Renal Single-Cell RNA Sequencing and Digital Cytometry in Dogs with X-Linked Hereditary Nephropathy
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Single-Cell Suspension Preparation
2.3. Single-Cell RNA Sequencing
2.4. Sample Size Calculation
2.5. Data and Statistical Analysis
3. Results
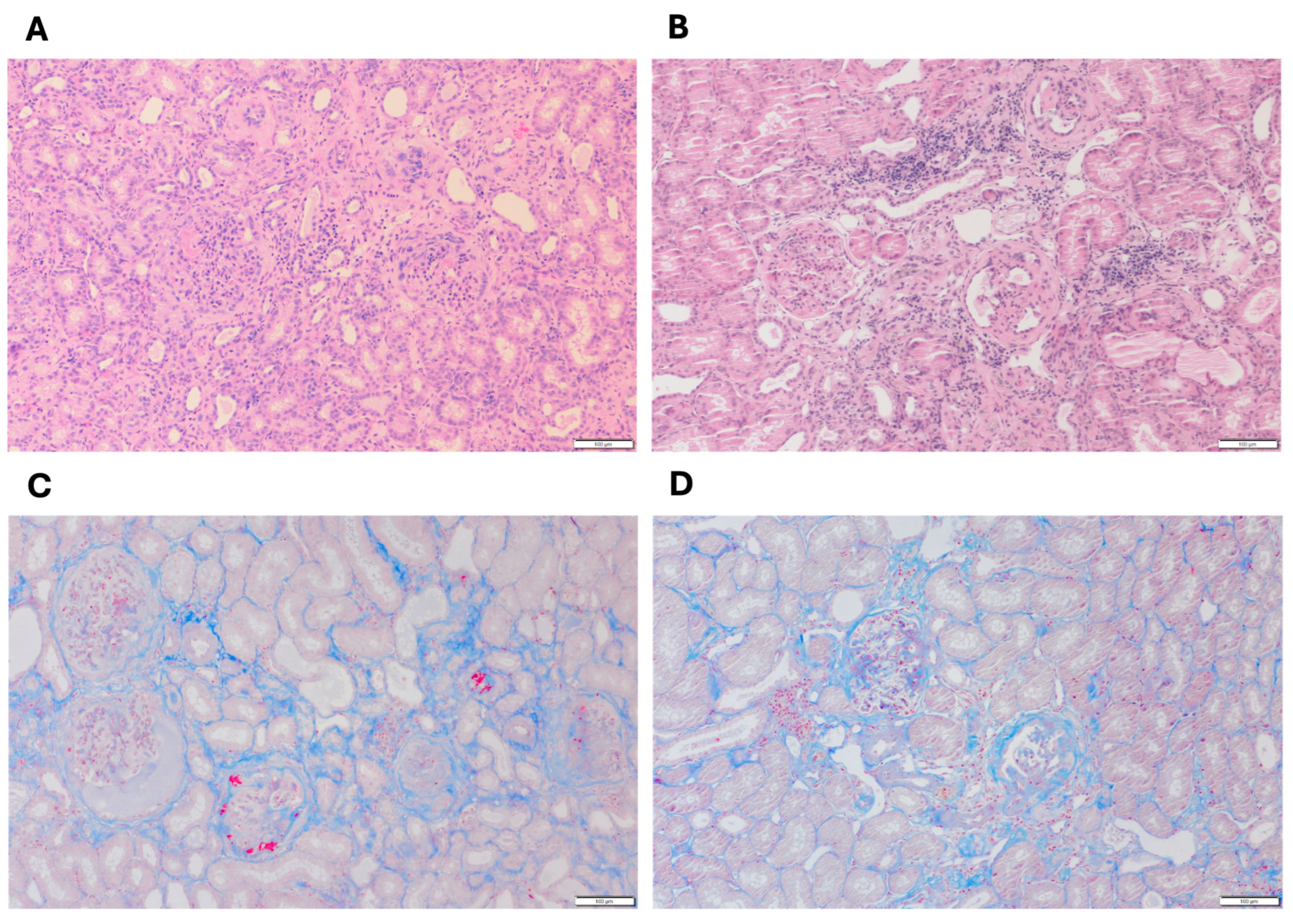
3.1. Histopathological Evaluation of Kidney Biopsies
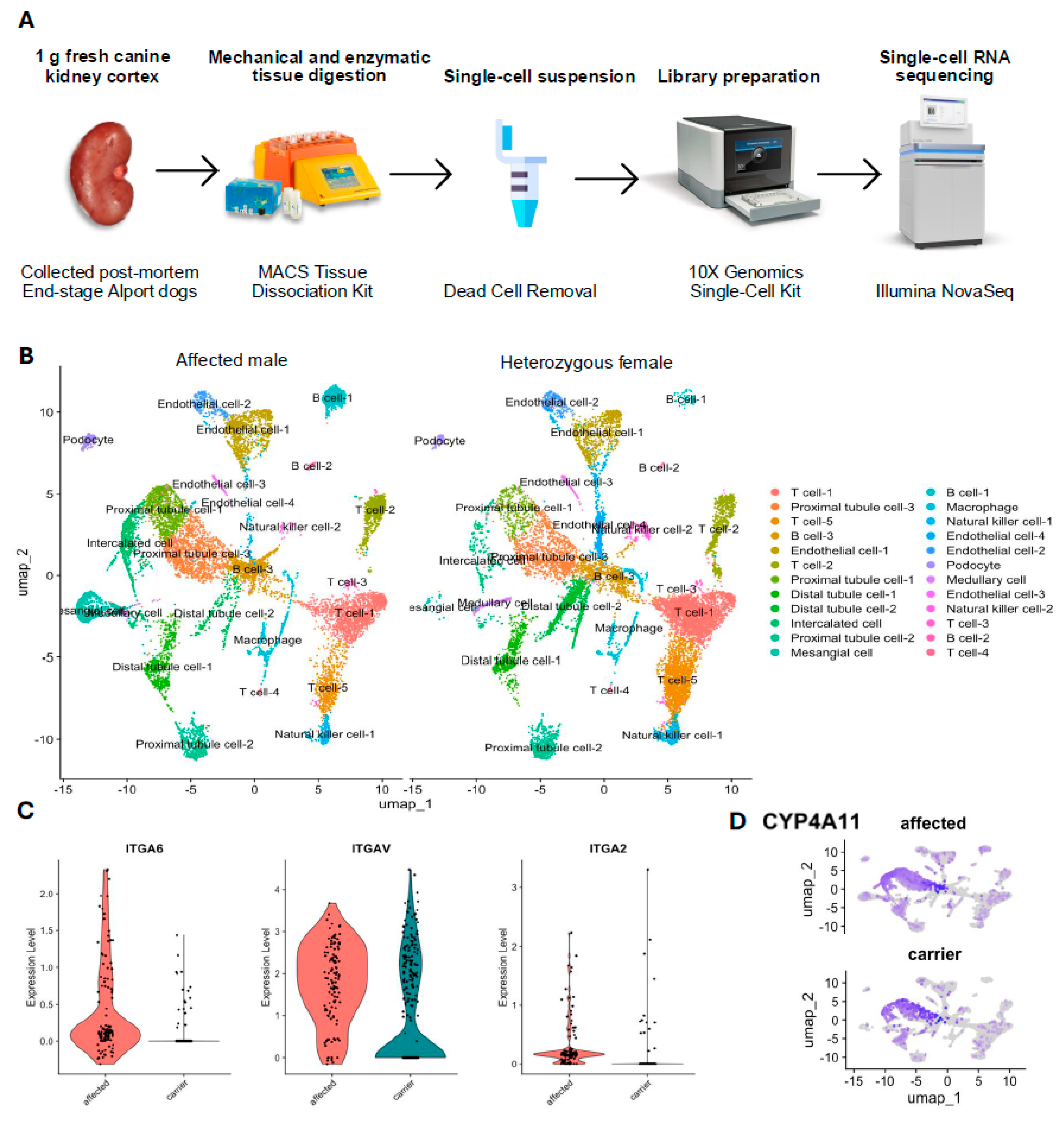
3.2. Unbiased Classification of Canine Renal Cells Using scRNA-Seq
3.3. Differential Gene Expression and Pathway Analysis
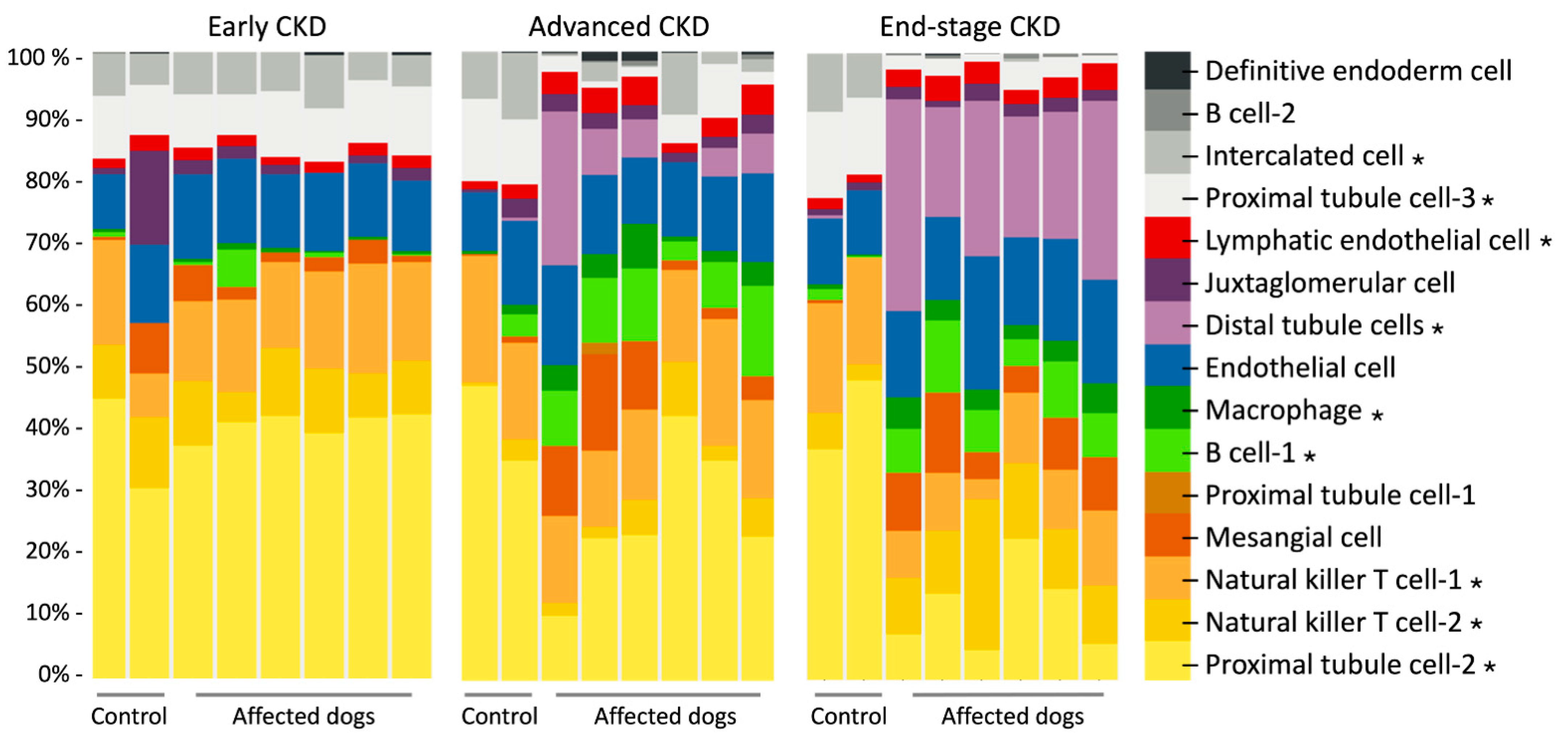
3.4. Digital Cytometry Using Bulk mRNA-Seq Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 20HETE | 20-hydroxyeicosatetraenoic acid |
| CKD | Chronic kidney disease |
| COL4A5 | α5 chain of the type IV collagen |
| CYP4A11 | Cytochrome P450 family 4 subfamily A member 11 |
| PANTHER | Protein Analysis THrough Evolutionary Relationships |
| RNA-seq | RNA sequencing |
| scRNA-seq | Single-cell RNA sequencing |
| XLHN | X-linked Hereditary Nephropathy |
References
- O’Neill, D.G.; Elliott, J.; Church, D.B.; McGreevy, P.D.; Thomson, P.C.; Brodbelt, D.C. Chronic Kidney Disease in Dogs in UK Veterinary Practices: Prevalence, Risk Factors, and Survival. J. Vet. Intern. Med. 2013, 27, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Thorner, P.S.; Marrano, P.; Baumal, R.; McInnes, R.R. Canine X Chromosome-Linked Hereditary Nephritis: A Genetic Model for Human X-Linked Hereditary Nephritis Resulting from a Single Base Mutation in the Gene Encoding the Alpha 5 Chain of Collagen Type IV. Proc. Natl. Acad. Sci. USA 1994, 91, 3989–3993. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.L.; Lees, G.E.; Kashtan, C.E.; Murphy, K.E. Genetic Cause of X-Linked Alport Syndrome in a Family of Domestic Dogs. Mamm. Genome 2003, 14, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Benali, S.L.; Lees, G.E.; Nabity, M.B.; Arico, A.; Drigo, M.; Gallo, E.; Giantin, M.; Aresu, L. X-Linked Hereditary Nephropathy in Navasota Dogs: Clinical Pathology, Morphology, and Gene Expression During Disease Progression. Vet. Pathol. 2016, 53, 803–812. [Google Scholar] [CrossRef]
- Greer, K.A.; Higgins, M.A.; Cox, M.L.; Ryan, T.P.; Berridge, B.R.; Kashtan, C.E.; Lees, G.E.; Murphy, K.E. Gene Expression Analysis in a Canine Model of X-Linked Alport Syndrome. Mamm. Genome 2006, 17, 976–990. [Google Scholar] [CrossRef]
- Rao, V.H.; Lees, G.E.; Kashtan, C.E.; Nemori, R.; Singh, R.K.; Meehan, D.T.; Rodgers, K.; Berridge, B.R.; Bhattacharya, G.; Cosgrove, D. Increased Expression of MMP-2, MMP-9 (Type IV Collagenases/Gelatinases), and MT1-MMP in Canine X-Linked Alport Syndrome (XLAS). Kidney Int. 2003, 63, 1736–1748. [Google Scholar] [CrossRef]
- Chu, C.P.; Hokamp, J.A.; Cianciolo, R.E.; Dabney, A.R.; Brinkmeyer-Langford, C.; Lees, G.E.; Nabity, M.B. RNA-Seq of Serial Kidney Biopsies Obtained during Progression of Chronic Kidney Disease from Dogs with X-Linked Hereditary Nephropathy. Sci. Rep. 2017, 7, 16776. [Google Scholar] [CrossRef]
- Chu, C.P.; Liu, S.; Song, W.; Xu, E.Y.; Nabity, M.B. Small RNA Sequencing Evaluation of Renal microRNA Biomarkers in Dogs with X-Linked Hereditary Nephropathy. Sci. Rep. 2021, 11, 17437. [Google Scholar] [CrossRef]
- Oliverio, A.L.; Bellomo, T.; Mariani, L.H. Evolving Clinical Applications of Tissue Transcriptomics in Kidney Disease: A Mini-Review. Front. Pediatr. 2019, 7, 306. [Google Scholar] [CrossRef]
- Saliba, A.E.; Westermann, A.J.; Gorski, S.A.; Vogel, J. Single-Cell RNA-Seq: Advances and Future Challenges. Nucleic Acids Res. 2014, 42, 8845–8860. [Google Scholar] [CrossRef]
- Park, J.; Shrestha, R.; Qiu, C.; Kondo, A.; Huang, S.; Werth, M.; Li, M.; Barasch, J.; Susztak, K. Single-Cell Transcriptomics of the Mouse Kidney Reveals Potential Cellular Targets of Kidney Disease. Science 2018, 360, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lee, J.W.; Chou, C.L.; Nair, A.V.; Battistone, M.A.; Paunescu, T.G.; Merkulova, M.; Breton, S.; Verlander, J.W.; Wall, S.M.; et al. Transcriptomes of Major Renal Collecting Duct Cell Types in Mouse Identified by Single-Cell RNA-Seq. Proc. Natl. Acad. Sci. USA 2017, 114, E9989–E9998. [Google Scholar] [CrossRef]
- Qiu, C.; Huang, S.; Park, J.; Park, Y.; Ko, Y.A.; Seasock, M.J.; Bryer, J.S.; Xu, X.X.; Song, W.C.; Palmer, M.; et al. Renal Compartment-Specific Genetic Variation Analyses Identify New Pathways in Chronic Kidney Disease. Nat. Med. 2018, 24, 1721–1731. [Google Scholar] [CrossRef]
- Der, E.; Ranabothu, S.; Suryawanshi, H.; Akat, K.M.; Clancy, R.; Morozov, P.; Kustagi, M.; Czuppa, M.; Izmirly, P.; Belmont, H.M.; et al. Single Cell RNA Sequencing to Dissect the Molecular Heterogeneity in Lupus Nephritis. JCI Insight 2017, 2, e93009. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Chew, C.; Brand, O.J.; Yamamura, T.; Lawless, C.; Morais, M.R.P.T.; Zeef, L.; Lin, I.-H.; Howell, G.; Lui, S.; Lausecker, F.; et al. Kidney Resident Macrophages Have Distinct Subsets and Multifunctional Roles. Matrix Biol. 2024, 127, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Wang, C.; Fan, X.; Fang, L.; Li, L.; Zhang, X.; Yu, K.; Liu, L.; Guo, L.; Yang, X. Novel Biomarkers Related to Oxidative Stress and Immunity in Chronic Kidney Disease. Heliyon 2024, 10, e27754. [Google Scholar] [CrossRef]
- Dhillon, P.; Mulholland, K.A.; Hu, H.; Park, J.; Sheng, X.; Abedini, A.; Liu, H.; Vassalotti, A.; Wu, J.; Susztak, K. Increased Levels of Endogenous Retroviruses Trigger Fibroinflammation and Play a Role in Kidney Disease Development. Nat. Commun. 2023, 14, 1–20. [Google Scholar] [CrossRef]
- Lees, G.E.; Helman, R.G.; Kashtan, C.E.; Michael, A.F.; Homco, L.D.; Millichamp, N.J.; Camacho, Z.T.; Templeton, J.W.; Ninomiya, Y.; Sado, Y.; et al. New Form of X-Linked Dominant Hereditary Nephritis in Dogs. Am. J. Vet. Res. 1999, 60, 373–383. [Google Scholar] [CrossRef]
- 10x Genomics Chromium Single Cell Applications—Guidelines for Optimal Sample Preparation. Available online: https://www.10xgenomics.com/support/single-cell-gene-expression/documentation/steps/sample-prep/chromium-single-cell-applications-guidelines-for-optimal-sample-preparation (accessed on 31 March 2025).
- Davis, A.; Gao, R.; Navin, N.E. SCOPIT: Sample Size Calculations for Single-Cell Sequencing Experiments. BMC Bioinform. 2019, 20, 566. [Google Scholar] [CrossRef]
- Du, Y.; Huang, Q.; Arisdakessian, C.; Garmire, L.X. Evaluation of STAR and Kallisto on Single Cell RNA-Seq Data Alignment. G3 (Bethesda) 2020, 10, 1775–1783. [Google Scholar] [CrossRef]
- Lun, A.T.L.; Riesenfeld, S.; Andrews, T.; Dao, T.P.; Gomes, T.; Participants in the 1st Human Cell Atlas Jamboree; Marioni, J.C. EmptyDrops: Distinguishing Cells from Empty Droplets in Droplet-Based Single-Cell RNA Sequencing Data. Genome Biol. 2019, 20, 63. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J.A.; Richard, A.C.; Bach, K.; Lun, A.T.L.; Marioni, J.C. Detection and Removal of Barcode Swapping in Single-Cell RNA-Seq Data. Nat. Commun. 2018, 9, 2667. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Luecken, M.D.; Theis, F.J. Current Best Practices in Single-Cell RNA-Seq Analysis: A Tutorial. Mol. Syst. Biol. 2019, 15, e8746. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Hu, C.; Li, T.; Xu, Y.; Zhang, X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.; Yang, K.; Ou, Q.; et al. CellMarker 2.0: An Updated Database of Manually Curated Cell Markers in Human/Mouse and Web Tools Based on scRNA-Seq Data. Nucleic Acids Res. 2023, 51, D870–D876. [Google Scholar] [CrossRef] [PubMed]
- Franzén, O.; Gan, L.-M.; Björkegren, J.L. PanglaoDB: A Web Server for Exploration of Mouse and Human Single-Cell RNA Sequencing Data. Database 2019, 2019, baz046. [Google Scholar] [CrossRef]
- Wu, H.; Kirita, Y.; Donnelly, E.L.; Humphreys, B.D. Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. J. Am. Soc. Nephrol. 2019, 30, 23–32. [Google Scholar] [CrossRef]
- Liao, J.; Yu, Z.; Chen, Y.; Bao, M.; Zou, C.; Zhang, H.; Liu, D.; Li, T.; Zhang, Q.; Li, J.; et al. Single-Cell RNA Sequencing of Human Kidney. Sci. Data 2020, 7, 4. [Google Scholar] [CrossRef]
- Combes, A.N.; Phipson, B.; Lawlor, K.T.; Dorison, A.; Patrick, R.; Zappia, L.; Harvey, R.P.; Oshlack, A.; Little, M.H. Single Cell Analysis of the Developing Mouse Kidney Provides Deeper Insight into Marker Gene Expression and Ligand-Receptor Crosstalk. Development 2019, 146, dev178673. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining Cell Type Abundance and Expression from Bulk Tissues with Digital Cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Jedlicka, J.; Soleiman, A.; Draganovici, D.; Mandelbaum, J.; Ziegler, U.; Regele, H.; Wuthrich, R.P.; Gross, O.; Anders, H.J.; Segerer, S. Interstitial Inflammation in Alport Syndrome. Hum. Pathol. 2010, 41, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Daga, S.; Donati, F.; Capitani, K.; Croci, S.; Tita, R.; Giliberti, A.; Valentino, F.; Benetti, E.; Fallerini, C.; Niccheri, F.; et al. New Frontiers to Cure Alport Syndrome: COL4A3 and COL4A5 Gene Editing in Podocyte-Lineage Cells. Eur. J. Hum. Genet. 2020, 28, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.; Wu, G. Molecular Mechanisms Involved in Podocyte EMT and Concomitant Diabetic Kidney Diseases: An Update. Ren. Fail. 2017, 39, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Hang, X.; Wei, Y.; Wang, H.; Zhang, L.; Zhao, L. Crosstalk among Podocytes, Glomerular Endothelial Cells and Mesangial Cells in Diabetic Kidney Disease: An Updated Review. Cell Commun. Signal 2024, 22, 136. [Google Scholar] [CrossRef]
- Li, Y.; Yang, J.; Dai, C.; Wu, C.; Liu, Y. Role for Integrin-Linked Kinase in Mediating Tubular Epithelial to Mesenchymal Transition and Renal Interstitial Fibrogenesis. J. Clin. Investig. 2003, 112, 503–516. [Google Scholar] [CrossRef]
- Wu, X.; Gao, Y.; Xu, L.; Dang, W.; Yan, H.; Zou, D.; Zhu, Z.; Luo, L.; Tian, N.; Wang, X.; et al. Exosomes from High Glucose-Treated Glomerular Endothelial Cells Trigger the Epithelial-Mesenchymal Transition and Dysfunction of Podocytes. Sci. Rep. 2017, 7, 9371. [Google Scholar] [CrossRef]
- Maile, L.A.; Gollahon, K.; Wai, C.; Dunbar, P.; Busby, W.; Clemmons, D. Blocking αVβ3 Integrin Ligand Occupancy Inhibits the Progression of Albuminuria in Diabetic Rats. J. Diabetes Res. 2014, 2014, 421827. [Google Scholar] [CrossRef]
- Maile, L.A.; Busby, W.H.; Gollahon, K.A.; Flowers, W.; Garbacik, N.; Garbacik, S.; Stewart, K.; Nichols, T.; Bellinger, D.; Patel, A.; et al. Blocking Ligand Occupancy of the αVβ3 Integrin Inhibits the Development of Nephropathy in Diabetic Pigs. Endocrinology 2014, 155, 4665–4675. [Google Scholar] [CrossRef]
- Rubel, D.; Frese, J.; Martin, M.; Leibnitz, A.; Girgert, R.; Miosge, N.; Eckes, B.; Müller, G.-A.; Gross, O. Collagen Receptors Integrin Alpha2beta1 and Discoidin Domain Receptor 1 Regulate Maturation of the Glomerular Basement Membrane and Loss of Integrin Alpha2beta1 Delays Kidney Fibrosis in COL4A3 Knockout Mice. Matrix Biol. 2014, 34, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, Y.; Gao, X.; Wang, G. Identification of Cell Subpopulations Associated with Disease Phenotypes from scRNA-Seq Data Using PACSI. BMC Biol. 2023, 21, 159. [Google Scholar] [CrossRef] [PubMed]
- Fustero-Torre, C.; Jiménez-Santos, M.J.; García-Martín, S.; Carretero-Puche, C.; García-Jimeno, L.; Ivanchuk, V.; Di Domenico, T.; Gómez-López, G.; Al-Shahrour, F. Beyondcell: Targeting Cancer Therapeutic Heterogeneity in Single-Cell RNA-Seq Data. Genome Med. 2021, 13, 187. [Google Scholar] [CrossRef] [PubMed]
- Haller, H.; Bertram, A.; Nadrowitz, F.; Menne, J. Monocyte Chemoattractant Protein-1 and the Kidney. Curr. Opin. Nephrol. Hypertens. 2016, 25, 42. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, T.; Mao, K.; Shao, X.; Xu, Y.; Zhu, M.; Zhou, H.; Wang, Q.; Li, Z.; Xie, Y.; et al. A Single-Cell Survey of the Human Glomerulonephritis. J. Cell Mol. Med. 2021, 25, 4684–4695. [Google Scholar] [CrossRef]
- Gerhardt, L.M.S.; Liu, J.; Koppitch, K.; Cippà, P.E.; McMahon, A.P. Single-Nuclear Transcriptomics Reveals Diversity of Proximal Tubule Cell States in a Dynamic Response to Acute Kidney Injury. Proc. Natl. Acad. Sci. USA 2021, 118, e2026684118. [Google Scholar] [CrossRef]
- Tajti, F.; Kuppe, C.; Antoranz, A.; Ibrahim, M.M.; Kim, H.; Ceccarelli, F.; Holland, C.H.; Olauson, H.; Floege, J.; Alexopoulos, L.G.; et al. A Functional Landscape of CKD Entities from Public Transcriptomic Data. Kidney Int. Rep. 2020, 5, 211–224. [Google Scholar] [CrossRef]
- Gainer, J.V.; Lipkowitz, M.S.; Yu, C.; Waterman, M.R.; Dawson, E.P.; Capdevila, J.H.; Brown, N.J.; Aask Study Group. Association of a CYP4A11 Variant and Blood Pressure in Black Men. J. Am. Soc. Nephrol. 2008, 19, 1606–1612. [Google Scholar] [CrossRef]
- Ho, Y.; Hu, P.; Peel, M.T.; Chen, S.; Camara, P.G.; Epstein, D.J.; Wu, H.; Liebhaber, S.A. Single-Cell Transcriptomic Analysis of Adult Mouse Pituitary Reveals Sexual Dimorphism and Physiologic Demand-Induced Cellular Plasticity. Protein Cell 2020, 11, 565–583. [Google Scholar] [CrossRef]
| Affected Male Dog | Heterozygous Female Dog | |
|---|---|---|
| Reads mapped to genome | 95.4% | 95.3% |
| Number of cells examined | 12,778 | 13,190 |
| Median genes per cell | 1575 | 1278 |
| Total genes detected | 25,438 | 24,208 |
| Cell Type | Marker Genes | Cell Number in Affected Male Dog | Cell Percentage in Affected Male Dog | Cell Number in Heterozygous Female Dog | Cell Percentage in Heterozygous Female Dog |
|---|---|---|---|---|---|
| B cell-1 | CD79B [31] | 364 | 2.85% | 394 | 2.99% |
| B cell-2 | CD79B [31] CTSS [32] | 9 | 0.07% | 142 | 1.08% |
| B cell-3 | (Determined by CellMarker [28]) | 908 | 7.11% | 853 | 6.47% |
| Distal tubule cell-1 | UMOD, DEFB1, TMEM213 [31] | 650 | 5.09% | 573 | 4.34% |
| Distal tubule cell-2 | DEFB1, ATP6V1G3 [31] | 205 | 1.60% | 918 | 6.96% |
| Endothelial cell-1 | PECAM1, EMCN, FLT1, TEK [30] FAM167B, SOX17, MYCT1, TIE1, ADGRL4, SOX18, PTPRB [32] | 910 | 7.12% | 717 | 5.44% |
| Endothelial cell-2 | PECAM1, EMCN, FLT1, TEK, ADGRL4, SOX18 [30] FAM167B, SOX17, MYCT1, TIE1, PTPRB [32] | 238 | 1.86% | 313 | 2.37% |
| Endothelial cell-3 | PECAM1, EMCN [30] GPHBP1, MYCT1, TIE1, SOX18 [32] | 160 | 1.25% | 150 | 1.14% |
| Endothelial cell-4 | PECAM1, EMCN [30] GPHBP1, MYCT1, TIE1, SOX18 [32] | 103 | 0.81% | 524 | 3.97% |
| Intercalated cell | ATP6V1G3, ATP6V0D2, TMEM213 [31] | 843 | 6.60% | 223 | 1.69% |
| Macrophage | LYZ, CD14 [31] C1QB, CTSS, CCL4, CCL3, AIF1, CX3CR1 [32] | 182 | 1.42% | 495 | 3.75% |
| Medullary cell | FBLN5 [28] | 71 | 0.56% | 268 | 2.03% |
| Mesangial cell | MYLL9, ITGA8 [30] | 909 | 7.11% | 37 | 0.28% |
| Natural killer cell-1 | NKG7 [31] CD52, CCL4 [32] | 857 | 6.71% | 50 | 0.38% |
| Natural killer cell-2 | NKG7 [31] CD52 [32] | 40 | 0.31% | 212 | 1.61% |
| Podocyte | NPHS2, NPHS1, WT1, SYNPO [30] | 281 | 2.20% | 66 | 0.50% |
| Proximal tubule cell-1 | SLC13A3, SLC34A1, GPX3 [31] RDH16 [32] | 1,031 | 8.07% | 347 | 2.63% |
| Proximal tubule cell-2 | SLC13A3, SLC34A1, GPX3 [31] RDH16 [32] | 458 | 3.58% | 523 | 3.97% |
| Proximal tubule cell-3 | GPX3 [31] RDH16 [32] | 1,431 | 11.20% | 1182 | 8.96% |
| T cell-1 | IL7R [31] CD52 [32] | 1,586 | 12.41% | 2863 | 21.71% |
| T cell-2 | IL7R [31] CD52 [32] | 781 | 6.11% | 620 | 4.70% |
| T cell-3 | IL7R [31] NKG7 [31] CCL4 [32] | 95 | 0.74% | 66 | 0.50% |
| T cell-4 | IL7R [31] CTSS, CX3CR1 [32] | 112 | 0.88% | 20 | 0.15% |
| T cell-5 | CCL4, CCL3 [32] | 554 | 4.34% | 1634 | 12.39% |
| Cell Types | PANTHER Pathways | Genes in the Pathway | Fold Enrichment | p Value |
|---|---|---|---|---|
| All cells | FAS signaling pathway | LMNA, ENSCAFG00845026419, JUN, GSN | 12.84 | 3.82 × 10−2 |
| CCKR signaling map | CPE, CREM, CCK, JUN, MMP9, IER3, CCK, MEF2C | 4.66 | 2.24 × 10−2 | |
| Integrin signaling pathway | COL1A1, COL1A2, ACAT2, COL6A3, COL1A2, FN1, COL3A1, COL1A1, FN1 | 4.23 | 4.62 × 10−2 | |
| Podocytes | Integrin signaling pathway | COL11A1, COL4A1, ITGAV, VCL, COL1A2, ACTA2, ASAP1, COL6A3, NTN4, CAV1, COL1A2, COL5A2, FN1, COL8A2, ITGA2, LAMB1, COL4A2, FN1, COL4A4, ITGA6, COL5A3 | 4.52 | 1.47 × 10−6 |
| Proximal tubule cell-2 | Inflammation mediated by chemokine and cytokine signaling pathway | CCL2, MYH11, ACTA2, ITGA2, CXCL10 | 8.45 | 1.14 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, C.P.; Osorio, D.; Nabity, M.B. Renal Single-Cell RNA Sequencing and Digital Cytometry in Dogs with X-Linked Hereditary Nephropathy. Animals 2025, 15, 2061. https://doi.org/10.3390/ani15142061
Chu CP, Osorio D, Nabity MB. Renal Single-Cell RNA Sequencing and Digital Cytometry in Dogs with X-Linked Hereditary Nephropathy. Animals. 2025; 15(14):2061. https://doi.org/10.3390/ani15142061
Chicago/Turabian StyleChu, Candice P., Daniel Osorio, and Mary B. Nabity. 2025. "Renal Single-Cell RNA Sequencing and Digital Cytometry in Dogs with X-Linked Hereditary Nephropathy" Animals 15, no. 14: 2061. https://doi.org/10.3390/ani15142061
APA StyleChu, C. P., Osorio, D., & Nabity, M. B. (2025). Renal Single-Cell RNA Sequencing and Digital Cytometry in Dogs with X-Linked Hereditary Nephropathy. Animals, 15(14), 2061. https://doi.org/10.3390/ani15142061








