Simple Summary
Analyzing the genetic basis of important economic traits of livestock and poultry and screening relevant molecular markers are the theoretical basis and premise of using molecular breeding technology to improve relevant traits. The FTO gene, which regulates adipogenesis and fat accumulation and is strongly associated with body fat index, was the first disease-associated gene to be identified using gene-wide association analysis (GWAS) in humans. However, the association between the FTO gene polymorphism and important traits of chicken is still poorly understood. It is not clear whether SNPs in the FTO gene can be used as markers for the molecular marker-assisted breeding of important economic traits in chickens. In the present study, SNPs in exons of the FTO gene were detected by PCR amplification and DNA sequencing.
Abstract
Fat volume and obesity-related genes (e.g., the FTO gene) are important candidate genes affecting energy metabolism. Single nucleotide polymorphisms (SNPs) in the FTO gene are associated with carcass, growth and meat quality traits of pigs, cattle, sheep, rabbits and ducks. The purpose of this study was to detect the single nucleotide polymorphisms in the chicken FTO gene coding region by DNA sequencing and analyze its association with the carcass and growth traits of Heying black chickens. We detected polymorphisms in exons 5, 7, 8 and 9, respectively, g.57337C>A, g.64757T>G, g.97213G>A and g.220985G>A, which are synonymous mutations. g.57337C>A mutation site CA and AA genotype individuals were significantly higher than CC genotype individuals in live weights, head weights, breast muscle weights and leg muscle weights (p < 0.05), AA genotype individuals were significantly higher than CC genotype individuals in slaughter live weights and liver weights (p < 0.05) and CA and AA genotype individuals were significantly higher than CC genotype individuals in heart weights (p < 0.01). In terms of growth traits, the weights of individuals with genotype CA at 8 weeks were significantly higher than that of individuals with genotype CC (p < 0.05), and the weights of individuals with genotype AA and CA at 10 weeks and 16 weeks were significantly higher than that of individuals with genotype CC (p < 0.05). For the g.64757T > G mutation, individuals with the TT genotype exhibited significantly higher values (p < 0.05) than those with the TG genotype across multiple traits, including slaughter weight, live weight, eviscerated weight and semi-eviscerated weight. Individuals with GG genotypes were significantly higher than individuals with TG genotypes (p < 0.05) in slaughter weights and wing weights. In terms of growth traits, the 16-week-old body weight of individuals with TT genotype was significantly higher than that of individuals with TG genotype (p < 0.01). The GA genotype exhibited significantly higher slaughter weights in the g.97213G>A variant compared to the GG genotype (p < 0.01), and in live weights, eviscerated weights, semi-eviscerated weights, leg muscle weights and wing weights, GA genotype was significantly higher than in GG genotype (p < 0.05). In terms of growth traits, GA genotype was significantly higher in individuals 8 weeks old, 10 weeks old and 16 weeks old than GG genotype (p < 0.05). g.220985G>A was significantly higher in individuals with GG genotype than GA genotype (p < 0.05). In terms of growth traits, the weight of GG genotype was significantly higher than that of GA genotype (p < 0.05). The results showed that the FTO gene may be a candidate gene related to chicken growth and slaughter traits and lays a foundation for Heying black chicken assisted breeding.
1. Introduction
The long-term goal of broiler genetic improvement is to enhance meat production performance and growth traits. In the 21st century, molecular breeding technology represented by functional genomics, molecular marker-assisted breeding and gene editing has become an important symbol and development direction of animal breeding. Analyzing the genetic basis of important economic traits of livestock and poultry and screening relevant molecular markers are the theoretical basis and premise of using molecular breeding technology to improve relevant traits. Single nucleotide polymorphism (SNP) was first proposed in 1997 [1]. As a third-generation genetic marker, SNPs have been widely used in population genetics and disease-related gene localization research, and play an important role in early diagnosis, prevention and treatment of diseases, study of the influence of genetic factors in drug metabolism and guidance for drug clinical use [2]. With the rapid development of molecular biology and cross-cutting disciplines, the variety of SNP detection techniques is increasing. In addition to the common sequencing, allele-specific amplification and high-resolution melting curve methods, there are also mass spectrometry, gene chips, oligonucleotide linkage analysis, dynamic allele-specific hybridization, and endonuclease-based restriction fragment length polymorphisms and random amplification polymorphisms [3]. SNP detection will become more efficient and rapid, and SNP-based research will become increasingly widespread, whether in molecular marker-assisted breeding, rapid identification of specific strains or even between individuals, research into resistance to disease in animal organisms or individualized drug delivery.
With the rapid development of molecular biology technology, SNP gene detection and typing technology and methods continue to innovate. Although some classical SNP detection technologies are still widely used in research, highly sensitive and high-throughput SNP detection methods are also increasingly valued. For example, the most widely used SNP detection method in research is PCR-DNA direct sequencing, namely DNA Sanger sequencing [4]. Ye used SNP arrays to genotype 450 roosters [5]. Zhang directly sequenced chickens and detected AMY1A polymorphisms in chickens. Seven SNPs in the 5′ flanking region, exon, intron, and 3′ UTR (3′ untranslated region) of AMY1A were significantly associated with daily weight gain, average daily feed intake, leg muscle weight and abdominal fat [6].
The Fat mass and obesity-associated (FTO) gene is an m6A demethylase belonging to the ALKB dioxygenase family [7]. The FTO protein is an AlkB-like DNA/RNA demethylase with a strong preference for 3-methylthymidine (3-meT) in single-stranded DNA or 3-methyluracil (3-meU) in single-stranded RNA [8,9,10]. The catalytic activity of the FTO gene is generated through the interaction of the amino-terminal and carboxy-terminal structural domains. The FTO gene possesses an additional loop that selectively competes with the unmethylated strand of the DNA double-stranded to bind the FTO gene, suggesting that it has an important role in the selection of duplex nucleic acids by the FTO gene [11]. The FTO gene, which regulates adipogenesis and fat accumulation and is strongly associated with body fat, was the first disease-associated gene to be identified using gene-wide association analysis (GWAS) [12]. The FTO gene has also been associated with type 2 diabetes [13], growth retardation, developmental delay [14], metabolic disorders [15] and hypertension [16]. In addition, the FTO gene also plays a crucial role in muscle growth, as it can promote the development of bovine muscle satellite cells, which are mainly responsible for muscle growth and regeneration [17].
The polymorphisms of the FTO gene and their associations with economic traits have been studied in pigs [18], cattle [19], sheep [20], rabbits [21] and ducks [22]. However, the association between the FTO gene polymorphism and important traits of chicken is still poorly understood. Muscle development is intricately linked to an animal’s growth rate, meat yield and quality, while also serving as an indicator of its health status and feed utilization efficiency. Slaughter characteristics, on the other hand, dictate the economic value, quality and safety of meat products, reflect breeding management standards and cater to market demands and consumer preferences. Both factors are pivotal in enhancing the efficiency of animal husbandry. It is not clear whether SNPs in the FTO gene can be used as markers for the molecular marker-assisted breeding of important economic traits in chickens. In the present study, SNPs in exons of the FTO gene were detected by PCR amplification and DNA sequencing. The association between the FTO gene polymorphism and carcass traits and growth traits of Heying black chicken was analyzed. We hope to provide a theoretical basis for the molecular marker-assisted breeding of Heying black chicken.
2. Materials and Methods
2.1. Animals
Heying black chicken was jointly developed by Yangzhou University and Jiangsu Sandali Animal Husbandry Development Co., Ltd., Changzhou, Jiangsu. Through several generations of continuous breeding and cultivation, the production performance has become stable. As a high-quality black-feathered chicken, Heying black chicken has the characteristics of low fat, high protein, rich nutrition and delicious meat. The experimental birds for this study were from the seventh generation of the Heying black chicken B line, which was named after a local chicken breed. A total of 176 chickens were raised under the same conditions in Jiangsu Heying Animal Husbandry Co., Ltd., Changzhou, Jiangsu. They were raised and then slaughtered at the age of 16 weeks. The living environment and nutrition level of chickens were the same. The 176 Heying black chickens (133 hens and 43 roosters) were randomly selected, and the body weight at 0, 2, 4, 8, 10 and 16 weeks of age were recorded. In total, 2.0 mL of blood were collected from the wing vein, anticoagulated with sodium citrate, and stored in a freezer at −20 °C. The 176 Heying black chickens were slaughtered and measured at 16 weeks, and 14 carcass traits including slaughter weights, live weights, semi-eviscerated weights (on the basis of slaughter weights, the weights after removing internal organs) and eviscerated weights (on the basis of semi-eviscerated weight, the weight after removing heart, liver and fat), breast muscle weights (bilateral pectoralis major muscles), leg muscle weights (all muscles from both thighs), head weights, foot weights (foot and shanks), heart weights, liver weights, spleen weights, abdominal fat weights, wing weights and stomach weights (muscle stomach and glandular stomach) were recorded.
For chicks in the age range of 1–3 days, the ambient temperature must be meticulously maintained within the interval of 33–35 °C. As the chicks reach 4–7 days of age, the temperature can be gradually decreased to 32–34 °C. During the second week of their growth, an optimal temperature range of 28–30 °C was sustained. In the third week, the temperature was regulated to fall between 25 and 28 °C. By the fourth week, the temperature was adjusted to 22–25 °C. From the fifth week until the time of slaughter, the temperature needed to be stabilized at 20–21 °C to ensure the proper growth and development of the chickens. For the experimental conditions concerning humidity, we maintained a relative humidity level of 65% to 70% for the chickens aged between 1 and 7 days. The humidity can be adjusted to 60–65% at 8–10 days of age. The relative humidity during the period of 11 to 28 days of age was generally maintained at 55–60%. After 28 days of age, the humidity was usually stabilized at around 55%. The chicken coop has a light exposure time of 12 h and a dark exposure time of 12 h. The chickens in the farm were provided with clean, sterile water via a fully automatic water dispenser, allowing them free access to drinking water at all times. The diet was supplemented with feed manually, and the feed was added three times a day. The feed formula consists of the following components: 3.2% wheat bran, 62% corn, 31% soybean meal, 1.3% calcium hydrogen phosphate, 1.2% stone powder, 1% additives and 0.3% salt.
2.2. Genomic DNA Extraction
Genomic DNA was extracted from blood by the phenol/chloroform method. An ultraviolet spectrophotometer was used to determine the concentration and purity of DNA. The DNA was stored at −20 °C for PCR amplification at a later date.
2.3. Primer Design
Based on the chicken FTO gene sequence published in GenBank, seven pairs of primers (P1–P7) were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 5 October 2022) to amplify the exons of the FTO gene. The primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China, and their sequences are presented in Table 1.

Table 1.
Sanger sequencing of PCR products.
2.4. PCR Amplification and Sequencing
The reaction mixtures were added in a 200 μL PCR tube plate for pre-denaturation at 94 °C for 3 min. The following PCR thermal profile was used: denaturation at 94 °C for 15 s, primer annealing at 58 °C for 15 s, DNA synthesis at 72 °C for 30 s, 30 cycles and extension at 72 °C for 5 min. PCR products were sent to Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China, for sequencing.
2.5. Analysis Software
DNAMAN 5.2 and Chromas 1.62 were used to visualize the sequencing peak map and detect polymorphic sites through sequence alignment.
2.6. Statistical Model and Analysis
- The data were statistically analyzed using the general linear model (GLM) and Multiple Comparisons (version SPSS 25). The model was Yijk = μ + Gj + Sk + eijk [23], where Yijk was the individual phenotypic record, μ was the population mean, Gj was the effect of genotype, Sk was the sex effect, and eijk was the random error. Differences among genotypes were indicated by superscript letters: different uppercase letters represented highly significant differences (p < 0.01), different lowercase letters represented significant differences (p < 0.05), and the same letters indicated no significant difference (p > 0.05)
- Polymorphism information content (PIC) was calculated using the formulawhere n is the number of alleles; Pi and Pj are the frequencies of the ith and jth alleles, respectively.
- Effective number of alleles (Ne) was calculated using the formula
- Average heterozygosity was calculated using the formula
2.7. Spatiotemporal Expression Differences and Tissue Expression Profiles
Leg and pectoral tissues were collected during the embryonic stage (d12, d14, d16, d18, and d21) and growth period (W2, W4, W8, W10, and W16), and heart, liver, spleen, lung, kidney, fat, breast muscles and leg muscles were collected at the 1st and 16th weeks of life. RNA extraction was performed using the VeZol-Pure Total RNA Isolation Kit (Vazyme, Nanjing, China), followed by reverse transcription with HiScript IV RT SuperMix for qPCR (including gDNA wiper, Vazyme, Nanjing, China) and subsequent real-time fluorescence quantification employing ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). The primer sequences used were as follows: FTO (forward: TTCACCAAGGCGACCTCTAC; reverse: GCTGAACCGAGGTGAAAAGC) and β-actin (forward: CAGCCATCTTTCTTGGGTAT; reverse: CTGTGATCTCCTTCTGCATCC). The following PCR protocol was applied: initial denaturation (1 min at 95 °C), followed by a three-step amplification program (20 s at 95 °C, 20 s at 60 °C, 20 s at 72 °C) that was repeated 42 times. The 2−ΔΔCt method was used to analyze the real-time PCR data relative to the average value of control. The quantitative real-time PCR (RT-qPCR) results for all genes were statistically tested using Student’s t-test.
3. Results
3.1. PCR Amplification Results
Four SNPs g.57337C>A, g.64757T>G, g.97213G>A, and g.220985G>A were detected in the exon regions of the FTO gene using DNA sequencing (Figure 1). The g.57337C>A SNP was in exon 5, and forms CC, AA and CA. The g.64757T>G mutation was in exon 7, and forms GG, TT and TG. The g.97213G>A mutation was in exon 8, and forms GG and GA. The g.220985G>A mutation was in exon 9, and forms GG and GA.
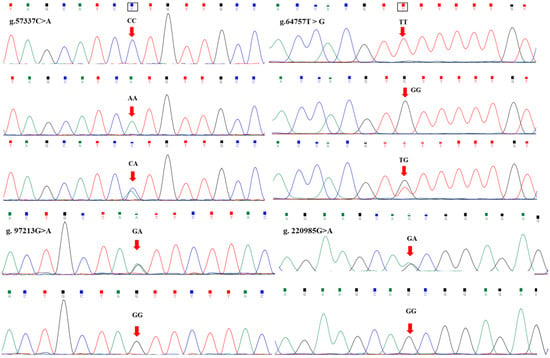
Figure 1.
Sequence analysis of mutations in exon regions of the FTO gene.
3.2. Genotyping and Allele Frequency Analysis of the FTO Gene
Polymorphism and genetic diversity analysis of four SNP loci are as shown in Table 2; the gene frequencies of alleles A and C are 0.236 and 0.764. The gene frequencies of alleles T and G were 0.261 and 0.739, respectively. The gene frequencies of alleles G and A were 0.952 and 0.048, respectively. The gene frequencies of alleles G and A were 0.949 and 0.051, respectively. The PICs of alleles g.97213G>A and g.220985G>A were less than 0.25, indicating that they were lowly polymorphic. g.57337C>A and g.64757T>G were moderately polymorphic with PIC values greater than 0.25.

Table 2.
Polymorphisms at SNPs of the FTO gene.
3.3. Association Analysis Between the FTO Gene Polymorphisms and Chicken Carcass Traits
3.3.1. Association Analysis of g.57337C>A SNP and Carcass Traits of Heying Black Chicken
SPSS 25.0 was used to analyze the association between the g.57337C>A site of the FTO gene and the carcass traits of the Heying black chicken. The analysis showed that the CA and AA genotype individuals were significantly higher than the CC genotype individuals in live weights, head weights, breast muscle weights and leg muscle weights (p < 0.05). The AA genotype individuals were significantly higher than the CA genotype individuals in live weights (p < 0.05). The heart weights of CA and AA genotype individuals were significantly higher than that of CC genotype individuals (p < 0.01) (Table 3).

Table 3.
Association analysis of g.57337C>A SNP and carcass traits of the Heying black chicken population.
3.3.2. Association Analysis of g.64757T>G SNP and Carcass Traits of Heying Black Chicken
The results showed that the TT genotype individuals were significantly higher than the TG genotype individuals in live weights, eviscerated weights and semi-eviscerated weights (p < 0.05). The slaughter weights and wing weights of TT and GG genotype individuals were significantly higher than those of TG genotype individuals (p < 0.05) (Table 4).

Table 4.
Association analysis of g.64757T>G SNP and carcass traits of the Heying black chicken population.
3.3.3. Association Analysis Between g.97213G>A SNP and Carcass Traits of Heying Black Chicken
The results showed that GA genotype individuals were significantly higher than GG genotype individuals in slaughter weights (p < 0.01), and GA genotype individuals were significantly higher than GG genotype individuals in live weights, eviscerated weights, semi-eviscerated weights, leg muscle weights and wing weights (p < 0.05) (Table 5).

Table 5.
Association analysis of g.97213G>A SNP and carcass traits of the Heying black chicken population.
3.3.4. Association Analysis Between g.220985G>A SNP and Carcass Traits of Heying Black Chicken
The GG genotype individuals were significantly higher than the GA genotype individuals in slaughter weights and live weights (p < 0.05) (Table 6). Although GG genotype in other tissues was higher than GA genotype, it was not significant.

Table 6.
Association analysis between g.220985G>A SNP and carcass traits of the Heying black chicken population.
3.4. Association Analysis of the FTO Gene Polymorphism and Growth Traits of Heying Black Chicken
3.4.1. Association Analysis of g.57337C>A SNP and Growth Traits of Heying Black Chicken
The association analysis between the FTO gene mutation and the growth traits of Heying black chicken showed that the body weights of individuals with the CA genotype at 8 weeks was significantly higher than that of individuals with the CC genotype (p < 0.05), Individuals with the AA genotype had body weights between that of individuals with the other two genotypes. The body weights of individuals with AA and CA genotypes at 10 weeks and 16 weeks were significantly higher than that of individuals with the CC genotype (p < 0.05) (Table 7).

Table 7.
Association analysis of g.57337C>A SNP and growth traits of the Heying black chicken population.
3.4.2. Association Analysis Between g.64757T>G SNP and Growth Traits of Heying Black Chicken
The body weight of TT genotype individuals at 16 weeks was significantly higher than that of TG genotype individuals (p < 0.05). Body weight of individuals with the GG genotype was between that of individuals with the other two genotypes (Table 8).

Table 8.
Association analysis between g.64757T>G SNP and growth traits of Heying black chicken population.
3.4.3. Association Analysis Between g.97213G>A SNP and Growth Traits of Heying Black Chicken
The weight of GA genotype individuals at 8, 10 and 16 weeks of age was significantly higher than that of GG genotype individuals (p < 0.05) (Table 9). Although GG genotype in other weeks was heavier than GA genotype, it was not significant.

Table 9.
Association analysis between g.97213G>A SNP and growth traits of the Heying black chicken population.
3.4.4. Association Analysis of g.2220985G>A SNP and Growth Traits of Heying Black Chicken
The weight of individuals with GG genotype at the age of 8 weeks, 10 weeks and 16 weeks was significantly higher than that of individuals with GA genotype (p < 0.05) (Table 10). Although GG genotype in other weeks was higher than GA genotype, it was not significant.

Table 10.
Association analysis between g.2220985G>A SNP and growth traits of the Heying black chicken population.
3.5. Spatiotemporal Expression Differences and Tissue Expression Profiles
The quantification of the FTO gene revealed that expression patterns in the breast muscle and leg muscle followed a similar temporal trajectory (Figure 2A). Notably, the FTO gene mRNA expression reached significant levels during the late embryonic and late growth stages (p < 0.05), and it was highly significant in the early growth stage (p < 0.01). Additionally, FTO gene mRNA expression varied across different tissues in an age-dependent manner (Figure 2B). Specifically, at 16 weeks of age, the heart and lungs exhibited significantly higher the FTO gene mRNA expression compared to their levels at 1 week of age (p < 0.01), while the liver and kidneys showed a significant increase (p < 0.05). Conversely, fat tissue and breast muscle displayed significantly higher FTO gene mRNA expression at 1 week of age than at 16 weeks (p < 0.05).
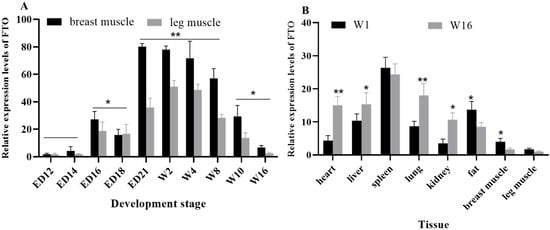
Figure 2.
The FTO gene mRNA spatiotemporal expression and tissue expression. (A) The expression levels of the FTO in chicken leg muscle and breast muscle at different stages. (B) The expression levels of the FTO gene in different tissues of chicken W1 and W16 stages. * p < 0.05 and ** p < 0.01.
4. Discussion
At present, the research on the FTO gene polymorphism is mainly focused on humans. Most studies focus on the relationship between the FTO gene variation and obesity [24], body mass index [25], metabolic disease and tumor [26] in different regions and ethnic groups. The FTO gene is also widely distributed among various tissues of animals. Chung [27] studied the association between the SNP site of the FTO gene and the meat quality traits of Korean cattle. The study showed that the mutation of g.125550A>T site has a significant association with the meat quality traits; Fan et al. [28] found a highly significant association between the pig’s total intramuscular fat content and the c.594C>G mutation of the FTO gene. Fu et al. [29] detected the FTO gene polymorphism in pigs and found that the G allele at A227G seems to have a beneficial effect on fat deposition. This indicates that the FTO gene may participate in the mechanism of fat deposition and is one of the main genes affecting meat quality traits. These studies further showed that the mutation site of the FTO gene was significantly related to the slaughter and growth traits of livestock, and may be one of the important candidate genes for the economic traits of livestock and poultry. Improvements in growth and carcass traits are the main objectives for chicken breeding.
To date, the association analysis between the FTO gene polymorphism and chicken carcass traits and growth traits has not been reported, and whether it can be used for molecular marker-assisted selection of chicken growth and carcass traits remains to be uncovered. Exons, as a part of eukaryotic genes, contain the core information required for protein synthesis. In this study, primers were designed to detect the SNP sites in the exons of the FTO gene of Heying black chicken by PCR synthesis and DNA sequencing technology. Four mutation sites were detected, which were g.57337C>A, g.64757T>G, g.97213G>A and g.220985G>A.
The genotypes of the four mutation sites are significantly related to live weights and slaughter weights. Live weight refers to the weight 12 h after feeding before slaughter. Some studies have shown that heavier broilers (>3.3 kg) produce less breast meat (<3.0 and 3.0 to 3.3 kg) [30]. The slaughter weight refers to the weight of a chicken after bloodletting, hair removal, foot removal (below the tarsal bones) and head removal. The ratio of slaughter weights to live weight determines the slaughter rate of poultry. It is well known that skeletal muscle, as the body’s largest organ, accounts for about 40~50% of the total body weight [31]. The semi-eviscerated weights, and leg and breast muscles are closely related to the development of skeletal muscle. This study showed that g.64757T>G and g.97213G>A were significantly associated with the semi-eviscerated weights (on the basis of slaughter weights, the weight after removing internal organs) and eviscerated weights (on the basis of semi clean bore weight, the weight after removing heart, liver, and fat). Polymorphisms g.57337C>A and g.97213G>A 356 were significantly associated with the leg muscle weights and breast muscle weights. In addition, g.57337C>A was significantly associated with head weights, while g.64757T>G and g.97213G>A were significantly associated with wing weights. Interestingly, our research found that in addition to the traits related to skeletal muscle development, the FTO gene also has different associations with some internal organs. For example, g.57337C>A is significantly related to liver weights and heart weights, which needs further exploration and research.
Growth traits are important variables to evaluate the profitability of broiler production. Carcass development often parallels changes in growth traits, with most carcass traits showing significant positive associations [32]. This study showed that four mutation sites (g.57337C>A, g.64757T>G, g.97213G>A and g.220985G>A) were significantly associated with body weights at 16 weeks of age. In addition, g.57337C>A, g.97213G>A and g.220985G>A were significantly associated with body weights at the age of 8 weeks and 10 weeks. Wang et al. [33] showed that the polymorphism formed by insertion/deletion (InDel) of the FTO gene was significantly related to the growth traits of Tongyang sheep and could be used to in marker-assisted selection for Tongyang sheep. Polymorphisms in the FTO gene showed significant associations with body weights of different breeds of rabbits at various ages, reflecting variations in the effects of the FTO gene among species [21].
In conclusion, the four SNPs of the FTO gene showed significant association with the slaughter traits related to the skeletal muscle development of Heying black chicken, and also showed significant association with the late growth stage (8, 10 and 16 weeks old). The findings suggest that the FTO gene may be a candidate gene related to chicken growth and slaughter traits.
It is quite intriguing to observe that the FTO gene expression remains relatively low during the early stages of embryonic development, then gradually increases in the later embryonic phases, persisting through the growth stage, before ultimately declining around 8 weeks of age in the later stages of life. However, research has demonstrated that the level of the FTO gene mRNA in the skeletal muscle of 8-week-old chickens is actually higher than that observed in 4-week-old chickens [34]. Wang found that the FTO gene is highly expressed in the hypothalamus, liver, visceral fat and cerebellum of chickens [35]. This study found that the FTO gene is expressed differently in chicken heart, liver, spleen, lung, kidney, fat, breast muscles and leg muscles. The observed differences in the temporal expression pattern of the FTO gene between the present study and previous research may be attributed to variations in chicken breeds.
5. Conclusions
This study detected a mutation site in exons 5, 7, 8 and 9 with g.57337C>A, g.64757T>g, g.97213G>A and g.220985G>A, which are homologous mutations. The SNPs have varying degrees of impact on carcass traits. The FTO gene may be a candidate gene related to chicken growth and slaughter traits and lays a foundation for Heying black chicken assisted breeding.
Author Contributions
Conceptualization, H.D., L.C., C.C. and T.Z.; Data curation, H.D.; Formal analysis, T.Z.; Funding acquisition, K.X. and. J.W.; Investigation, H.D., W.C. and T.Z.; Methodology, H.D.; Software, H.D.; Supervision, G.Z.; Writing—original draft, T.Z. and H.D.; Writing—review and editing, T.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the China Agriculture Research System of MOF and MARA (CARS-41), This work was funded by the National Key Research and Development Program of China (2024YFF1000200), Jiangsu Agriculture Science and Technology Innovation Fund (JASTIF) (CX (24)3081).
Institutional Review Board Statement
This study was performed following the Chinese guidelines for animal welfare, and the animal protocol was approved by the Animal Welfare Committee of Yangzhou University (permit number SYXK [Su] 2016–0020).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Collins, F.S.; Guyer, M.S.; Charkravarti, A. Variations on a theme: Cataloging human DNA sequence variation. Science 1997, 278, 1580–1581. [Google Scholar] [CrossRef] [PubMed]
- Shastry, B.S. SNPs: Impact on gene function and phenotype. Methods Mol. Biol. 2009, 578, 3–22. [Google Scholar] [CrossRef]
- Kim, S.; Misra, A. SNP genotyping: Technologies and biomedical applications. Annu. Rev. Biomed. Eng. 2007, 9, 289–320. [Google Scholar] [CrossRef]
- Vossen, R.H. Genotyping DNA Variants with High-Resolution Melting Analysis. Methods Mol. Biol. 2017, 1492, 17–28. [Google Scholar] [CrossRef]
- Ye, S.; Yuan, X.; Lin, X.; Gao, N.; Luo, Y.; Chen, Z.; Li, J.; Zhang, X.; Zhang, Z. Imputation from SNP chip to sequence: A case study in a Chinese indigenous chicken population. J. Anim. Sci. Biotechnol. 2018, 9, 30. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhong, H.; Lin, S.; Liang, L.; Ye, S.; Xu, Z.; Ji, C.; Zhang, Z.; Zhang, D.; Zhang, X. Polymorphisms of AMY1A gene and their association with growth, carcass traits and feed intake efficiency in chickens. Genomics 2021, 113, 583–594. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.; Girard, C.A.; Tung, Y.C.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.; McDonough, M.A.; Cunliffe, S.; McNeill, L.A.; et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007, 318, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pulido, L.; Andrade-Navarro, M.A. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 2007, 8, 23. [Google Scholar] [CrossRef]
- Jia, G.; Yang, C.G.; Yang, S.; Jian, X.; Yi, C.; Zhou, Z.; He, C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008, 582, 3313–3319. [Google Scholar] [CrossRef]
- Han, Z.; Niu, T.; Chang, J.; Lei, X.; Zhao, M.; Wang, Q.; Cheng, W.; Wang, J.; Feng, Y.; Chai, J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 2010, 464, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.; Yeo, G.S. The bigger picture of FTO: The first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 2014, 10, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Naaz, K.; Kumar, A.; Choudhury, I. Assessment of FTO Gene Polymorphism and its Association with Type 2 Diabetes Mellitus in North Indian Populations. Indian J. Clin. Biochem. 2019, 34, 479–484. [Google Scholar] [CrossRef]
- Daoud, H.; Zhang, D.; McMurray, F.; Yu, A.; Luco, S.M.; Vanstone, J.; Jarinova, O.; Carson, N.; Wickens, J.; Shishodia, S.; et al. Identification of a pathogenic FTO mutation by next-generation sequencing in a newborn with growth retardation and developmental delay. J. Med. Genet. 2016, 53, 200–207. [Google Scholar] [CrossRef]
- Peng, S.; Xiao, W.; Ju, D.; Sun, B.; Hou, N.; Liu, Q.; Wang, Y.; Zhao, H.; Gao, C.; Zhang, S.; et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 2019, 11, eaau7116. [Google Scholar] [CrossRef]
- He, D.; Fu, M.; Miao, S.; Hotta, K.; Chandak, G.R.; Xi, B. FTO gene variant and risk of hypertension: A meta-analysis of 57,464 hypertensive cases and 41,256 controls. Metabolism 2014, 63, 633–639. [Google Scholar] [CrossRef]
- Ma, Z.; Chai, Z.; Yang, H.; Zhang, X.; Zhao, H.; Luo, X.; Zhong, J.; Wu, Z. Comprehensive analysis of the expression patterns and function of the FTO-LINE1 axis in yak tissues and muscle satellite cells. Front. Vet. Sci. 2024, 11, 1448587. [Google Scholar] [CrossRef] [PubMed]
- Dvořáková, V.; Bartenschlager, H.; Stratil, A.; Horák, P.; Stupka, R.; Cítek, J.; Sprysl, M.; Hrdlicová, A.; Geldermann, H. Association between polymorphism in the FTO gene and growth and carcass traits in pig crosses. Genet. Sel. Evol. 2012, 44, 13. [Google Scholar] [CrossRef]
- Jevsinek Skok, D.; Kunej, T.; Kovac, M.; Malovrh, S.; Potocnik, K.; Petric, N.; Zgur, S.; Dovc, P.; Horvat, S. FTO gene variants are associated with growth and carcass traits in cattle. Anim. Genet. 2016, 47, 219–222. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, D.; Zhang, X.; Li, F.; Xu, D.; Zhao, L.; Li, X.; Zhang, Y.; Wang, J.; Yang, X.; et al. Expression features of the ovine FTO gene and association between FTO polymorphism and tail fat deposition related-traits in Hu sheep. Gene 2022, 826, 146451. [Google Scholar] [CrossRef]
- Zhang, G.W.; Gao, L.; Chen, S.Y.; Zhao, X.B.; Tian, Y.F.; Wang, X.; Deng, X.S.; Lai, S.J. Single nucleotide polymorphisms in the FTO gene and their association with growth and meat quality traits in rabbits. Gene 2013, 527, 553–557. [Google Scholar] [CrossRef]
- Gan, W.; Song, Q.; Zhang, N.N.; Xiong, X.P.; Wang, D.M.; Li, L. Association between FTO polymorphism in exon 3 with carcass and meat quality traits in crossbred ducks. Genet. Mol. Res. 2015, 14, 6699–6714. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, G.X.; Zhang, T.; Wang, J.Y.; Fan, Q.C.; Tang, Y.; Ding, F.X.; Zhang, L. Myf5 and MyoG gene SNPs associated with Bian chicken growth trait. Genet. Mol. Res. 2016, 15, 10–4238. [Google Scholar] [CrossRef] [PubMed]
- Reuter, É.M.; Reuter, C.P.; de Castro Silveira, J.F.; Carroll, S.; Hobkirk, J.P.; Todendi, P.F.; de Moura Valim, A.R.; de Mello, E.D. FTO gene polymorphism and longitudinal changes in nutritional/obesity status in children and adolescents: Schoolchildren’s health cohort study. Eur. J. Pediatr. 2021, 180, 3325–3333. [Google Scholar] [CrossRef]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Velazquez-Roman, J.; Angulo-Zamudio, U.A.; León-Sicairos, N.; Medina-Serrano, J.; DeLira-Bustillos, N.; Villamil-Ramírez, H.; Canizales-Quinteros, S.; Macías-Kauffer, L.; Campos-Romero, A.; Alcántar-Fernández, J.; et al. Association of FTO, ABCA1, ADRB3, and PPARG variants with obesity, type 2 diabetes, and metabolic syndrome in a Northwest Mexican adult population. J. Diabetes Complicat. 2021, 35, 108025. [Google Scholar] [CrossRef]
- Chung, E.R. Novel SNP in the coding region of the FTO gene is associated with marbling score in Hanwoo (Korean cattle). J. Anim. Sci. Technol. 2014, 56, 27. [Google Scholar] [CrossRef]
- Fan, B.; Onteru, S.K.; Plastow, G.S.; Rothschild, M.F. Detailed characterization of the porcine MC4R gene in relation to fatness and growth. Anim. Genet. 2009, 40, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, L.; Ren, S. Effect of FTO Expression and Polymorphism on Fat Deposition in Suzhong Pigs. Asian-Australas J. Anim. Sci. 2013, 26, 1365–1373. [Google Scholar] [CrossRef]
- Kennedy, O.B.; Stewart-Knox, B.J.; Mitchell, P.C.; Thurnham, D.I. Flesh colour dominates consumer preference for chicken. Appetite 2005, 44, 181–186. [Google Scholar] [CrossRef]
- Ju, H.; Yang, Y.; Sheng, A.; Jiang, X. Role of microRNAs in skeletal muscle development and rhabdomyosarcoma (review). Mol. Med. Rep. 2015, 11, 4019–4024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Li, Y.; Wu, J.; Wang, X.; Bian, C.; Tian, Y.; Sun, G.; Han, R.; Liu, X.; et al. Genome-wide association study reveals the genetic determinism of growth traits in a Gushi-Anka F(2) chicken population. Heredity 2021, 126, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, S.; Yuan, T.; Sun, X. Genetic effects of FTO gene insertion/deletion (InDel) on fat-tail measurements and growth traits in Tong sheep. Anim. Biotechnol. 2021, 32, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Krzysik-Walker, S.M.; Ramachandran, R. Cloning and characterization of chicken fat mass and obesity associated (Fto) gene: Fasting affects Fto expression. Domest. Anim. Endocrinol. 2012, 42, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.; Rao, K.; Yuan, L.; Everaert, N.; Buyse, J.; Grossmann, R.; Zhao, R. Chicken FTO gene: Tissue-specific expression, brain distribution, breed difference and effect of fasting. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2012, 163, 246–252. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).