Mixed Ensiling Increases Degradation Without Altering Attached Microbiota Through In Situ Ruminal Incubation Technique
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Silage Preparation and Sample Collection
2.2. Animals
2.3. In Situ Ruminal Incubation
2.4. Analytical Methods
2.5. DNA Extraction and High-Throughput Sequencing
2.6. Calculations
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaballah, E.S.; Abomohra, A.E.; Xu, C.; Elsayed, M.; Abdelkader, T.K.; Lin, J.; Yuan, Q. Enhancement of biogas production from rape straw using different co-pretreatment techniques and anaerobic co-digestion with cattle manure. Bioresour. Technol. 2020, 309, 123311. [Google Scholar] [CrossRef] [PubMed]
- Griffith, C.; Ribeiro, G.O.; Oba, M.; McAllister, T.A.; Beauchemin, K.A. Potential for improving fiber digestion in the rumen of cattle (Bos taurus) through microbial inoculation from bison (Bison bison): In situ fiber degradation. J. Anim. Sci. 2017, 95, 2156–2167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, M.; Wang, X.; Yu, Z.; Na, R. Ensiling alfalfa with whole crop corn improves the silage quality and in vitro digestibility of the silage mixtures. Grassl. Sci. 2017, 63, 211–217. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, L.; Ma, G.; Jiang, X.; Yang, J.; Lv, J.; Zhang, Y. Cellulase Interacts with Lactate Bacteria to Affect Fermentation Quality, Microbial Community, and Ruminal Degradability in Mixed Silage of Soybean Residue and Corn Stover. Animals 2021, 11, 334. [Google Scholar] [CrossRef] [PubMed]
- Xing, L. Study on the Effects of Lactobacillus and Cellulase Additives on the Quality of Different Silage. Master’s Thesis, China Agricultural University, Beijing, China, 2004. [Google Scholar]
- Jiang, B.W. Effects of Enzyme Bacteria Mixed Treatment of Roughage on Fiber Structure, Growth Performance and Rumen Microflora of Tan Sheep. Ph.D. Thesis, Ningxia University, Ningxia, China, 2021. [Google Scholar]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, Y.; Li, Y.; Zhang, Y.; Liu, T.; Wang, Y.; Sharpton, T.J.; Zhu, W. Progressive colonization of bacteria and disappearance of rice straw in the rumen by Illumina Sequencing. Front. Microbiol. 2017, 8, 2165. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, M.; Xue, C.; Zhu, W.; Mao, S. Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J. Dairy Sci. 2016, 99, 9668–9681. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.; Yabuki, H.; Kobayashi, Y. Interaction of rumen bacteria as assumed by colonization patterns on untreated and alkali-treated rice straw. J. Anim. Sci. 2014, 85, 524–531. [Google Scholar] [CrossRef]
- Meale, S.J.; Beauchemin, K.A.; Hristov, A.N.; Chaves, A.V.; McAllister, T.A. Board-invited review: Opportunities and challenges in using exogenous enzymes to improve ruminant production. J. Anim. Sci. 2014, 92, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.X.; Zhang, X.M.; Yi, S.Y.; Wang, R.; Li, Q.S.; Zhang, W.Q.; Qu, J.J.; Huo, J.B.; Lin, B.; Tan, B.; et al. Mixed ensiling plus nitrate destroy fiber structure of rape straw, increase degradation and reduce methanogenesis through in vitro ruminal fermentation. J. Sci. Food Agric. 2024, 104, 3428–3436. [Google Scholar] [CrossRef] [PubMed]
- NY/T 815-2004; Beef Cattle Feeding Standards. The Standardization Administration of the People’s Republic of China: Beijing, China, 2004.
- Zhang, X.M.; Gruninger, R.J.; Alemu, A.W.; Wang, M.; Tan, Z.L.; Kindermann, M.; Beauchemin, K.A. 3-Nitrooxypropanol supplementation had little effect on fiber degradation and microbial colonization of forage particles when evaluated using the in situ ruminal incubation technique. J. Dairy Sci. 2020, 103, 8986–8997. [Google Scholar] [CrossRef] [PubMed]
- Association of Official Analytical Chemists (AOAC). Official Methods of Analysis, 18th ed.; AOAC International: Gaithersburg, MD, USA, 2005. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.A.C.C. A simple colorimetric assay for muramic acid and lactic acid. Appl. Biochem. Biotech. 1996, 56, 49–58. [Google Scholar] [CrossRef]
- Wang, M.; Sun, X.Z.; Janssen, P.H.; Tang, S.X.; Tan, Z.L. Responses of methane production and fermentation pathways to the increased dissolved hydrogen concentration generated by eight substrates in in vitro ruminal cultures. Anim. Feed. Sci. Technol. 2014, 194, 1–11. [Google Scholar] [CrossRef]
- Weatherburn, M. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 1967, 39, 971–974. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, R.; Wang, M.; Zhang, X.; Mao, H.; Tan, Z. Short communication: Variability in fermentation end-products and methanogen communities in different rumen sites of dairy cows. J. Dairy Sci. 2018, 101, 5153–5158. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. J. ISME 2013, 7, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Ørskov, E.; McDonald, I. The estimation of protein degradability in the rumen from incubation measurements weighted according to rate of passage. J. Agric. Sci. 1979, 92, 499–503. [Google Scholar] [CrossRef]
- McDonald, I. A revised model for the estimation of protein degradability in the rumen. J. Agric. Sci. 1981, 96, 251–252. [Google Scholar] [CrossRef]
- Borreani, G.; Tabacco, E.; Schmidt, R.J.; Holmes, B.J.; Muck, R.E. Silage review: Factors affecting dry matter and quality losses in silages. J. Dairy Sci. 2018, 101, 3952–3979. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.F.; Sales, G.F.C.; Schwan, R.F.; Ávila, C.L.S. Criteria for lactic acid bacteria screening to enhance silage quality. J. Appl. Microbiol. 2021, 130, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Ni, K.; Wang, F.; Zhu, B.; Yang, J.; Zhou, G.; Pan, Y.; Tao, Y.; Zhong, J. Effects of Lactate bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresour. Technol. 2017, 238, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Bonaldi, D.S.; Carvalho, B.F.; Ávila, C.L.D.S.; Silva, C.F. Effects of Bacillus subtilis and its metabolites on corn silage quality. Lett. Appl. Microbiol. 2021, 73, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Ni, K.; Wang, T.; Yang, X.; Zhang, J.; Liu, Y.; Shi, W.; Yan, L.; Jie, C.; Zhong, J. Effects of ferulic acid esterase-producing Lactobacillus fermentum and cellulase additives on the fermentation quality and microbial community of alfalfa silage. PeerJ 2019, 7, e7712. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Shao, T.; Li, J.; Yang, L.; Yuan, X. Effect of alfalfa microbiota on fermentation quality and bacterial community succession in fresh or sterile Napier grass silages. J. Dairy Sci. 2020, 103, 4288–4301. [Google Scholar] [CrossRef] [PubMed]
- Muck, R.E.; Nadeau, E.M.G.; McAllister, T.A.; Contreras-Govea, F.E.; Santos, M.C.; Kung, L., Jr. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hu, Z.; Wei, M.; Yong, M.; Niu, H. Effects of inoculation of Lactiplantibacillus plantarum and Lentilactobacillus buchneri on fermentation quality, aerobic stability, and microbial community dynamics of wilted Leymus chinensis silage. Front. Microbiol. 2022, 13, 928731. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part IV beneficial effects. Ann. Agric. Environ. Med. 2016, 23, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Huang, R.; Zhang, Z.; Shi, S. Seed-Borne Erwinia persicina Affects the Growth and Physiology of Alfalfa (Medicago sativa L.). Front. Microbiol. 2022, 13, 891188. [Google Scholar] [CrossRef] [PubMed]
- Kaparullina, E.; Doronina, N.; Chistyakova, T.; Trotsenko, Y. Stenotrophomonas chelatiphaga sp. nov., a new aerobic EDTA-degrading bacterium. Syst. Appl. Microbiol. 2009, 32, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Q.; Hou, J.; Yu, Z. Effects of biological additives on ultrastructure and fiber content of Leymus chinensis silage. Acta Pratacult. Sin. 2016, 25, 94–101. [Google Scholar]
- Hu, Z.; Niu, H.; Tong, Q.; Chang, J.; Yu, J.; Li, S.; Zhang, S.; Ma, D. The Microbiota Dynamics of Alfalfa Silage During Ensiling and After Air Exposure, and the Metabolomics After Air Exposure Are Affected by Lactobacillus casei and Cellulase Addition. Front. Microbiol. 2020, 11, 519121. [Google Scholar] [CrossRef] [PubMed]
- Ávila, C.L.S.; Carvalho, B.F. Silage fermentation-updates focusing on the performance of micro-organisms. J. Appl. Microbiol. 2020, 28, 966–984. [Google Scholar] [CrossRef] [PubMed]
- Balan, V.; Bals, B.; Chundawat, S.P.; Marshall, D.; Dale, B.E. Lignocellulosic biomass pretreatment using AFEX. Methods Mol. Biol. 2009, 581, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Adesogan, A.T.; Arriola, K.G.; Jiang, Y.; Oyebade, A.; Paula, E.M.; Pech-Cervantes, A.A.; Romero, J.J.; Ferraretto, L.F.; Vyas, D. Symposium review: Technologies for improving fiber utilization. J. Dairy Sci. 2019, 102, 5726–5755. [Google Scholar] [CrossRef] [PubMed]
- Moraïs, S.; Mizrahi, I. Islands in the stream: From individual to communal fiber degradation in the rumen ecosystem. FEMS Microbiol. Rev. 2019, 43, 362–379. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.E.; Foster, J.L.; McCuistion, K.C.; Redmon, L.A.; Jessup, R.W. Nutritive value, fermentation characteristics, and in situ disappearance kinetics of sorghum silage treated with inoculants. J. Dairy Sci. 2013, 96, 7120–7131. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, K.A.; Ribeiro, G.O.; Ran, T.; Marami, M.R.; Yang, W.Z.; Khanaki, H.; Gruninger, R.; Tsang, A.; McAllister, T.A. Recombinant fibrolytic feed enzymes and ammonia fibre expansion (AFEX) pretreatment of crop residues to improve fibre degradability in cattle. Anim. Feed. Sci. Technol. 2019, 256, 114260. [Google Scholar] [CrossRef]
- Zhang, X.M.; Wang, M.; Yu, Q.; Ma, Z.Y.; Beauchemin, K.A.; Wang, R.; Wen, J.N.; Lukuyu, B.A.; Tan, Z.L. Liquid hot water treatment of rice straw enhances anaerobic degradation and inhibits methane production during in vitro ruminal fermentation. J. Dairy Sci. 2020, 103, 4252–4261. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.B.; Pell, A.N.; Chase, L.E. Characteristics of neutral detergent-soluble fiber fermentation by mixed ruminal microbes. Anim. Feed. Sci. Technol. 1988, 70, 23–39. [Google Scholar] [CrossRef]
- Robinson, P.H.; Brice, R.E.; Morrison, I.M. Effect of straw treatment and laboratory of incubation on degradation of neutral detergent residue in the rumen. Anim. Feed. Sci. Technol. 1987, 18, 75–82. [Google Scholar] [CrossRef]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing Global Ruminant Agricultural Challenges Through Understanding the Rumen Microbiome: Past, Present, and Future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.X.; Zhang, X.M.; Li, Q.S.; Wang, R.; Zhang, M.; Zhang, S.Z.; Lin, B.; Tan, B.; Tan, Z.L.; Wang, M. Comparison of in situ ruminal straw fiber degradation and bacterial community between buffalo and Holstein fed with high-roughage diet. Front. Microbiol. 2023, 13, 1079056. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Hu, J.; Shi, B.; Dingkao, R.; Wang, J.; Li, S.; Zhang, W.; Luo, Y.; Liu, X. Characteristics and Functions of the Rumen Microbial Community of Cattle-Yak at Different Ages. Biomed. Res. Int. 2020, 2020, 3482692. [Google Scholar] [CrossRef] [PubMed]
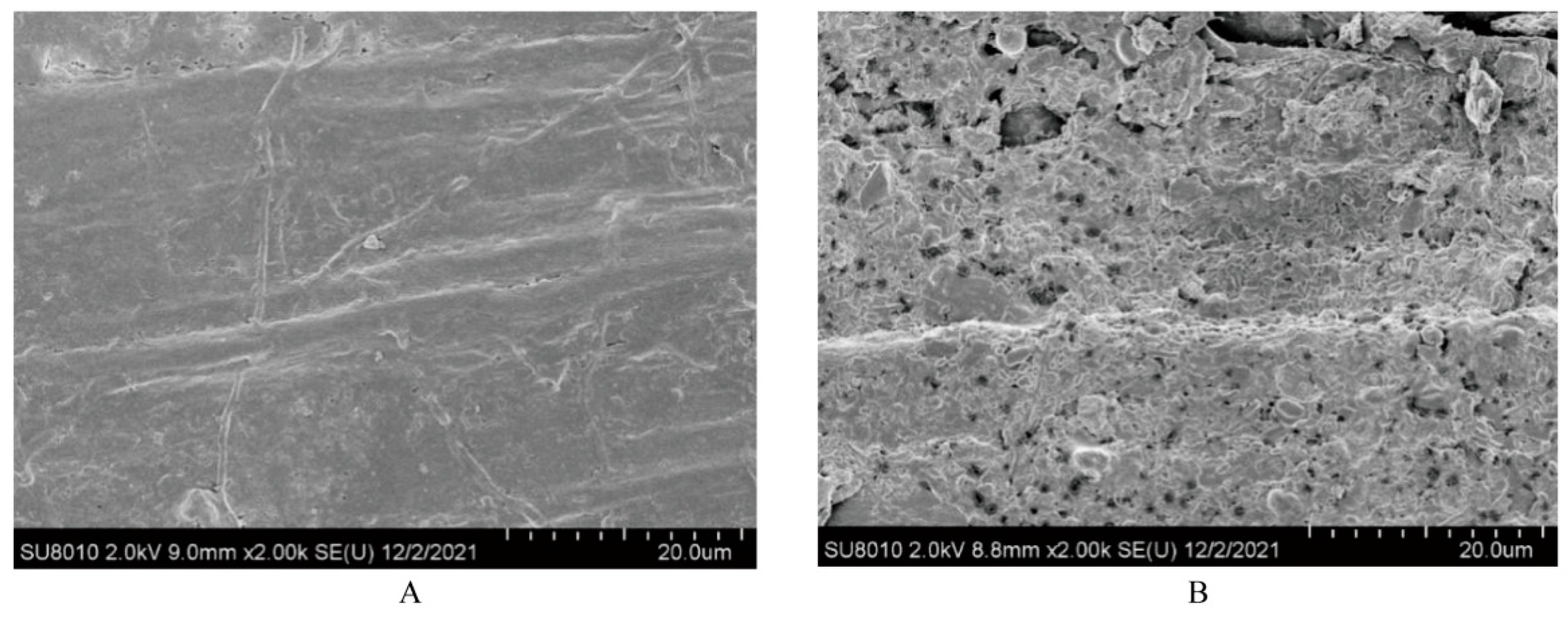
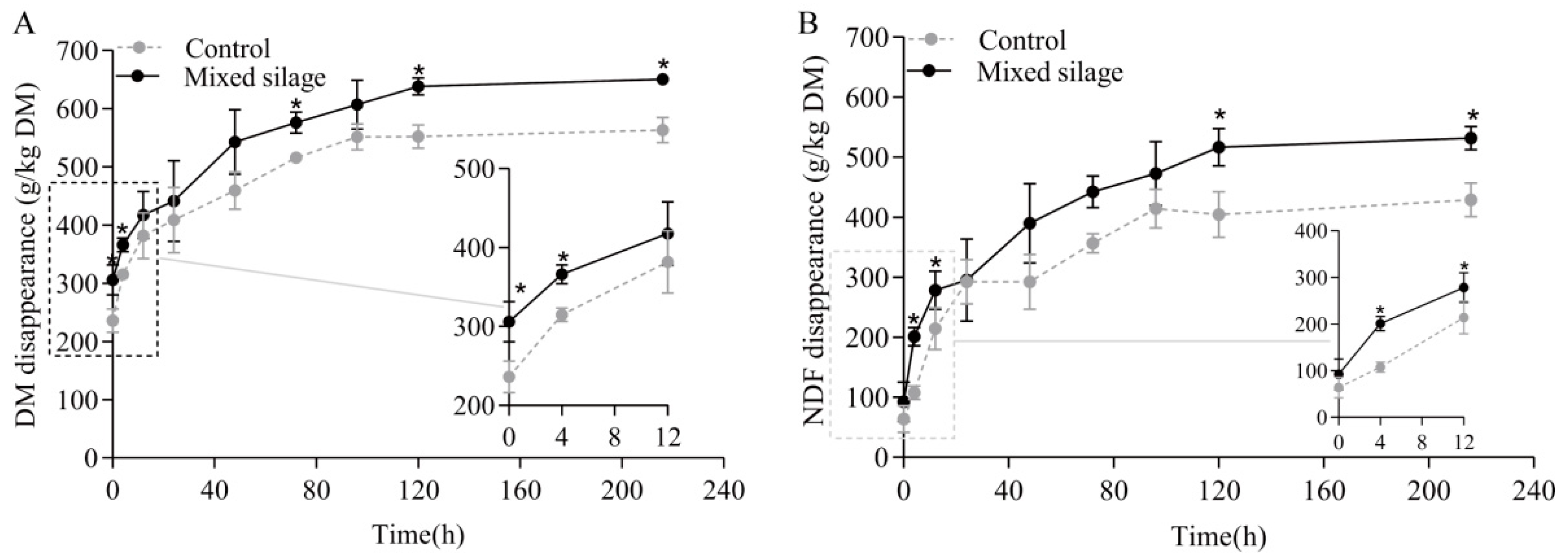
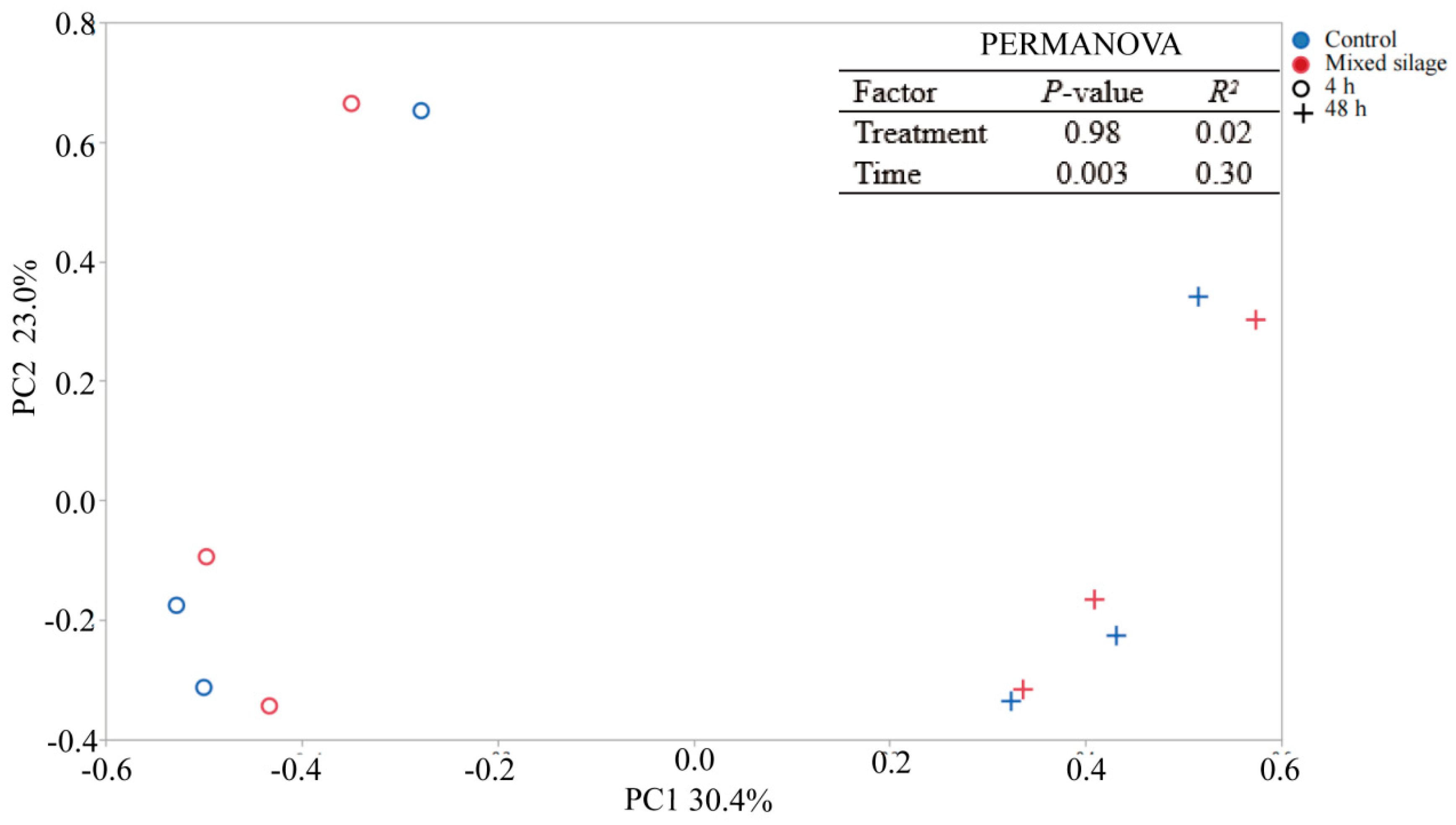
| Item * | Value |
|---|---|
| Ingredients, g/kg | |
| Wheat straw | 800 |
| Corn | 115.9 |
| Soybean meal | 13.3 |
| Cottonseed meal | 15.2 |
| Wheat bran | 15.2 |
| Soybean hull | 11.4 |
| DDGS | 9.5 |
| NaHCO3 | 1.9 |
| NaCl | 1.9 |
| Premix | 5.7 |
| Urea | 10.0 |
| Chemical composition, g/kg | |
| DM | 921 |
| OM | 902 |
| CP | 82 |
| NDF | 634 |
| ADF | 377 |
| NEm, MJ/kg | 4.19 |
| NEf, MJ/kg | 2.18 |
| Items * | Control | Mixed Silage |
|---|---|---|
| Fermentation end products, g/kg DM | ||
| pH | - | 4.22 ± 0.15 |
| Ammonia-N, g/kg DM | 3.52 ± 0.32 | 2.66 ± 0.18 |
| Lactate, g/kg DM | 20.5 ± 1.22 | 32.3 ± 1.36 |
| Acetate, g/kg DM | 9.23 ± 1.23 | 24.2 ± 1.56 |
| Propionate, g/kg DM | 0.26 ± 0.19 | 2.29 ± 0.22 |
| Butyrate, g/kg DM | 0.05 ± 0.02 | 0.01 ± 0.01 |
| Nutrient composition, g/kg DM | ||
| DM | 408 ± 1.25 | 405 ± 2.00 |
| CP | 60.5 ± 0.78 | 61.3 ± 0.89 |
| NDS | 322 ± 2.12 | 377 ± 1.56 |
| NDF | 678 ± 2.12 | 623 ± 1.56 |
| ADF | 440 ± 1.56 | 413 ± 0.98 |
| Hemicellulose | 238 ± 1.76 | 210 ± 1.35 |
| Bacterial compositions (genus), % | ||
| Lentilactobacillus | - | 52.6 ± 2.35 |
| Pediococcus | - | 23.6 ± 2.89 |
| Pantoea | - | 5.29 ± 1.23 |
| Stenotrophomonas | - | 3.23 ± 0.98 |
| Levilactobacillus | - | 2.34 ± 0.78 |
| Erwinia | - | 2.17 ± 1.35 |
| Secundilactobacillus | - | 1.23 ± 0.46 |
| Escherichia | - | 0.82 ± 0.50 |
| Salmonella | - | 0.61 ± 0.23 |
| Sphingobacterium | - | 0.61 ± 0.12 |
| Sarcina | - | 0.53 ± 0.13 |
| others | - | 6.97 ± 2.67 |
| Items a | Treatments b | SEM | p-Value | |
|---|---|---|---|---|
| Control | Mixed Silage | |||
| DM | ||||
| a, g/kg | 289 | 340 | 12.6 | 0.01 |
| b, g/kg | 272 | 318 | 13.0 | 0.08 |
| a + b, g/kg | 561 | 659 | 22.3 | 0.01 |
| c,% h−1 | 3.18 | 2.37 | 0.49 | 0.29 |
| Lag, h | 2.82 | 2.46 | 0.232 | 0.08 |
| ED2, g/kg | 449 | 507 | 17.5 | 0.02 |
| ED6, g/kg | 379 | 427 | 16.2 | 0.03 |
| NDF | ||||
| a, g/kg | 108 | 159 | 12.1 | 0.01 |
| b, g/kg | 310 | 370 | 14.5 | 0.04 |
| a + b, g/kg | 418 | 529 | 26.0 | 0.01 |
| c,% h−1 | 3.16 | 2.51 | 0.42 | 0.31 |
| lag, h | 3.13 | 2.36 | 0.292 | 0.31 |
| ED2, g/kg | 293 | 361 | 19.4 | 0.02 |
| ED6, g/kg | 212 | 266 | 16.4 | 0.04 |
| μNDF, g/kg | 582 | 471 | 26.0 | 0.01 |
| Items * | Treatment | Incubation Time (h) | SEM | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| Control | Mixed Silage | 4 | 48 | Treatment | Incubation Time | Treatment × Incubation Time | ||
| Phylum Firmicutes | 43.0 | 45.5 | 43.5 | 45.0 | 1.12 | 0.33 | 0.55 | 0.48 |
| Papillibacter | 2.29 | 2.31 | 0.98 | 3.62 | 0.414 | 0.92 | <0.001 | 0.47 |
| Kineothrix | 1.46 | 1.69 | 1.29 | 1.86 | 0.151 | 0.29 | 0.03 | 0.06 |
| Lacrimispora | 1.58 | 2.11 | 1.06 | 2.63 | 0.351 | 0.26 | 0.01 | 0.27 |
| Negativibacillus | 0.59 | 0.54 | 0.29 | 0.83 | 0.120 | 0.70 | 0.01 | 0.66 |
| Phylum Bacteroidetes | 32.2 | 31.2 | 38.7 | 24.6 | 2.27 | 0.59 | <0.001 | 0.66 |
| Prevotella | 16.3 | 16.8 | 23.0 | 10.2 | 2.36 | 0.88 | 0.01 | 0.93 |
| Paraprevotella | 5.89 | 4.73 | 7.12 | 3.50 | 0.752 | 0.21 | 0.01 | 0.55 |
| Phylum Proteobacteria | 16.4 | 14.9 | 11.3 | 20.0 | 1.50 | 0.23 | <0.001 | 0.73 |
| Stenotrophomonas | 9.98 | 8.54 | 4.54 | 14.0 | 1.51 | 0.14 | <0.001 | 0.13 |
| Phylum Fibrobacteres | 1.79 | 2.00 | 0.73 | 3.06 | 0.459 | 0.63 | 0.001 | 0.15 |
| Fibrobacter | 1.75 | 1.94 | 0.68 | 3.01 | 0.458 | 0.66 | 0.001 | 0.14 |
| Phylum Actinobacteria | 0.61 | 0.49 | 0.30 | 0.79 | 0.094 | 0.37 | 0.01 | 0.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, X.; Zhang, M.; Zhang, J.; Zhang, X.; Zhang, S.; Lin, B.; Wang, T.; Tan, Z.; Wang, M. Mixed Ensiling Increases Degradation Without Altering Attached Microbiota Through In Situ Ruminal Incubation Technique. Animals 2025, 15, 2131. https://doi.org/10.3390/ani15142131
Pu X, Zhang M, Zhang J, Zhang X, Zhang S, Lin B, Wang T, Tan Z, Wang M. Mixed Ensiling Increases Degradation Without Altering Attached Microbiota Through In Situ Ruminal Incubation Technique. Animals. 2025; 15(14):2131. https://doi.org/10.3390/ani15142131
Chicago/Turabian StylePu, Xuanxuan, Min Zhang, Jianjun Zhang, Xiumin Zhang, Shizhe Zhang, Bo Lin, Tianwei Wang, Zhiliang Tan, and Min Wang. 2025. "Mixed Ensiling Increases Degradation Without Altering Attached Microbiota Through In Situ Ruminal Incubation Technique" Animals 15, no. 14: 2131. https://doi.org/10.3390/ani15142131
APA StylePu, X., Zhang, M., Zhang, J., Zhang, X., Zhang, S., Lin, B., Wang, T., Tan, Z., & Wang, M. (2025). Mixed Ensiling Increases Degradation Without Altering Attached Microbiota Through In Situ Ruminal Incubation Technique. Animals, 15(14), 2131. https://doi.org/10.3390/ani15142131





