Characterizing the Fermentation of Oat Grass (Avena sativa L.) in the Rumen: Integrating Degradation Kinetics, Ultrastructural Examination with Scanning Electron Microscopy, Surface Enzymatic Activity, and Microbial Community Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Raw Materials
2.2. Animal Feeding
2.3. Experimental Design and Sample Collection
2.4. Sample Determination and Analysis
2.4.1. Chemical Analysis
2.4.2. Ruminal Degradability Analysis
2.4.3. Scanning Electron Microscopy (SEM)
2.4.4. Cellulase Activity Assay
2.4.5. Rumen Microbiome Analysis Methods
2.5. Statistical Analysis Method
3. Results
3.1. Ruminal Degradability
3.2. Scanning Electron Microscopy (SEM) Observations
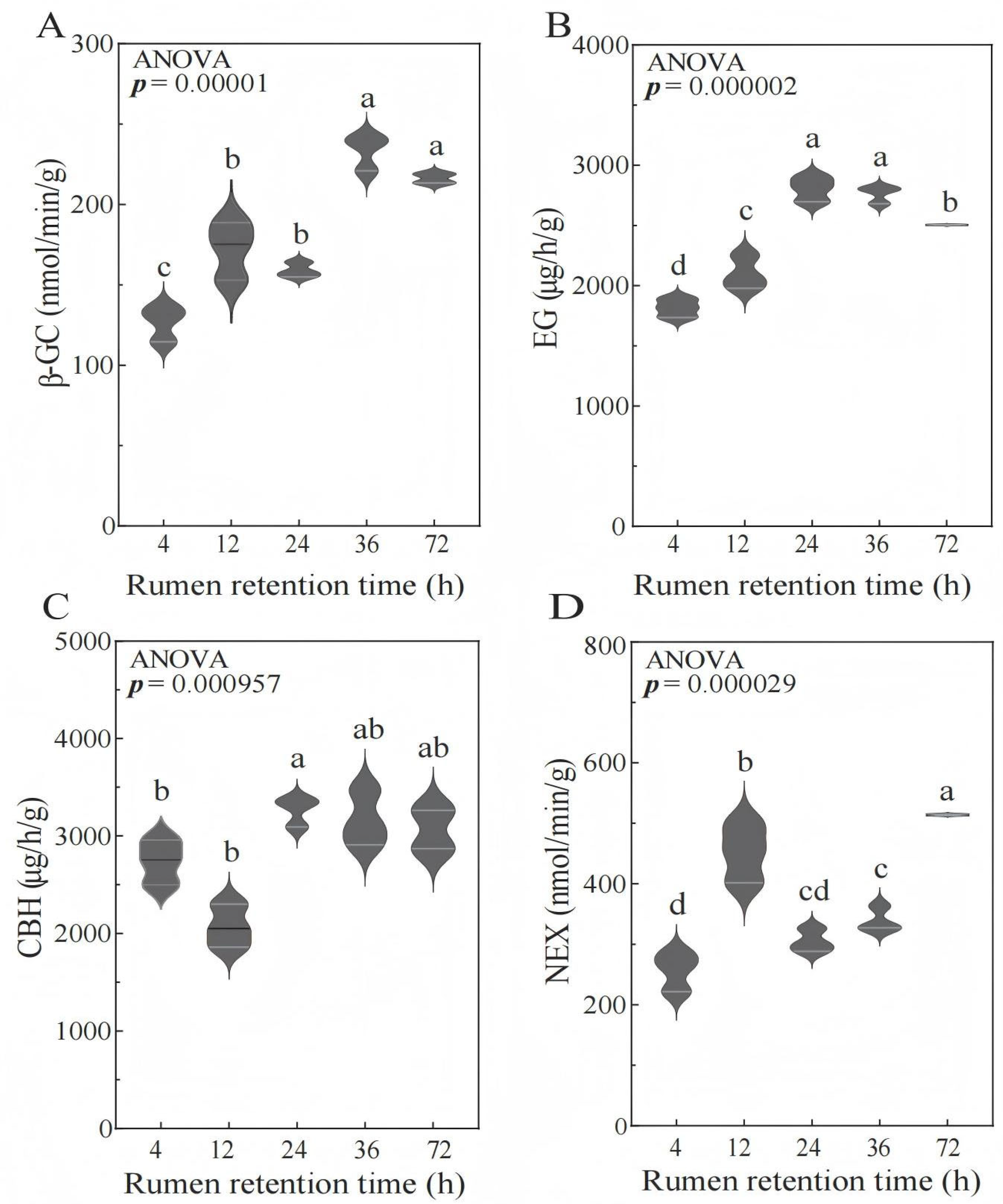
3.3. Cellulase Activity Analysis
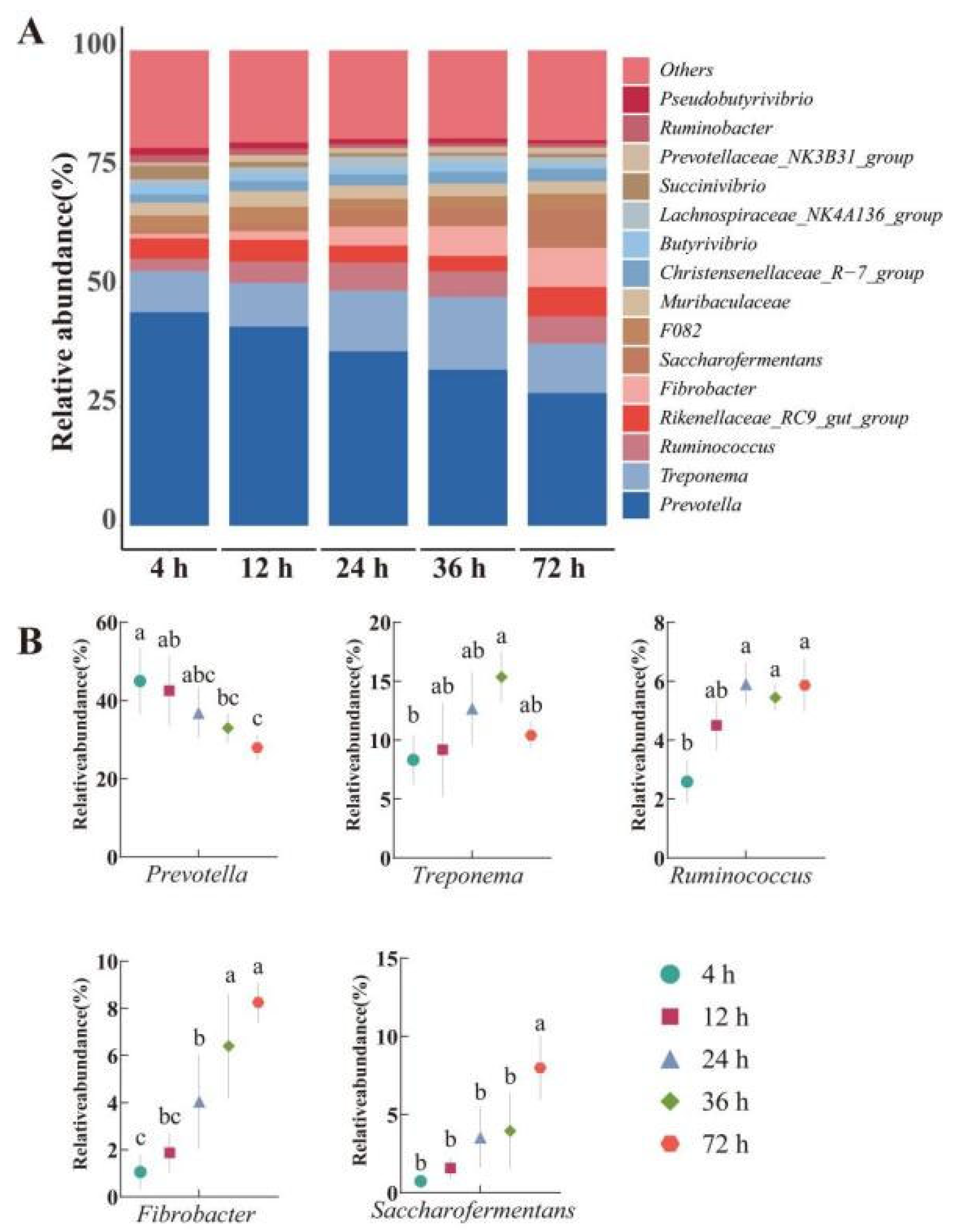
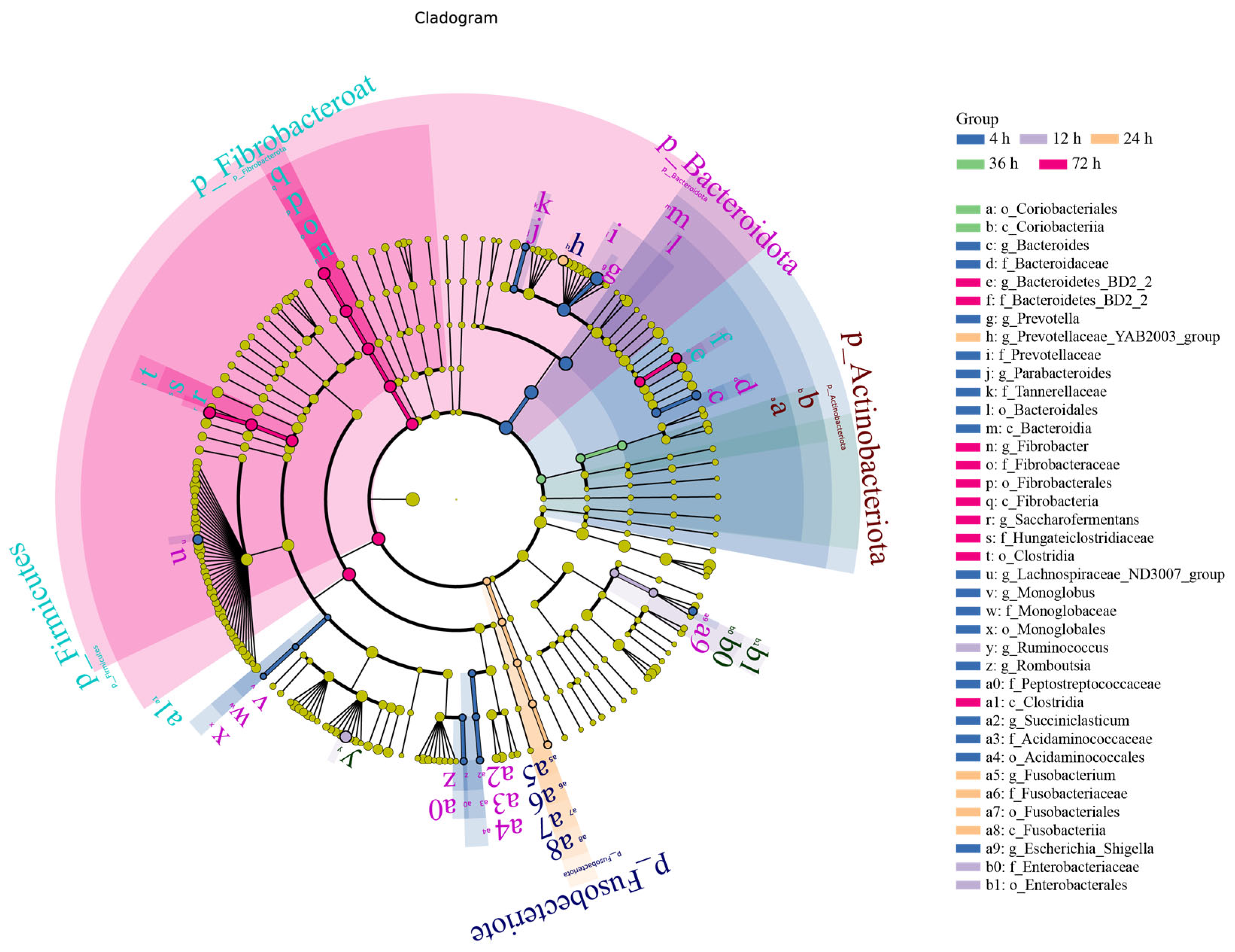
3.4. Rumen Microbiome Analysis
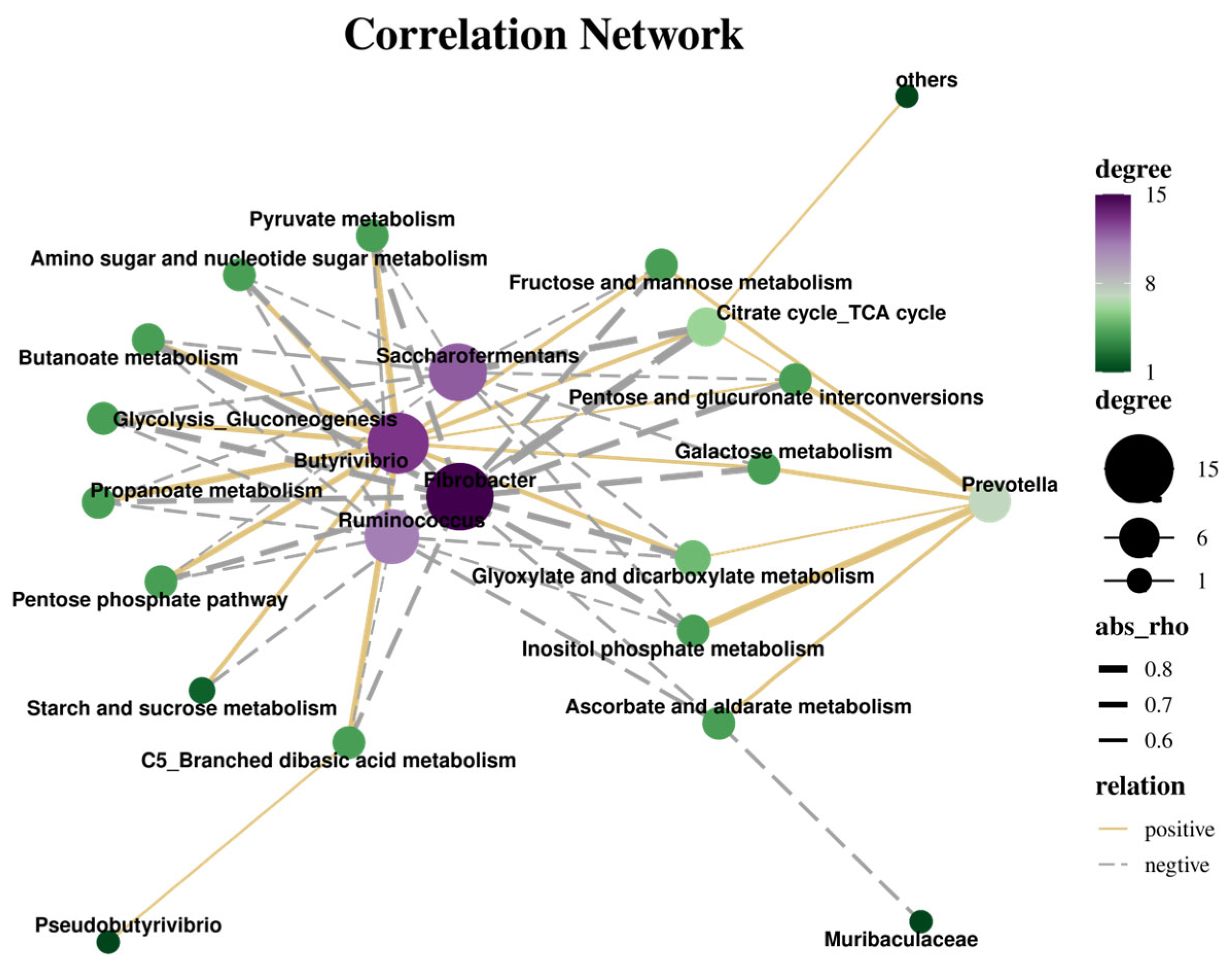
3.5. The Correlation Between Nutrient Degradation Rate, Cellulase Activity, and Rumen Bacteria
3.6. Functional Prediction
3.7. Correlation Analysis Between KEGG and Nutrition, Enzyme Activity, and Bacteria
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Takizawa, S.; Asano, R.; Fukuda, Y.; Feng, M.; Baba, Y.; Abe, K.; Tada, C.; Nakai, Y. Change of Endoglucanase Activity and Rumen Microbial Community During Biodegradation of Cellulose Using Rumen Microbiota. Front. Microbiol. 2020, 11, 603818. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Li, R.; Wang, Y.; Cui, X.; Niu, D.; Yang, F.; Xu, C. Ensiling with rumen fluid promoted Irpex lacteus colonization on the non-sterile naked oat straw for enhanced lignocellulose degradation and enzymatic hydrolysis. Biochem. Eng. J. 2022, 183, 108462. [Google Scholar] [CrossRef]
- Wang, Y.L.; Wang, W.K.; Wu, Q.C.; Zhang, F.; Li, W.J.; Li, S.L.; Wang, W.; Cao, Z.J.; Yang, H.J. In Situ Rumen Degradation Characteristics and Bacterial Colonization of Corn Silages Differing in Ferulic and p-Coumaric Acid Contents. Microorganisms 2022, 10, 2269. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Q.; Sun, S.; Sun, H.; Wang, Y.; Shen, X.; Zhang, L. Exploring AI-2-mediated interspecies communications within rumen microbial communities. Microbiome 2022, 10, 167. [Google Scholar] [CrossRef]
- Vahidi, M.F.; Gharechahi, J.; Behmanesh, M.; Ding, X.Z.; Han, J.L.; Hosseini Salekdeh, G. Diversity of microbes colonizing forages of varying lignocellulose properties in the sheep rumen. Peer J. 2021, 9, e10463. [Google Scholar] [CrossRef]
- Christopherson, M.R.; Suen, G. Nature’s bioreactor: The rumen as a model for biofuel production. Biofuels 2013, 4, 511–521. [Google Scholar] [CrossRef]
- Craig, W.M.; Broderick, G.A.; Ricker, D.B. Quantitation of microorganisms associated with the particulate phase of ruminal ingesta. J. Nutr. 1987, 117, 56–62. [Google Scholar] [CrossRef]
- Wang, L.W.; Su, S.F.; Zhao, J.; He, X.L.; Fu, S.Y.; Wang, B.; Wang, Y.F.; Wang, D.Q.; Yun, N.N.; Chen, X.; et al. Effects of dietary oat supplementation on carcass traits, muscle metabolites, amino acid profiles, and its association with meat quality of Small-tail Han sheep. Food Chem. 2023, 411, 135456. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, M.; Xue, C.; Zhu, W.; Mao, S. Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J. Dairy Sci. 2016, 99, 9668–9681. [Google Scholar] [CrossRef]
- Gu, M.; Liu, H.; Jiang, X.; Qiu, S.; Li, K.; Lu, J.; Zhang, M.; Qiu, Y.; Wang, B.; Ma, Z.; et al. Analysis of Rumen Degradation Characteristics, Attached Microbial Community, and Cellulase Activity Changes of Garlic Skin and Artemisia argyi Stalk. Animals 2024, 14, 169. [Google Scholar] [CrossRef]
- Jin, W.; Wang, Y.; Li, Y.; Cheng, Y.; Zhu, W. Temporal changes of the bacterial community colonizing wheat straw in the cow rumen. Anaerobe 2018, 50, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, D.; Labandera, M. Determination of dry matter and crude protein contents of undried forage by near infrared reflectance spectroscopy. J. Sci. Food Agric. 2002, 82, 380–384. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Ørskov, E.R.; McDonald, I. The estimation of protein degradability in the rumen from incubation measurements weighted according to rate of passage. J. Agric. Sci. 1979, 92, 499–503. [Google Scholar] [CrossRef]
- Kennedy, P.M.; Murphy, M.R. The nutritional implications of differential passage of particles through the ruminant alimentary tract. Nutr. Res. Rev. 1988, 1, 189–208. [Google Scholar] [CrossRef][Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author Correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Rehemujiang, H.; Yusuf, H.A.; Ma, T.; Diao, Q.; Kong, L.; Kang, L.; Tu, Y. Evaluating Fermentation Quality, Aerobic Stability, and Rumen-Degradation (In Situ) Characteristics of Various Protein-Based Total Mixed Rations. Animals 2023, 13, 2730. [Google Scholar] [CrossRef]
- Zhang, C.T.; Tu, Y.; Ma, T.; Diao, Q.Y. Connection between Circadian Rhythm and Rumen Digestibility of Concentrate and Roughage in Sheep. Agriculture 2022, 12, 2152. [Google Scholar] [CrossRef]
- Katongole, C.B.; Lumu, R.; Lindberg, J.E. Comparative chemical composition and rumen degradation of common reed and elephant grass in urban/peri-urban dairying systems in Uganda. Agroecol. Sustain. Food Syst. 2021, 45, 892–906. [Google Scholar] [CrossRef]
- Li, S.; He, L.; Mo, F.; Zhang, W. In Situ Degradation Kinetics of 25 Feedstuffs and the Selection of Time Points in Mathematical Statistics. Animals 2023, 13, 947. [Google Scholar] [CrossRef] [PubMed]
- Turgut, L.; Yanar, M. In situ dry matter and crude protein degradation kinetics of some forages in Eastern Turkey. Small Rumin. Res. 2004, 52, 217–222. [Google Scholar] [CrossRef]
- Ma, Y.; Khan, M.Z.; Liu, Y.; Xiao, J.; Chen, X.; Ji, S.; Cao, Z.; Li, S. Analysis of Nutrient Composition, Rumen Degradation Characteristics, and Feeding Value of Chinese Rye Grass, Barley Grass, and Naked Oat Straw. Animals 2021, 11, 2486. [Google Scholar] [CrossRef]
- Byskov, M.V.; Nadeau, E.; Johansson, B.E.O.; Norgaard, P. Variations in automatically recorded rumination time as explained by variations in intake of dietary fractions and milk production, and between-cow variation. J. Dairy Sci. 2015, 98, 3926–3937. [Google Scholar] [CrossRef]
- Guan, H.; Xu, D.; Li, H.P.; Jia, Z.F.; Ma, X.; Liu, W.H.; Chen, Y.J.; Li, X.Y.; Huang, Y.L.; Zhou, Q.P.; et al. A study of nutritional quality and rumen degradation characteristics of 17 oat varieties in high cold regions. Acta Pratacult. Sin. 2024, 33, 185–198. [Google Scholar] [CrossRef]
- Gharechahi, J.; Salekdeh, G.H. A metagenomic analysis of the camel rumen’s microbiome identifies the major microbes responsible for lignocellulose degradation and fermentation. Biotechnol. Biofuels 2018, 11, 216. [Google Scholar] [CrossRef]
- Jiao, T.; Wu, J.P.; Casper, D.P.; Davis, D.I.; Brown, M.A.; Zhao, S.G.; Liang, J.Y.; Lei, Z.M.; Holloway, B. Feeding Sheep Cobalt and Oregano Essential Oil Alone or in Combination on Ruminal Nutrient Digestibility, Fermentation, and Fiber Digestion Combined With Scanning Electron Microscopy. Front. Vet. Sci. 2021, 8, 639432. [Google Scholar] [CrossRef]
- Huws, S.A.; Edwards, J.E.; Creevey, C.J.; Rees Stevens, P.; Lin, W.; Girdwood, S.E.; Pachebat, J.A.; Kingston-Smith, A.H. Temporal dynamics of the metabolically active rumen bacteria colonizing fresh perennial ryegrass. FEMS Microbiol. Ecol. 2016, 92, fiv137. [Google Scholar] [CrossRef]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms 2023, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Shinkai, T.; Ueki, T.; Kobayashi, Y. Detection and identification of rumen bacteria constituting a fibrolytic consortium dominated by Fibrobacter succinogenes. Anim. Sci. J. 2010, 81, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Gharechahi, J.; Vahidi, M.F.; Ding, X.Z.; Han, J.L.; Salekdeh, G.H. Temporal changes in microbial communities attached to forages with different lignocellulosic compositions in cattle rumen. FEMS Microbiol. Ecol. 2020, 96, fiaa069. [Google Scholar] [CrossRef]
- He, S.C.; Zhang, R.Y.; Wang, R.J.; Wu, D.W.; Dai, S.F.; Wang, Z.B.; Chen, T.; Mao, H.M.; Li, Q. Responses of nutrient utilization, rumen fermentation and microorganisms to different roughage of dairy buffaloes. BMC Microbiol. 2024, 24, 188. [Google Scholar] [CrossRef]
- Dong, C.; Wei, M.; Sun, F.; Bao, H.; Bao, M.; Ju, J.; Du, L. Effect of lactic acid bacteria preparations on calf fecal flora. Rev. Bras. De. Zootec.Braz. J. Anim. Sci. 2023, 52, e20210199. [Google Scholar] [CrossRef]
- Wang, R.J.; He, S.C.; Huang, D.; Wu, D.W.; He, S.Y.; Guo, T.Q.; Chen, C.G.; Mao, H.M. Effects of king grass and rice straw hay on apparent digestibility and ruminal microorganisms of buffalo. Anim. Biotechnol. 2023, 34, 1514–1523. [Google Scholar] [CrossRef]
- Saro, C.; Ranilla, M.J.; Tejido, M.L.; Carro, M.D. Influence of forage type in the diet of sheep on rumen microbiota and fermentation characteristics. Livest. Sci. 2014, 160, 52–59. [Google Scholar] [CrossRef]
- Saminathan, M.; Gan, H.M.; Abdullah, N.; Wong, C.M.V.L.; Ramiah, S.K.; Tan, H.Y.; Sieo, C.C.; Ho, Y.W. Changes in rumen protozoal community by condensed tannin fractions of different molecular weights from a Leucaena leucocephala hybrid invitro. J. Appl. Microbiol. 2017, 123, 41–53. [Google Scholar] [CrossRef]
- Woodruff, K.L.; Hummel, G.L.; Austin, K.J.; Lake, S.L.; Cunningham-Hollinger, H.C. Calf rumen microbiome from birth to weaning and shared microbial properties to the maternal rumen microbiome. J. Anim. Sci. 2022, 100, skac264. [Google Scholar] [CrossRef]
- Qiu, S.; Li, K.; He, X.; Gu, M.; Jiang, X.; Lu, J.; Ma, Z.; Liang, X.; Gan, Q. The Effects of Composite Alkali-Stored Spent Hypsizygus marmoreus Substrate on Carcass Quality, Rumen Fermentation, and Rumen Microbial Diversity in Goats. Animals 2024, 14, 166. [Google Scholar] [CrossRef]
- Moharrery, A.; Das, T.K. Correlation between microbial enzyme activities in the rumen fluid of sheep under different treatments. Reprod. Nutr. Dev. 2001, 41, 513–529. [Google Scholar] [CrossRef] [PubMed]
- Rubino, F.; Carberry, C.; Waters, S.M.; Kenny, D.; McCabe, M.S.; Creevey, C.J. Divergent functional isoforms drive niche specialisation for nutrient acquisition and use in rumen microbiome. ISME J. 2017, 11, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, M.; Patel, S.H.; Pandit, R.J.; Jakhesara, S.J.; Rank, D.N.; Joshi, C.G.; Kunjadiya, A.P. Rumen and fecal microbial profiles in cattle fed high lignin diets using metagenome analysis. Anaerobe 2022, 73, 102508. [Google Scholar] [CrossRef] [PubMed]
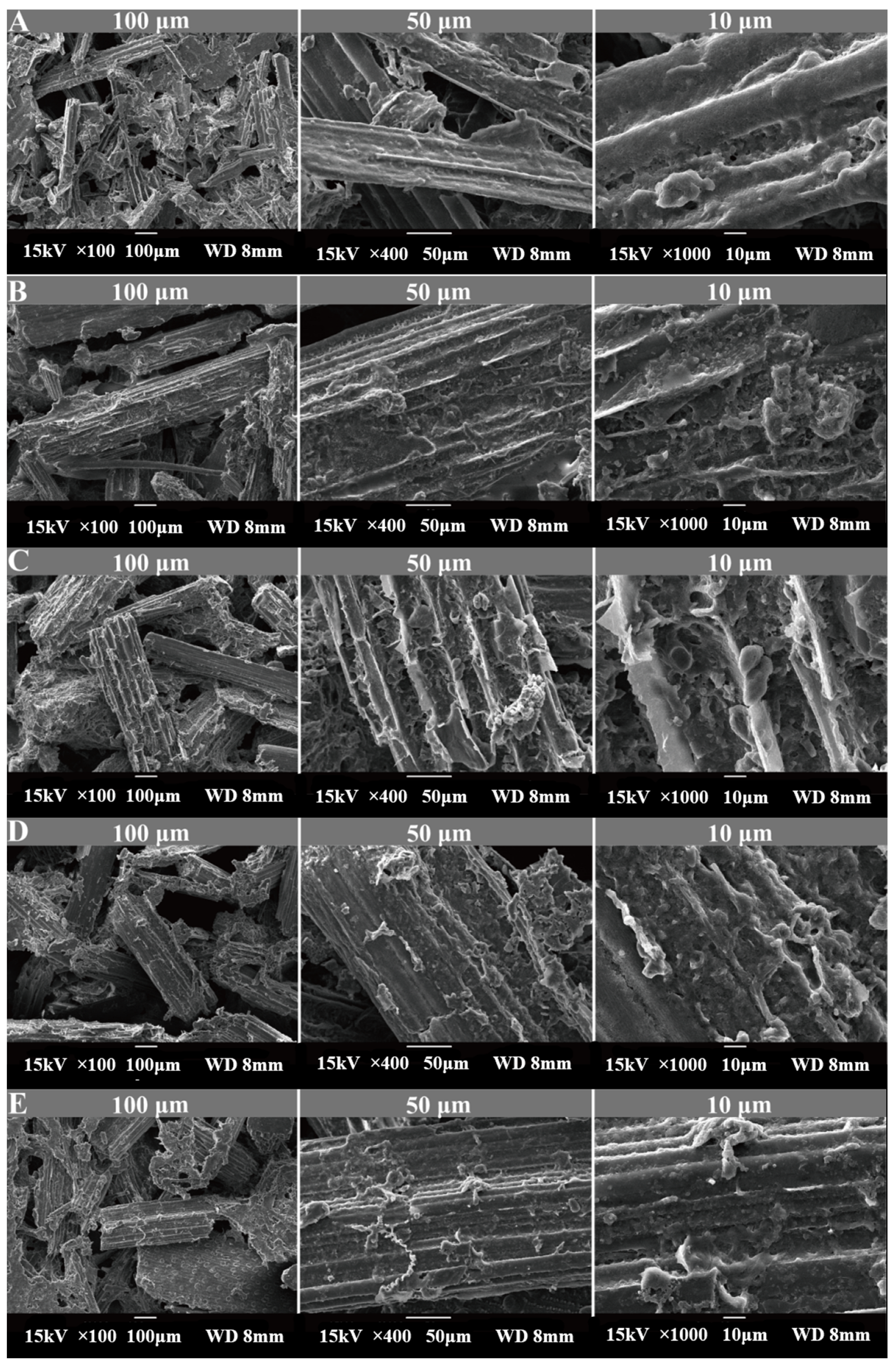






| Items | Content (%) | Nutrient Composition 2 | Content (%) |
|---|---|---|---|
| Leymus chinensis | 55 | Dry matter | 88.69 |
| Corn | 31 | Crude protein | 10.94 |
| Soybean meal | 12.2 | Neutral detergent fiber | 48.66 |
| NaCl | 0.5 | Acid detergent fiber | 24.83 |
| Limestone powder | 0.1 | Ether extract | 2.36 |
| Calcium bicarbonate | 0.2 | Calcium | 0.62 |
| Premix 1 | 1 | Phosphorus | 0.35 |
| Total | 100 | Net energy for lactation (MJ/kg DM) | 11.67 |
| Items | DM | CP | NDF | ADF |
|---|---|---|---|---|
| Ruminal degradation rate/% | ||||
| 4 h | 20.80 ± 4.74 e | 24.47 ± 4.91 e | 13.37 ± 4.04 e | 9.30 ± 2.00 e |
| 8 h | 27.60 ± 4.78 de | 30.39 ± 5.51 de | 19.52 ± 4.15 de | 13.04 ± 1.14 e |
| 12 h | 34.00 ± 3.93 d | 35.75 ± 4.82 d | 27.24 ± 4.33 d | 18.97 ± 2.53 d |
| 24 h | 45.82 ± 3.61 c | 48.64 ± 3.32 c | 36.93 ± 6.16 c | 28.13 ± 2.60 c |
| 36 h | 54.72 ± 3.57 b | 55.50 ± 4.82 bc | 44.88 ± 5.73 bc | 34.69 ± 2.05 b |
| 48 h | 61.38 ± 4.28 ab | 60.56 ± 3.89 ab | 50.74 ± 4.65 ab | 41.68 ± 2.75 a |
| 72 h | 65.76 ± 3.35 a | 64.16 ± 3.53 a | 55.24 ± 3.92 a | 45.32 ± 2.05 a |
| p-value | <0.0031 | <0.001 | <0.0019 | <0.0068 |
| Degradation parameters/% | ||||
| “a” | 13.12 ± 2.72 A | 16.31 ± 3.14 A | 6.27 ± 3.29 B | 3.33 ± 1.5 B |
| “b” | 57.11 ± 3.13 A | 50.7 ± 3.10 AB | 52.26 ± 3.55 B | 47.42 ± 2.23 B |
| “c” (%/h) | 0.037 ± 0.007 | 0.042 ± 0.009 | 0.038 ± 0.009 | 0.031 ± 0.005 |
| “ED” | 47.94 ± 7.23 A | 48.69 ± 7.65 A | 38.41 ± 8.44 AB | 30.24 ± 4.66 B |
| Items | Rumen Retention Time | p-Value | ||||
|---|---|---|---|---|---|---|
| 4 h | 12 h | 24 h | 36 h | 72 h | ||
| β-GC nmol/min/g | 126.72 ± 11.00 c | 172.25 ± 18.04 b | 159.00 ± 4.69 b | 233.50 ± 11.01 a | 216.16 ± 3.91 a | <0.001 |
| EG nmol/min/g | 186.61 ± 8.09 d | 215.96 ± 14.39 c | 288.54 ± 10.59 a | 284.18 ± 7.29 a | 257.63 ± 0.64 b | <0.001 |
| CBH nmol/min/g | 281.57 ± 23.71 b | 315.83 ± 22.81 b | 335.69 ± 15.33 a | 325.35 ± 29.36 ab | 315.33 ± 28.53 ab | <0.001 |
| NEX nmol/min/g | 256.79 ± 32.19 d | 451.33 ± 48.33 b | 305.04 ± 19.39 cd | 339.20 ± 20.71 c | 513.76 ± 1.94 a | <0.001 |
| Items | Rumen Retention Time | p-Value | ||||
|---|---|---|---|---|---|---|
| 4 h | 12 h | 24 h | 36 h | 72 h | ||
| Chao1 | 539.31 ± 20.59 | 484.61 ± 158.5 | 521.41 ± 44.71 | 515.51 ± 99.08 | 424.95 ± 163.54 | 0.649 |
| ACE | 540.17 ± 20.51 | 482.63 ± 157.85 | 519.74 ± 43.72 | 516.05 ± 99.35 | 423.96 ± 162.57 | 0.634 |
| Shannon | 7.07 ± 0.24 | 7.15 ± 0.65 | 7.26 ± 0.40 | 7.32 ± 0.41 | 7.12 ± 0.63 | 0.944 |
| Simpson | 0.98 ± 0.01 | 0.98 ± 0.01 | 0.98 ± 0.01 | 0.99 ± 0.01 | 0.98 ± 0.01 | 0.573 |
| Goods_coverage | 0.9996 ± 0.0001 | 0.9997 ± 0.0002 | 0.9996 ± 0.0001 | 0.9997 ± 0.0001 | 0.9998 ± 0.0001 | 0.415 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, L.; Qiu, Y.; Zhang, M.; Wei, S.; Qiu, S.; Ma, Z.; Gu, M.; Wang, B.; Zhang, X.; Gu, M.; et al. Characterizing the Fermentation of Oat Grass (Avena sativa L.) in the Rumen: Integrating Degradation Kinetics, Ultrastructural Examination with Scanning Electron Microscopy, Surface Enzymatic Activity, and Microbial Community Analysis. Animals 2025, 15, 2049. https://doi.org/10.3390/ani15142049
Zhong L, Qiu Y, Zhang M, Wei S, Qiu S, Ma Z, Gu M, Wang B, Zhang X, Gu M, et al. Characterizing the Fermentation of Oat Grass (Avena sativa L.) in the Rumen: Integrating Degradation Kinetics, Ultrastructural Examination with Scanning Electron Microscopy, Surface Enzymatic Activity, and Microbial Community Analysis. Animals. 2025; 15(14):2049. https://doi.org/10.3390/ani15142049
Chicago/Turabian StyleZhong, Liepeng, Yujun Qiu, Mingrui Zhang, Shanchuan Wei, Shuiling Qiu, Zhiyi Ma, Mingming Gu, Benzhi Wang, Xinyue Zhang, Mingke Gu, and et al. 2025. "Characterizing the Fermentation of Oat Grass (Avena sativa L.) in the Rumen: Integrating Degradation Kinetics, Ultrastructural Examination with Scanning Electron Microscopy, Surface Enzymatic Activity, and Microbial Community Analysis" Animals 15, no. 14: 2049. https://doi.org/10.3390/ani15142049
APA StyleZhong, L., Qiu, Y., Zhang, M., Wei, S., Qiu, S., Ma, Z., Gu, M., Wang, B., Zhang, X., Gu, M., Shen, N., & Gan, Q. (2025). Characterizing the Fermentation of Oat Grass (Avena sativa L.) in the Rumen: Integrating Degradation Kinetics, Ultrastructural Examination with Scanning Electron Microscopy, Surface Enzymatic Activity, and Microbial Community Analysis. Animals, 15(14), 2049. https://doi.org/10.3390/ani15142049





