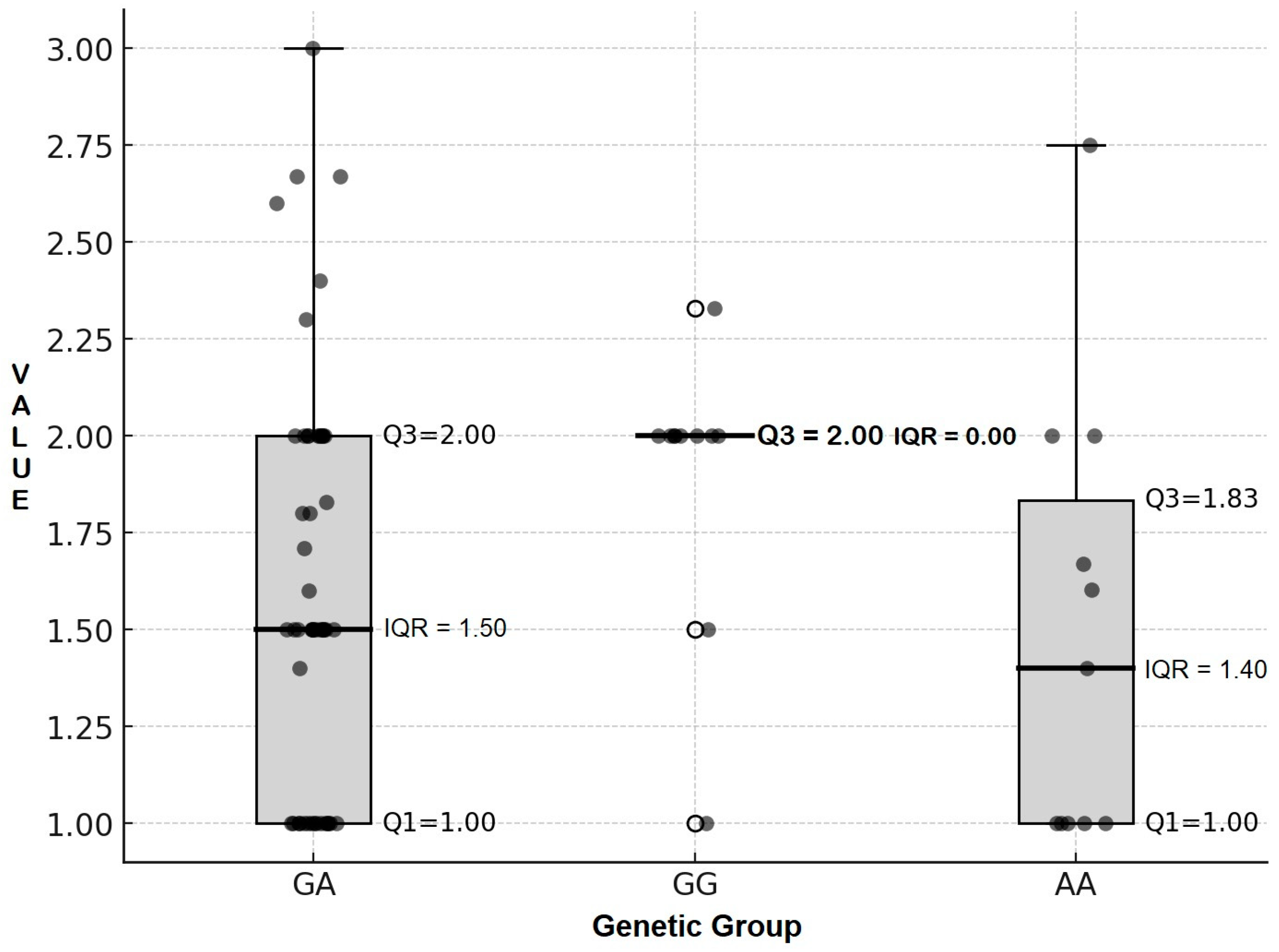
| Genetic Group | n | Mean | Median | IQR | Shapiro–Wilk Normality Test | Kruskal–Wallis Test |
|---|---|---|---|---|---|---|
| p-Value # | p-Value ## | |||||
| GG | 11 | 1.89 ± 0.35 | 2.00 | 0.00 | 0.0003 | |
| GA | 51 | 1.60 ± 0.52 | 1.50 | 1.00 | 0.0002 | 0.056 |
| AA | 11 | 1.49 ± 0.58 | 1.40 | 0.84 | 0.027 |
| Comparison | Median Differences (%) | Pairwise Mann–Whitney Tests (p-Value #) |
|---|---|---|
| GG vs. GA | +33.30 | 0.029 |
| GG vs. AA | +42.90 | 0.040 |
| GA vs. AA | +7.10 | 0.508 |
| Allele Frequency | Genetic Group Frequency | Genetic Group | No. of Does | |||||
|---|---|---|---|---|---|---|---|---|
| Gene | n | A | G | AA | AG | GG | GG | 53 |
| BMPRIB | 385 | 0.55 | 0.45 | 0.55 | 0.45 | 0.14 | AG | 316 |
| (16) | (316) | (53) | AA | 16 | ||||
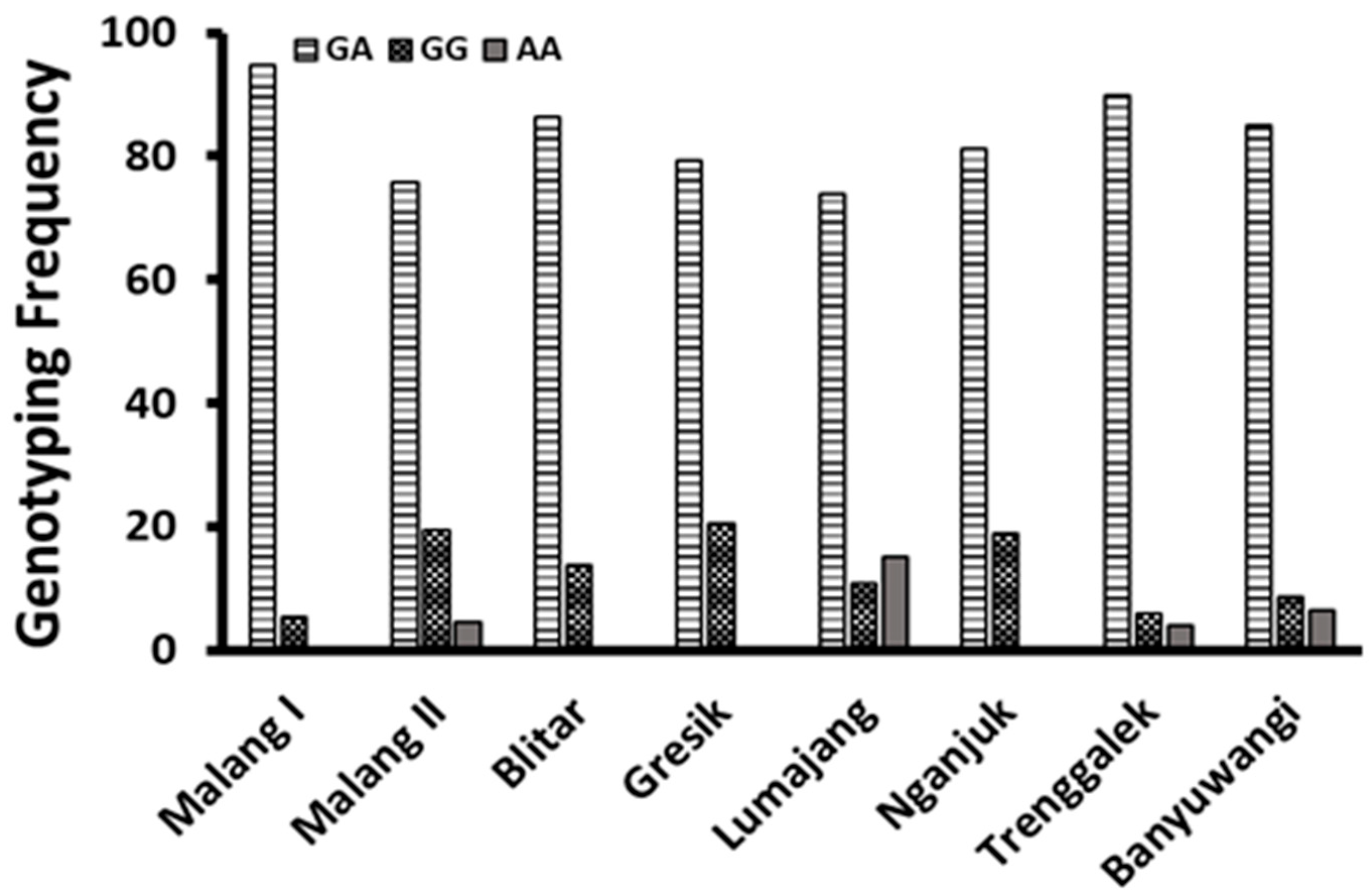
| Breeding Villages | n | Amount (Head) | Genetic Group Frequencies (%) | ||||
|---|---|---|---|---|---|---|---|
| Heterozygous | Homozygous for Allele G | Homozygous for Allele A | GA | GG | AA | ||
| Malang I | 38 | 36 | 2 | 0 | 94.74 | 5.26 | 0.00 |
| Malang II | 87 | 66 | 17 | 4 | 75.86 | 19.54 | 4.60 |
| Blitar | 22 | 19 | 3 | 0 | 86.36 | 13.64 | 0.00 |
| Gresik | 63 | 50 | 13 | 0 | 79.37 | 20.63 | 0.00 |
| Lumajang | 46 | 34 | 5 | 7 | 73.91 | 10.87 | 15.22 |
| Nganjuk | 32 | 26 | 6 | 0 | 81.25 | 18.75 | 0.00 |
| Trenggalek | 50 | 45 | 3 | 2 | 90.00 | 6.00 | 4.00 |
| Banyuwangi | 47 | 40 | 4 | 3 | 85.11 | 8.51 | 6.38 |
| Total | 385 | 316 b | 53 a | 16 a | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mudawamah, M.; Fadli, M.Z.; Ciptadi, G.; Fatchiyah, F.; Lestari, M.W.; Oktanella, Y.; Susiati, S.; Charles, A.L. Polymorphism of the BMPR1B Variants for Prolific Traits in the Indonesian Local Ettawah Goat. Animals 2025, 15, 2781. https://doi.org/10.3390/ani15192781
Mudawamah M, Fadli MZ, Ciptadi G, Fatchiyah F, Lestari MW, Oktanella Y, Susiati S, Charles AL. Polymorphism of the BMPR1B Variants for Prolific Traits in the Indonesian Local Ettawah Goat. Animals. 2025; 15(19):2781. https://doi.org/10.3390/ani15192781
Chicago/Turabian StyleMudawamah, Mudawamah, Muhammad Zainul Fadli, Gatot Ciptadi, Fatchiyah Fatchiyah, Mahayu Woro Lestari, Yudith Oktanella, Susiati Susiati, and Albert Linton Charles. 2025. "Polymorphism of the BMPR1B Variants for Prolific Traits in the Indonesian Local Ettawah Goat" Animals 15, no. 19: 2781. https://doi.org/10.3390/ani15192781
APA StyleMudawamah, M., Fadli, M. Z., Ciptadi, G., Fatchiyah, F., Lestari, M. W., Oktanella, Y., Susiati, S., & Charles, A. L. (2025). Polymorphism of the BMPR1B Variants for Prolific Traits in the Indonesian Local Ettawah Goat. Animals, 15(19), 2781. https://doi.org/10.3390/ani15192781







