Weighted Single-Step GWAS Reveals Genomic Regions Associated with Female Fertility in the Spanish Retinta Beef Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Data Recording
2.3. Animal Sampling for the Genomic Assays
2.4. Genotyping and Quality Control
2.5. Weighted Single-Step GREML Method
2.6. Genome-Wide Association Study Analysis
2.7. Identification of Candidate Genes and Gene Network
3. Results and Discussion
3.1. Descriptive Phenotypic Statistics
3.2. Estimation of Variance Components and Heritability
3.3. Genome-Wide Association Studies
3.4. Common Genes Between Traits
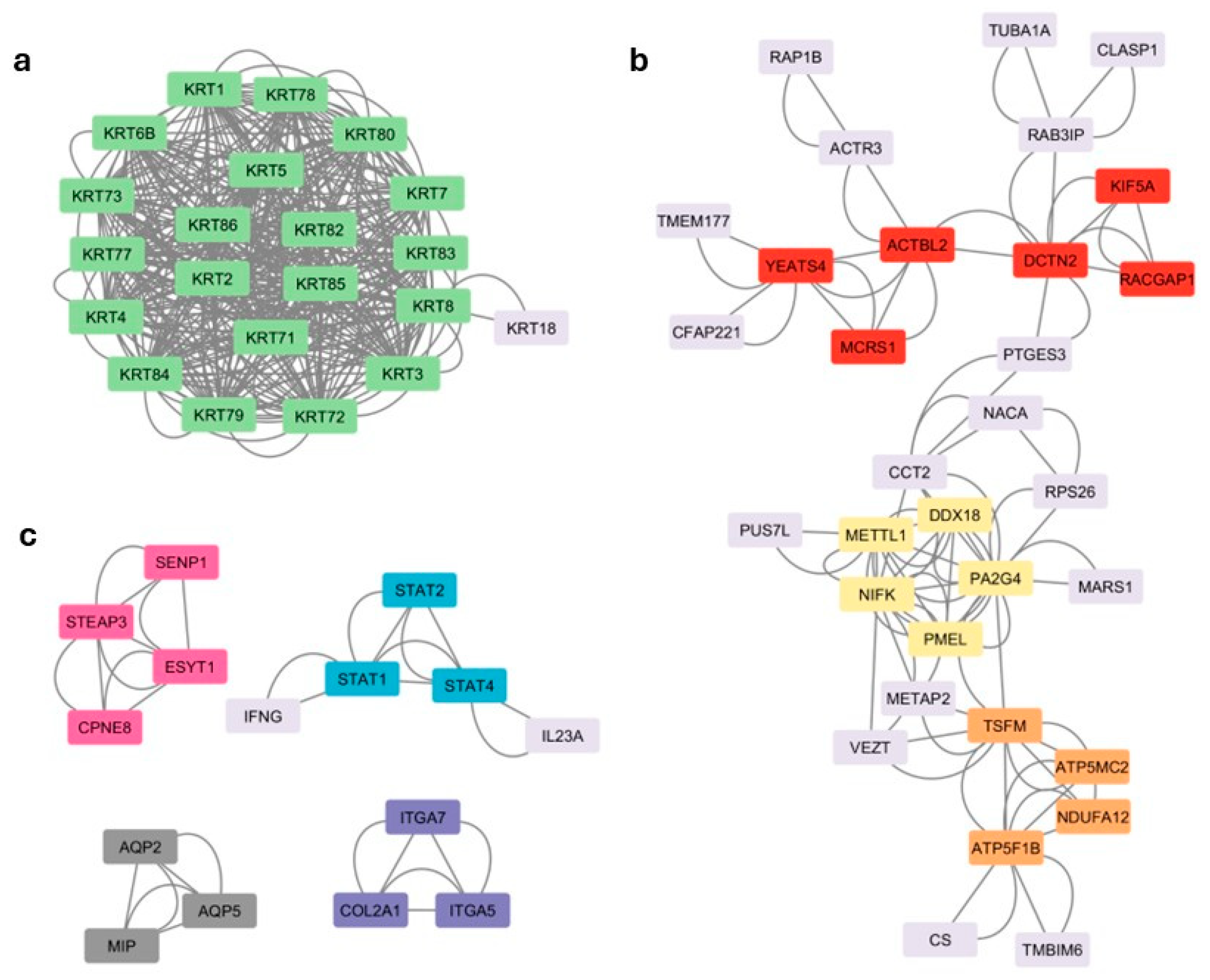
3.5. Cluster Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MAPA. El sector de la carne de vacuno en cifras: Principales indicadores económicos. In Catálogo de Publicaciones de la Administración General del Estado; Ministerio de Agricultura, Pesca y Alimentación. Secretaría General Técnica, Centro de Publicaciones: Madrid, Spain, 2023. [Google Scholar]
- de Macêdo Carvalho, C.B.; de Mello, A.C.L.; da Cunha, M.V.; de Oliveira Apolinário, V.X.; Dubeux Júnior, J.C.B.; da Silva, V.J.; Silva Medeiros, A.; Izidro, J.L.P.S.; Bretas, I.L. Ecosystem services provided by silvopastoral systems: A review. J. Agric. Sci. 2024, 162, 417–432. [Google Scholar] [CrossRef]
- Bellido, M.M. Producción animal en el suroeste español. Pastos 2011, 309–333. Available online: https://polired.upm.es/index.php/pastos/article/view/1642 (accessed on 30 July 2025).
- Jiménez, J.M.; Morales, R.M.; Menéndez-Buxadera, A.; Demyda-Peyrás, S.; Laseca, N.; Molina, A. Estimation of the genetic components of (co)variance and preliminary genome-wide association study for reproductive efficiency in retinta beef cattle. Animals 2023, 13, 501. [Google Scholar] [CrossRef]
- Jiménez, J.M.; Morales, R.M.; Demyda-Peyrás, S.; Molina, A. Genetic relationships between male and female reproductive traits in retinta beef cattle. Livest. Sci. 2025, 291, 105610. [Google Scholar] [CrossRef]
- Cammack, K.M.; Thomas, M.G.; Enns, R.M. Reproductive traits and their heritabilities in beef cattle. Prof. Anim. Sci. 2009, 25, 517–528. [Google Scholar] [CrossRef]
- Berry, D.; Evans, R. Genetics of reproductive performance in seasonal calving beef cows and its association with performance traits. J. Anim. Sci. 2014, 92, 1412–1422. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Alvarez, I.; Fernández, I.; Royo, L.J.; Díez, J.; Goyache, F. Genetic relationships between calving date, calving interval, age at first calving and type traits in beef cattle. Livest. Prod. Sci. 2002, 78, 215–222. [Google Scholar] [CrossRef]
- Bourdon, R.M.; Brinks, J.S. Genetic, environmental and phenotypic relationships among gestation length, birth weight, growth traits and age at first calving in beef cattle. J. Anim. Sci. 1982, 55, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Ponzoni, R.W.; Newman, S. Developing breeding objectives for australian beef cattle production. Anim. Sci. 2010, 49, 35–47. [Google Scholar] [CrossRef]
- Amer, P.R.; Simm, G.; Keane, M.G.; Diskin, M.G.; Wickham, B.W. Breeding objectives for beef cattle in ireland. Livest. Prod. Sci. 2001, 67, 223–239. [Google Scholar] [CrossRef]
- Jiménez, J.M.; Morales, R.; Molina, A.; Moreno-Millán, M.; Demyda-Peyrás, S. Effect of the rob(1;29) translocation on the fertility of beef cattle reared under extensive conditions: A 30-year retrospective study. Reprod. Domest. Anim. 2022, 57, 349–356. [Google Scholar] [CrossRef]
- Laseca, N.; Demyda-Peyrás, S.; Valera, M.; Ramón, M.; Escribano, B.; Perdomo-González, D.I.; Molina, A. A genome-wide association study of mare fertility in the pura raza español horse. Animal 2022, 16, 100476. [Google Scholar] [CrossRef] [PubMed]
- Ziadi, C.; Muñoz-Mejías, E.; Sánchez, M.; López, M.D.; González-Casquet, O.; Molina, A. Selection criteria for improving fertility in spanish goat breeds: Estimation of genetic parameters and designing selection indices for optimal genetic responses. Animals 2021, 11, 409. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Duan, X.; An, B.; Chang, T.; Liang, M.; Xu, L.; Zhang, L.; Li, J.; E, G.; Gao, H. Genome-wide association study based on random regression model reveals candidate genes associated with longitudinal data in chinese simmental beef cattle. Animals 2021, 11, 2524. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Zhang, T.; Xu, L.; Wang, T.; Wang, Z.; Zhu, B.; Zhang, L.; Gao, H.; Song, J.; Li, J.; et al. Integration of selection signatures and multi-trait gwas reveals polygenic genetic architecture of carcass traits in beef cattle. Genomics 2021, 113, 3325–3336. [Google Scholar] [CrossRef]
- Amorim, S.T.; Stafuzza, N.B.; Kluska, S.; Peripolli, E.; Pereira, A.S.C.; Muller da Silveira, L.F.; de Albuquerque, L.G.; Baldi, F. Genome-wide interaction study reveals epistatic interactions for beef lipid-related traits in nellore cattle. Anim. Genet. 2022, 53, 35–48. [Google Scholar] [CrossRef]
- Berton, M.P.; de Lemos, M.V.A.; Stafuzza, N.B.; Simielli Fonseca, L.F.; Silva, D.B.d.S.; Peripolli, E.; Pereira, A.S.C.; Magalhães, A.F.B.; Albuquerque, L.G.; Baldi, F. Integration analyses of structural variations and differential gene expression associated with beef fatty acid profile in nellore cattle. Anim. Genet. 2022, 53, 570–582. [Google Scholar] [CrossRef]
- Fonseca, P.A.S.; Schenkel, F.S.; Cánovas, A. Genome-wide association study using haplotype libraries and repeated-measures model to identify candidate genomic regions for stillbirth in holstein cattle. J. Dairy Sci. 2022, 105, 1314–1326. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Wilson, M.L.; Nilson, S.M.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E.; Seabury, C.M. Genome-wide association and genotype by environment interactions for growth traits in u.S. Red angus cattle. BMC Genom. 2022, 23, 517. [Google Scholar] [CrossRef]
- Li, S.; Ge, F.; Chen, L.; Liu, Y.; Chen, Y.; Ma, Y. Genome-wide association analysis of body conformation traits in chinese holstein cattle. BMC Genom. 2024, 25, 1174. [Google Scholar] [CrossRef]
- Purfield, D.C.; Evans, R.D.; Berry, D.P. Breed- and trait-specific associations define the genetic architecture of calving performance traits in cattle. J. Anim. Sci. 2020, 98, skaa151. [Google Scholar] [CrossRef]
- Purfield, D.C.; Evans, R.D.; Carthy, T.R.; Berry, D.P. Genomic regions associated with gestation length detected using whole-genome sequence data differ between dairy and beef cattle. Front. Genet. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Keogh, K.; Carthy, T.R.; McClure, M.C.; Waters, S.M.; Kenny, D.A. Genome-wide association study of economically important traits in charolais and limousin beef cows. Animal 2021, 15, 100011. [Google Scholar] [CrossRef] [PubMed]
- Stegemiller, M.R.; Murdoch, G.K.; Rowan, T.N.; Davenport, K.M.; Becker, G.M.; Hall, J.B.; Murdoch, B.M. Genome-wide association analyses of fertility traits in beef heifers. Genes 2021, 12, 217. [Google Scholar] [CrossRef]
- Butler, M.L.; Hartman, A.R.; Bormann, J.M.; Weaber, R.L.; Grieger, D.M.; Rolf, M.M. Genome-wide association study of beef bull semen attributes. BMC Genom. 2022, 23, 74. [Google Scholar] [CrossRef]
- Carvalho Filho, I.; Arikawa, L.M.; Mota, L.F.M.; Campos, G.S.; Fonseca, L.F.S.; Fernandes Júnior, G.A.; Schenkel, F.S.; Lourenco, D.; Silva, D.A.; Teixeira, C.S.; et al. Genome-wide association study considering genotype-by-environment interaction for productive and reproductive traits using whole-genome sequencing in nellore cattle. BMC Genom. 2024, 25, 623. [Google Scholar] [CrossRef]
- Misztal, I.; Legarra, A.; Aguilar, I. Computing procedures for genetic evaluation including phenotypic, full pedigree, and genomic information. J. Dairy Sci. 2009, 92, 4648–4655. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Luo, Z. Computing inbreeding coefficients in large populations. Genet. Sel. Evol. 1992, 24, 305–313. [Google Scholar] [CrossRef]
- Wellmann, R. Optimum contribution selection for animal breeding and conservation: The r package optisel. BMC Bioinform. 2019, 20, 25. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing. R Environment v4.5 “How About a Twenty-Six”; Viena, Austria. 11 04 2025. 2025. Available online: https://www.r-project.org/ (accessed on 10 July 2025).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Barrett, T.; Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Hocking, T.; Krylov, S.B. data.table: Extension of ‘data.frame’, R package version 1.17.99; 2025. Available online: https://github.com/rdatatable/data.table (accessed on 30 July 2025).
- Thermofisher. Axiom cnv Summary Tool User Manual. Available online: https://tools.thermofisher.com/content/sfs/manuals/axiom_cnv_summary_tool_usermanual.pdf (accessed on 15 August 2020).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Marschner, I. Glm2: Fitting generalized linear models with convergence problems. R J. 2011, 3, 12–15. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing v4.4.2 “Pile of Leaves”; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.r-project.org/ (accessed on 10 July 2025).
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of holstein final score1. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Lourenco, D.; Tsuruta, S.; Aguilar, I.; Masuda, Y.; Bermann, M.; Legarra, A.; Misztal, I. Recent updates in the blupf90 software suite. In Proceedings of the 12th World Congress on Genetics Applied to Livestock Production (Wcgalp), Rotterdam, The Netherlands, 3–8 July, 2022; Wageningen Academic Publishers: Wageningen, The Netherlands, 2022; pp. 1530–1533. [Google Scholar]
- Zhang, Z.; Liu, J.; Ding, X.; Bijma, P.; de Koning, D.-J.; Zhang, Q. Best linear unbiased prediction of genomic breeding values using a trait-specific marker-derived relationship matrix. PLoS ONE 2010, 5, e12648. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. String v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Roughsedge, T.; Amer, P.R.; Thompson, R.; Simm, G. Genetic parameters for a maternal breeding goal in beef production. J. Anim. Sci. 2005, 83, 2319–2329. [Google Scholar] [CrossRef]
- Márton, J.; Bene, S.A.; Szabó, F. Heritability estimates of age at first calving and correlation analysis in angus cows bred in hungary. Animals 2024, 14, 3715. [Google Scholar] [CrossRef]
- Meneses, C.; Carabaño, M.J.; Morales, R.M.; Molina, A.; Diaz, C. Modelling fertility traits under natural mating conditions in beef cattle. Interbull Bull. 2014, 48, 50–53. [Google Scholar]
- Smith, B.A.; Brinks, J.S.; Richardson, G.V. Estimation of genetic parameters among reproductive and growth traits in yearling heifers. J. Anim. Sci. 1989, 67, 2886–2891. [Google Scholar] [CrossRef]
- Martínez-Velázquez, G.; Gregory, K.E.; Bennett, G.L.; Van Vleck, L.D. Genetic relationships between scrotal circumference and female reproductive traits. J. Anim. Sci. 2003, 81, 395–401. [Google Scholar] [CrossRef]
- Brzáková, M.; Čítek, J.; Svitáková, A.; Veselá, Z.; Vostrý, L. Genetic parameters for age at first calving and first calving interval of beef cattle. Animals 2020, 10, 2122. [Google Scholar] [CrossRef]
- Cortés, O.; Carleos, C.; Baro, J.A.; Fernández, M.A.; Villa, J.; Menéndez-Buxadera, A.; Cañon, J. Realized genetic parameters of growth and reproductive traits after 25 years of selection in the asturiana de los valles beef cattle breed 3 4 25 años de selección en la raza de carne asturiana de los valles: Análisis de 5 los parámetros genéticos en caracteres de crecimiento y reproductivos. Actas Iberoam. Conserv. Anim. 2015, 5, 78–86. [Google Scholar]
- Lopez, B.I.; Son, J.-H.; Seo, K.; Lim, D. Estimation of genetic parameters for reproductive traits in hanwoo (Korean cattle). Animals 2019, 9, 715. [Google Scholar] [CrossRef] [PubMed]
- Veselá, Z.; Vostrý, L.; Svitáková, A. Genetic analysis of female fertility traits in beef cattle in the czech republic. Interbull Bull. 2013, 47, 172–175. [Google Scholar]
- Koots, K.R.; Gibson, J.; Smith, C.; Wilton, J. Analyses of published genetic parameter estimates for beef production traits. I. Heritability. Anim. Breed. Abstr. 1994, 62, 309–338. [Google Scholar]
- Tiezzi, F.; Maltecca, C. Accounting for trait architecture in genomic predictions of us holstein cattle using a weighted realized relationship matrix. Genet. Sel. Evol. 2015, 47, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lourenco, D.; Aguilar, I.; Legarra, A.; Misztal, I. Weighting strategies for single-step genomic blup: An iterative approach for accurate calculation of gebv and gwas. Front. Genet. 2016, 7, 151. [Google Scholar] [CrossRef]
- Martinez-Castillero, M.; Then, C.; Altarriba, J.; Srihi, H.; López-Carbonell, D.; Díaz, C.; Martinez, P.; Hermida, M.; Varona, L. Detection of genomic regions with pleiotropic effects for growth and carcass quality traits in the rubia gallega cattle breed. Animals 2021, 11, 1682. [Google Scholar] [CrossRef]
- Laseca, N.; Molina, A.; Perdomo-González, D.; Ziadi, C.; Azor, P.J.; Valera, M. Exploring the genetic landscape of vitiligo in the pura raza español horse: A genomic perspective. Animals 2024, 14, 2420. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding rnas: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Wang, P.; Paquet, É.R.; Robert, C. Comprehensive transcriptomic analysis of long non-coding rnas in bovine ovarian follicles and early embryos. PLoS ONE 2023, 18, e0291761. [Google Scholar] [CrossRef] [PubMed]
- Leung, L.; Kwong, M.; Hou, S.; Lee, C.; Chan, J.Y. Deficiency of the nrf1 and nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 2003, 278, 48021–48029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-M.; Deng, M.-T.; Lei, Z.-H.; Wan, Y.-J.; Nie, H.-T.; Wang, Z.-Y.; Fan, Y.-X.; Wang, F.; Zhang, Y.-L. Effects of nrf1 on steroidogenesis and apoptosis in goat luteinized granulosa cells. Reproduction 2017, 154, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Lamas-Toranzo, I.; Fonseca Balvís, N.; Querejeta-Fernández, A.; Izquierdo-Rico, M.J.; González-Brusi, L.; Lorenzo, P.L.; García-Rebollar, P.; Avilés, M.; Bermejo-Álvarez, P. Zp4 confers structural properties to the zona pellucida essential for embryo development. eLife 2019, 8, e48904. [Google Scholar] [CrossRef]
- Reding, J.J.; van der Westhuizen, R.R.; Berry, D.P.; van Marle-Köster, E. Understanding the underlying genetic mechanisms for age at first calving, inter-calving period and scrotal circumference in bonsmara cattle. BMC Genom. 2023, 24, 480. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Y.; Xu, S.; Qi, G.; Wu, X. Bioinformatics identification of micrornas involved in polycystic ovary syndrome based on microarray data. Mol. Med. Rep. 2019, 20, 281–291. [Google Scholar] [CrossRef]
- Yagi, T.; Takeichi, M. Cadherin superfamily genes: Functions, genomic organization, and neurologic diversity. Genes Dev. 2000, 14, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Zielak-Steciwko, A.E.; Browne, J.A.; McGettigan, P.A.; Gajewska, M.; Dzięcioł, M.; Szulc, T.; Evans, A.C. Expression of micrornas and their target genes and pathways associated with ovarian follicle development in cattle. Physiol. Genom. 2014, 46, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.; Tesfaye, D.; Rings, F.; Schellander, K.; Holker, M.; Van Poucke, M.; Van Zeveren, A.; Lemahieu, I.; Van Soom, A.; Peelman, L.J. Suppression of keratin 18 gene expression in bovine blastocysts by rna interference. Reprod. Fertil. Dev. 2010, 22, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Kassim, Y.; Sheng, H.; Xu, G.; Jin, H.; Iqbal, T.; Elashry, M.; Zhang, K. Integrated multi-omics analysis reveals key regulators of bovine oocyte maturation. Int. J. Mol. Sci. 2025, 26, 3973. [Google Scholar] [CrossRef] [PubMed]
- Viana, J.H.M.; Silva, B.D.M.; de Moura, R.M.; Féres, L.F.R.; Figueiredo, R.A. Oocyte developmental potential and embryo production before puberty in cattle. Anim. Reprod. 2024, 21, e20240069. [Google Scholar] [CrossRef]
- Vitorino Carvalho, A.; Eozenou, C.; Healey, G.D.; Forde, N.; Reinaud, P.; Chebrout, M.; Gall, L.; Rodde, N.; Padilla, A.L.; Delville, C.G.; et al. Analysis of stat1 expression and biological activity reveals interferon-tau-dependent stat1-regulated socs genes in the bovine endometrium. Reprod. Fertil. Dev. 2016, 28, 459–474. [Google Scholar] [CrossRef]
- Johnson, G.A.; Burghardt, R.C.; Bazer, F.W.; Seo, H.; Cain, J.W. Integrins and their potential roles in mammalian pregnancy. J. Anim. Sci. Biotechnol. 2023, 14, 115. [Google Scholar] [CrossRef] [PubMed]
- Sisco, B.; Pfeffer, P.L. Expression of activin pathway genes in granulosa cells of dominant and subordinate bovine follicles. Theriogenology 2007, 68, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Umer, S.; Zhao, S.J.; Sammad, A.; Weldegebriall Sahlu, B.; Pang, Y.; Zhu, H. Amh: Could it be used as a biomarker for fertility and superovulation in domestic animals? Genes 2019, 10, 1009. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.E.; Skinner, M.K. Bone morphogenetic protein-4 acts as an ovarian follicle survival factor and promotes primordial follicle development1. Biol. Reprod. 2003, 69, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Hara, S.; Kamei, D.; Sasaki, Y.; Tanemoto, A.; Nakatani, Y.; Murakami, M. Prostaglandin e synthases: Understanding their pathophysiological roles through mouse genetic models. Biochimie 2010, 92, 651–659. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, W.; Ding, J.; Song, D.; Cheng, W.; Yu, J.; Sun, S.; Mei, S.; Liang, X.; Zhao, Q.; et al. Integrative single-cell analysis reveals iron overload-induced senescence and metabolic reprogramming in ovarian endometriosis-associated infertility. Adv. Sci. 2025, 12, e17528. [Google Scholar] [CrossRef]
- Convissar, S.; Winston, N.J.; Fierro, M.A.; Scoccia, H.; Zamah, A.M.; Stocco, C. Sp1 regulates steroidogenic genes and lhcgr expression in primary human luteinized granulosa cells. J. Steroid Biochem. Mol. Biol. 2019, 190, 183–192. [Google Scholar] [CrossRef]
- Ortega, S.; Malumbres, M.; Barbacid, M. Cyclin d-dependent kinases, ink4 inhibitors and cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2002, 1602, 73–87. [Google Scholar] [CrossRef]
- Nelson, C.D.; Reinhardt, T.A.; Lippolis, J.D.; Sacco, R.E.; Nonnecke, B.J. Vitamin d signaling in the bovine immune system: A model for understanding human vitamin d requirements. Nutrients 2012, 4, 181–196. [Google Scholar] [CrossRef]
- Grzesiak, M.; Burzawa, G.; Kurowska, P.; Blaszczyk, K.; Szlaga, A.; Blasiak, A.; Sechman, A.; Rak, A. Altered vitamin d3 metabolism in the ovary and periovarian adipose tissue of rats with letrozole-induced pcos. Histochem. Cell Biol. 2021, 155, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Sağsöz, H.; Ketani, M.A.; Saruhan, B.G. Expression of the erbb/her receptor family in the bovine uterus during the sexual cycle and the relation of this family to serum sex steroids. Biotech. Histochem. 2012, 87, 105–116. [Google Scholar] [CrossRef]
- Lim, J.J.; Lima, P.D.A.; Salehi, R.; Lee, D.R.; Tsang, B.K. Regulation of androgen receptor signaling by ubiquitination during folliculogenesis and its possible dysregulation in polycystic ovarian syndrome. Sci. Rep. 2017, 7, 10272. [Google Scholar] [CrossRef]
- Tetsuka, M.; Nishimoto, H.; Miyamoto, A.; Okuda, K.; Hamano, S. Gene expression of 11β-hsd and glucocorticoid receptor in the bovine (bos taurus) follicle during follicular maturation and atresia: The role of follicular stimulating hormone. J. Reprod. Dev. 2010, 56, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Hatzirodos, N.; Hummitzsch, K.; Irving-Rodgers, H.F.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells from bovine ovarian follicles during atresia. BMC Genom. 2014, 15, 40. [Google Scholar] [CrossRef]
- Li, M.; Xue, K.; Ling, J.; Diao, F.-Y.; Cui, Y.-G.; Liu, J.-Y. The orphan nuclear receptor nr4a1 regulates transcription of key steroidogenic enzymes in ovarian theca cells. Mol. Cell. Endocrinol. 2010, 319, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.P.; Tayade, C.; Ashkar, A.A.; Hatta, K.; Zhang, J.; Croy, B.A. Interferon gamma in successful pregnancies. Biol. Reprod. 2009, 80, 848–859. [Google Scholar] [CrossRef] [PubMed]
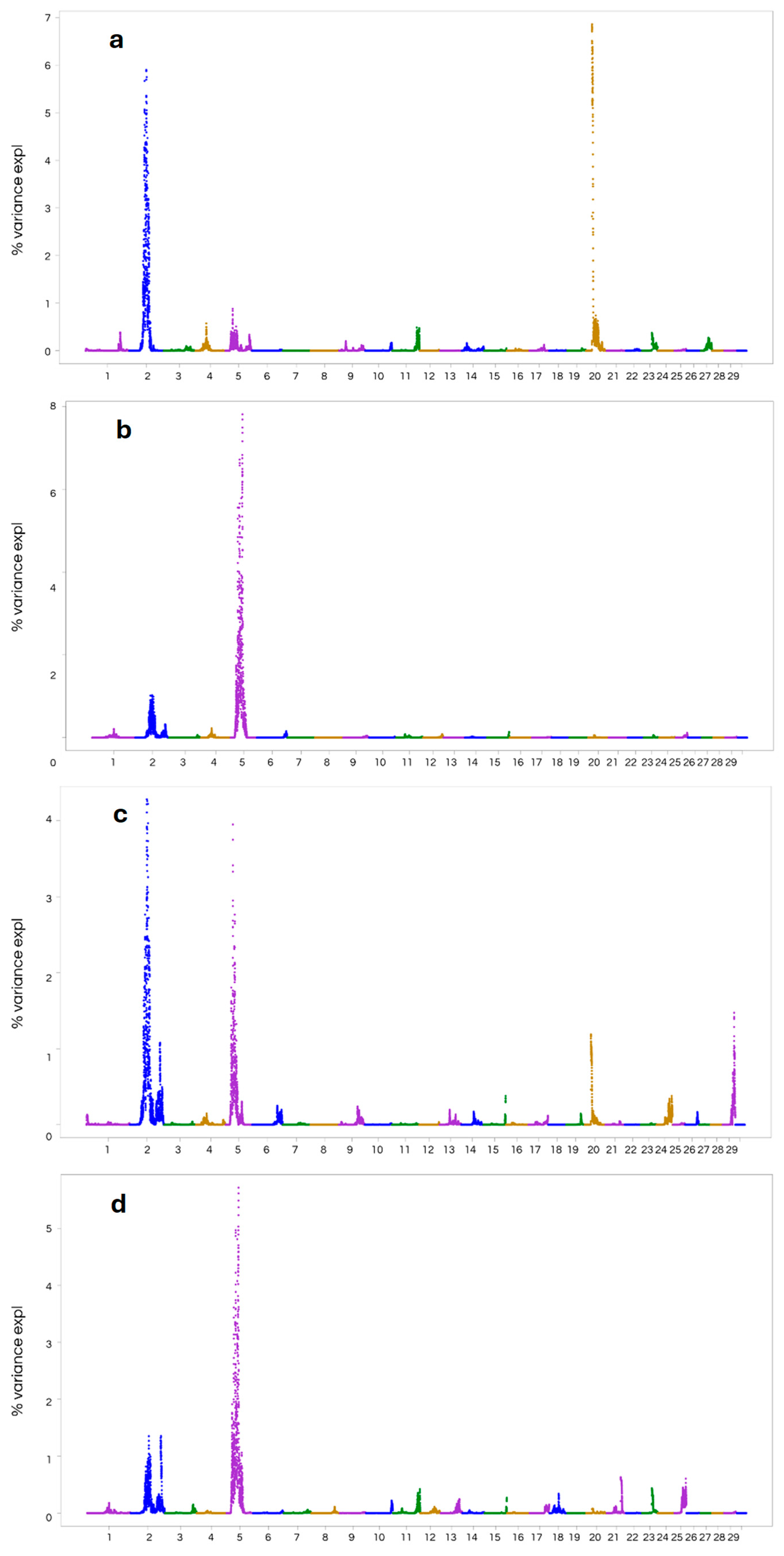
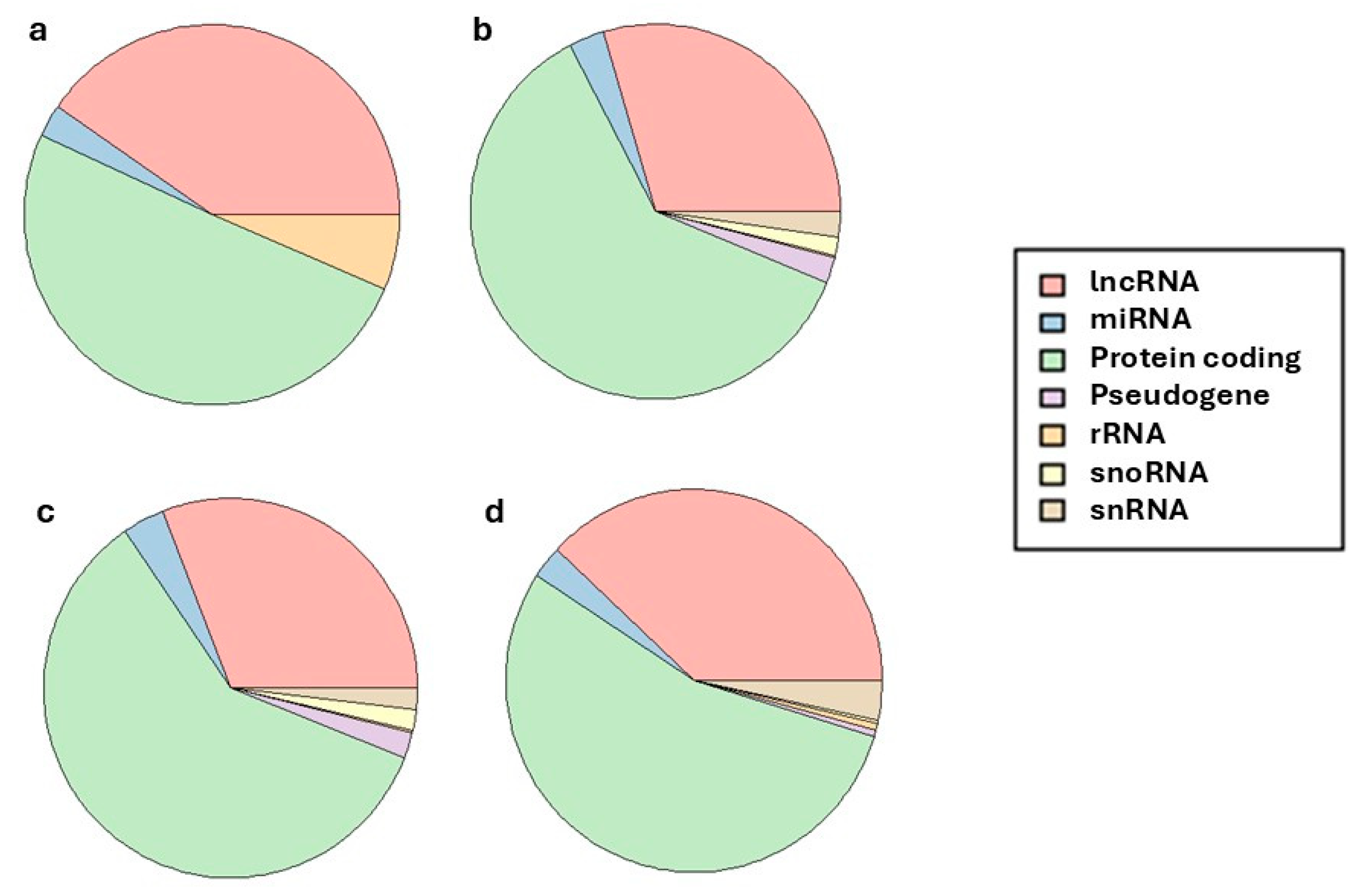
| Trait | Mean | Minimum | Maximum | Std.Dev. | Coef.Var. |
|---|---|---|---|---|---|
| AFC | 34.94 ± 0.032 | 17.03 | 48.9 | 6.72 | 19.24 |
| IC12 | 15.12 ± 0.027 | 9.031 | 24.9 | 3.86 | 25.5 |
| ACI | 15.74 ± 0.019 | 9.03 | 25.5 | 3.30 | 20.9 |
| RE | 72.45 ± 0.105 | 22.0 | 100.0 | 21.74 | 30.0 |
| Trait | (SE) | (SE) | (SE) | (SE) |
|---|---|---|---|---|
| AFC | 6.29 (0.34) | 15.90 (0.35) | 21.01 (0.28) | 0.15 (0.008) |
| IC12 | 3.65 (0.24) | 2.49 (0.13) | 8.90 (0.19) | 0.24 (0.015) |
| ACI | 2.91 (0.14) | 1.45 (0.07) | 6.26 (0.11) | 0.27 (0.012) |
| RE | 53.38 (2.16) | 73.79 (1.81) | 133.94 (1.71) | 0.20 (0.005) |
| Traits | Nº of Genes | Candidate Gene | Gene Name |
|---|---|---|---|
| AFC, IC12 | 32 | INSIG2 | Insulin induced gene 2 |
| STAT1 | Signal transducer and activator of transcription 1 | ||
| RE, IC12, ACI | 111 | ACVR1B | Activin A receptor type 1B |
| FRS2 | Fibroblast growth factor receptor substrate 2 | ||
| IFNG | Interferon gamma | ||
| KRT18 | Keratin 18 | ||
| KRT7 | Keratin 7 | ||
| KRT8 | Keratin 8 | ||
| NR4A1 | Nuclear receptor subfamily 4 group A member 1 | ||
| VDR | Vitamin D receptor | ||
| RE, ACI | 145 | AMHR2 | Anti-Müllerian hormone receptor type 2 |
| CDK2 | Cyclin-dependent kinase 2 | ||
| CDK4 | Cyclin-dependent kinase 4 | ||
| CYP27B1 | Cytochrome P450 family 27 subfamily B member 1 | ||
| ERBB3 | Erb-B2 receptor tyrosine kinase 3 | ||
| GDF11 | Growth differentiation factor 11 | ||
| HSD17B6 | Hydroxysteroid 17-beta dehydrogenase 6 | ||
| ITGA5 | Integrin subunit alpha 5 | ||
| PTGES3 | Prostaglandin E synthase 3 | ||
| SLC11A2 | Solute carrier family 11 member 2 | ||
| SP1 | Sp1 transcription factor |
| Cluster | Description | Gene Symbol |
|---|---|---|
| 1 | Keratinization | KRT78, KRT73, KRT84, KRT7, KRT86, KRT4, KRT6B, KRT77, KRT83, KRT71, KRT80, KRT2, KRT5, KRT1, KRT85, KRT3, KRT82, KRT79, KRT72, KRT8 |
| 2 | Cell cycle, Cytoskeletal organization, Chromatin remodeling | MCRS1, ACTBL2, KIF5A, RACGAP1, YEATS4, DCTN2 |
| 3 | RNA processing, Pigmentation | NIFK, PA2G4, PMEL, METTL1, DDX18 |
| 4 | ATP production | TSFM, NDUFA12, ATP5MC2, ATP5F1B |
| 5 | Protein regulation, Metal ion binding | CPNE8, ESYT1, STEAP3, SENP1 |
| 6 | Cytokine-mediated signaling pathway | STAT4, STAT2, STAT1 |
| 7 | Water channel activity | MIP, AQP2, AQP5 |
| 8 | Cell adhesion, Extracellular matrix organization | ITGA7, COL2A1, ITGA5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales, R.M.; Calvo-Rubio, G.A.; Ziadi, C.; Vargas-Pérez, M.Á.; Demyda-Peyrás, S.; Molina, A. Weighted Single-Step GWAS Reveals Genomic Regions Associated with Female Fertility in the Spanish Retinta Beef Cattle. Animals 2025, 15, 2665. https://doi.org/10.3390/ani15182665
Morales RM, Calvo-Rubio GA, Ziadi C, Vargas-Pérez MÁ, Demyda-Peyrás S, Molina A. Weighted Single-Step GWAS Reveals Genomic Regions Associated with Female Fertility in the Spanish Retinta Beef Cattle. Animals. 2025; 15(18):2665. https://doi.org/10.3390/ani15182665
Chicago/Turabian StyleMorales, Rosa María, Gabriel Anaya Calvo-Rubio, Chiraz Ziadi, María Ángeles Vargas-Pérez, Sebastián Demyda-Peyrás, and Antonio Molina. 2025. "Weighted Single-Step GWAS Reveals Genomic Regions Associated with Female Fertility in the Spanish Retinta Beef Cattle" Animals 15, no. 18: 2665. https://doi.org/10.3390/ani15182665
APA StyleMorales, R. M., Calvo-Rubio, G. A., Ziadi, C., Vargas-Pérez, M. Á., Demyda-Peyrás, S., & Molina, A. (2025). Weighted Single-Step GWAS Reveals Genomic Regions Associated with Female Fertility in the Spanish Retinta Beef Cattle. Animals, 15(18), 2665. https://doi.org/10.3390/ani15182665






