Taxonomic Profile of Cultivable Microbiota from Adult Sheep Follicular Fluid and Its Effects on In Vitro Development of Prepubertal Lamb Oocytes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Transport of Ovaries
2.1.1. Cumulus–Oocyte Complex (COC) Retrieval
2.1.2. Follicular Fluid (FF) Collection, Microbiota Propagation, and Supernatant Preparation
2.2. 16S rRNA Gene Sequencing
2.2.1. Prokaryotic DNA Extraction
2.2.2. 16S rRNA Gene Library Preparation and Sequencing
2.2.3. Bioinformatic Analysis
2.3. (Targeted) Culturomics
2.4. In Vitro Embryo Culture (IVC) of Prepubertal Lamb Oocytes
2.4.1. In Vitro Oocyte Maturation (IVM)
2.4.2. In Vitro Fertilization (IVF)
2.5. Oocyte and Embryo Quality Assessment
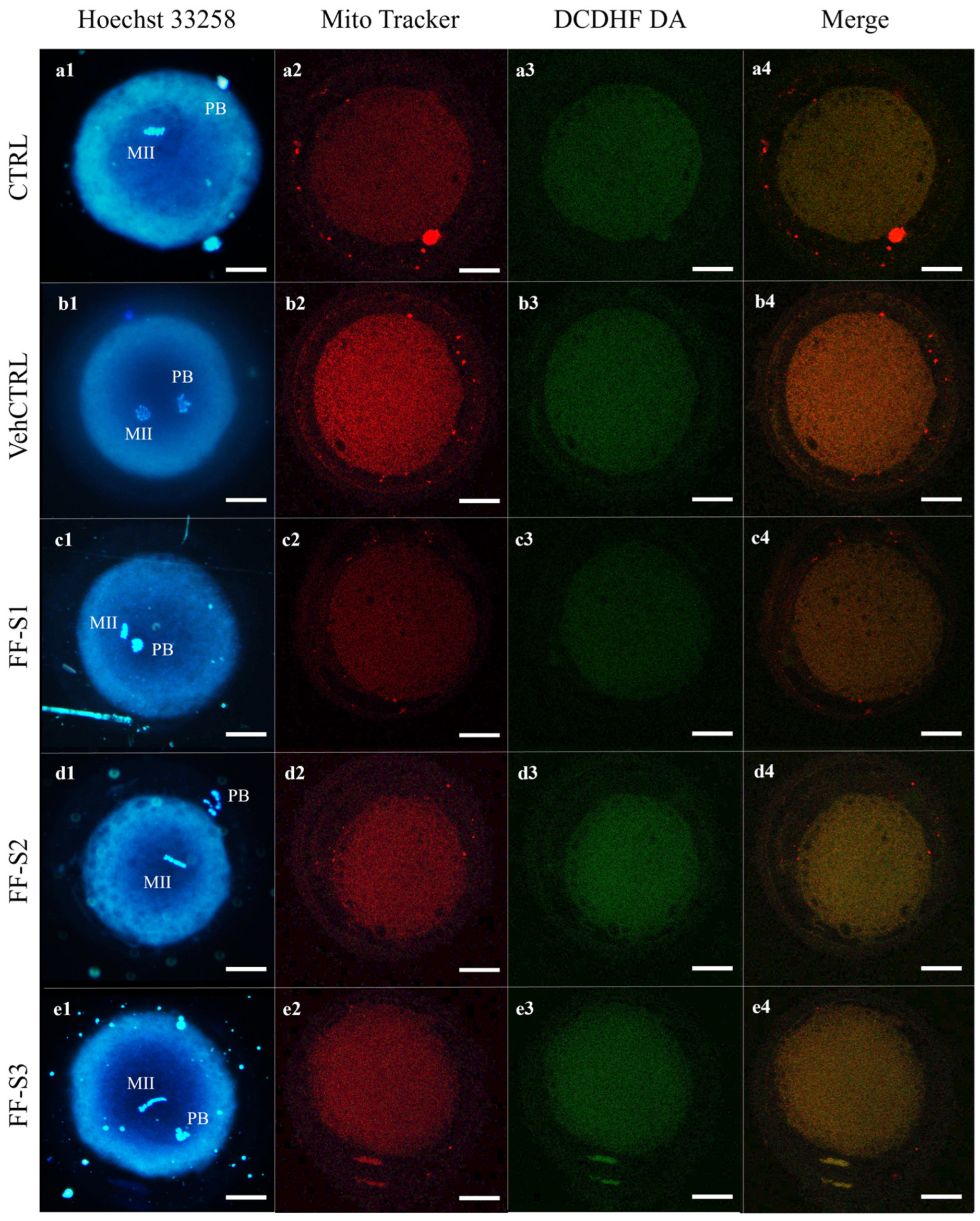
2.5.1. Oocyte Staining for Mitochondria and Reactive Oxygen Species (ROS)
2.5.2. Nuclear Chromatin Evaluation of Oocytes and Embryos
2.5.3. Assessment of Ooplasmic Mitochondrial Distribution Pattern
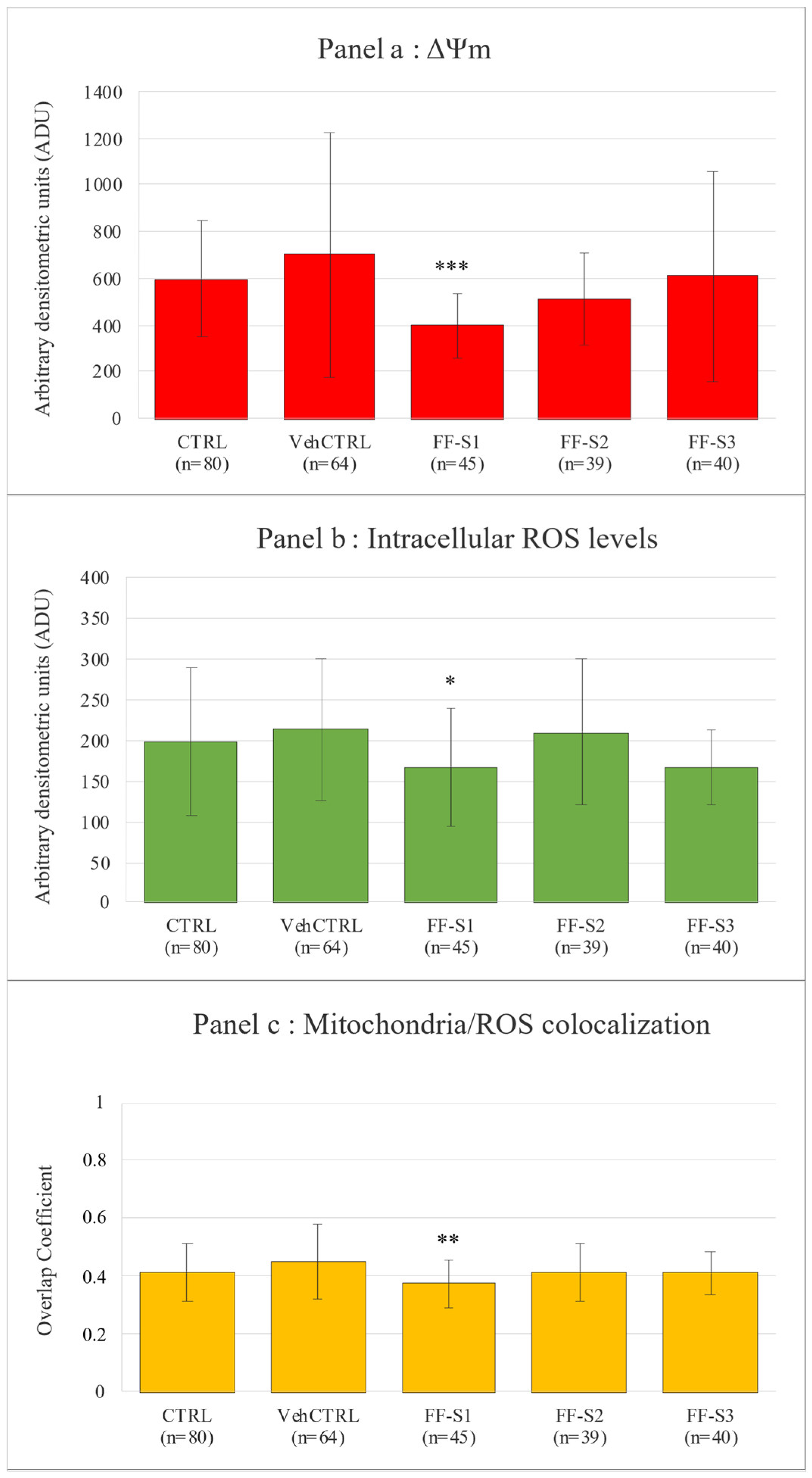
2.5.4. Quantification of Bioenergetic/Oxidative Variables and Mitochondria/ROS Colocalization Analysis
2.5.5. Embryo Development Evaluation
2.5.6. Statistical Analysis
3. Results
3.1. 16S rRNA Gene Sequencing (Prokaryotic Diversity Analysis)
3.2. (Targeted) Culturomics
3.3. Effects of In Vitro Oocyte Exposure to FF Microbiota Metabolites During IVM on Oocyte Maturation and Embryo Development
Evaluation of Oocyte Maturation and Bioenergetic/Oxidative Status
3.4. Evaluation of Embryo Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garcia-Garcia, R.M.; Arias-Álvarez, M.; Jordán-Rodríguez, D.; Rebollar, P.G.; Lorenzo, P.L.; Herranz, C.; Rodríguez, J.M. Female Reproduction and the Microbiota in Mammals: Where Are We? Theriogenology 2022, 194, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Campisciano, G.; Iebba, V.; Zito, G.; Luppi, S.; Martinelli, M.; Fischer, L.; De Seta, F.; Basile, G.; Ricci, G.; Comar, M. Lactobacillus Iners and Gasseri, Prevotella Bivia and HPV Belong to the Microbiological Signature Negatively Affecting Human Reproduction. Microorganisms 2020, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Cottell, E.; McMorrow, J.; Lennon, B.; Fawsy, M.; Cafferkey, M.; Harrison, R.F. Microbial Contamination in an In Vitro Fertilization-Embryo Transfer System. Fertil. Steril. 1996, 66, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Hamad, l.T.A.; Ahmed, A.T.; Sadeq, S.M.; Ismaeel, B. Microbial Colonization of Human Follicular Fluid and Adverse Outcome on In Vitro Fertilization Cases in Kamal Al-Samarrai’s Hospital for Fertility and In Vitro Fertilization Treatment in Baghdad, Iraq 2016. IOSR J. Dent. Med. Sci. (IOSR-JDMS) 2018, 17, 80–87. [Google Scholar]
- Kim, S.M.; Won, K.H.; Hong, Y.H.; Kim, S.K.; Lee, J.R.; Jee, B.C.; Suh, C.S. Microbiology of Human Follicular Fluid and the Vagina and Its Impact on In Vitro Fertilization Outcomes. Yonsei Med. J. 2022, 63, 941–947. [Google Scholar] [CrossRef]
- Noor, S.O. Bacterial Isolation of Human Follicular Fluid and Potential Impact on In Vitro Fertilization Outcomes. Med. Sci. 2020, 24, 2782–2791. [Google Scholar]
- Pelzer, E.S.; Allan, J.A.; Theodoropoulos, C.; Ross, T.; Beagley, K.W.; Knox, C.L. Hormone-Dependent Bacterial Growth, Persistence and Biofilm Formation—A Pilot Study Investigating Human Follicular Fluid Collected during IVF Cycles. PLoS ONE 2012, 7, e49965. [Google Scholar] [CrossRef]
- Pelzer, E.S.; Allan, J.A.; Waterhouse, M.A.; Ross, T.; Beagley, K.W.; Knox, C.L. Microorganisms Within Human Follicular Fluid: Effects on IVF. PLoS ONE 2013, 8, e59062. [Google Scholar] [CrossRef]
- Pelzer, E.S.; Harris, J.E.; Allan, J.A.; Waterhouse, M.A.; Ross, T.; Beagley, K.W.; Knox, C.L. TUNEL Analysis of DNA Fragmentation in Mouse Unfertilized Oocytes: The Effect of Microorganisms Within Human Follicular Fluid Collected During IVF Cycles. J. Reprod. Immunol. 2013, 99, 69–79. [Google Scholar] [CrossRef]
- Pelzer, E.S.; Allan, J.A.; Cunningham, K.; Mengersen, K.; Allan, J.M.; Launchbury, T.; Beagley, K.; Knox, C.L. Microbial Colonization of Follicular Fluid: Alterations in Cytokine Expression and Adverse Assisted Reproduction Technology Outcomes. Hum. Reprod. 2011, 26, 1799–1812. [Google Scholar] [CrossRef]
- Schenk, M.; Voroshilina, E.; Boldyreva, M.; Koranda, M.; Reinschissler, N.; Weiss, G. P–196 Bacterial Influence on Oocyte Quality—The Secret of a Successful Fertilization. Hum. Reprod. 2021, 36, deab130.195. [Google Scholar] [CrossRef]
- Usman, S.F.; Shuaibu, I.R.; Durojaiye, K.; Medugu, N.; Iregbu, K.C. The Presence of Microorganisms in Follicular Fluid and Its Effect on the Outcome of In Vitro Fertilization-Embryo Transfer (IVF-ET) Treatment Cycles. PLoS ONE 2021, 16, e0246644. [Google Scholar] [CrossRef] [PubMed]
- Richert, N.D.; Ryan, R.J. Specific Gonadotropin Binding to Pseudomonas Maltophilia. Proc. Natl. Acad. Sci. USA 1977, 74, 878–882. [Google Scholar] [CrossRef] [PubMed]
- Sluss, P.M.; Reichert, L.E. Secretion of an Inhibitor of Follicle-Stimulating Hormone Binding to Receptor by the Bacteria Serratia, Including a Strain Isolated from Porcine Follicular Fluid. Biol. Reprod. 1984, 31, 520–530. [Google Scholar] [CrossRef]
- Sluss, P.M.; Reichert, L.E. Presence of Bacteria in Porcine Follicular Fluid and Their Ability to Generate an Inhibitor of Follicle-Stimulating Hormone Binding to Receptor. Biol. Reprod. 1983, 29, 335–341. [Google Scholar] [CrossRef]
- Salary, A.; Kafi, M.; Derakhshandeh, A.; Moezzi, M.S. Detection of Bacteria in Bovine Ovarian Follicular Fluid. Lett. Appl. Microbiol. 2020, 70, 137–142. [Google Scholar] [CrossRef]
- Morton, K. Developmental Capabilities of Embryos Produced In Vitro from Prepubertal Lamb Oocytes. Reprod. Domest. Anim. 2008, 43, 137–143. [Google Scholar] [CrossRef]
- Paramio, M.; Izquierdo, D. Current Status of In Vitro Embryo Production in Sheep and Goats. Reprod. Domest. Anim. 2014, 49, 37–48. [Google Scholar] [CrossRef]
- Berean, D.; Blaga-Petrean, A.; Bogdan, I.; Bogdan, S.; Tamas-Krumpe, O.M.; Cimpean, R.; Pall, E.; Nap, M.E.; Bogdan, L.M. Effect of L-Arginine and Eugenol on Ram Semen Kinematic Parameters and Post Thawed Fertility Rate After Trans-Cervical Artificial Insemination. Indian. J. Anim. Res. 2022, 57, 552–557. [Google Scholar] [CrossRef]
- Falchi, L.; Ledda, S.; Zedda, M.T. Embryo Biotechnologies in Sheep: Achievements and New Improvements. Reprod. Domest. Anim. 2022, 57, 22–33. [Google Scholar] [CrossRef]
- Souza-Fabjan, J.M.G.; Oliveira, M.E.F.; Guimarães, M.P.P.; Brandão, F.Z.; Bartlewski, P.M.; Fonseca, J.F. Review: Non-Surgical Artificial Insemination and Embryo Recovery as Safe Tools for Genetic Preservation in Small Ruminants. Animal 2023, 17, 100787. [Google Scholar] [CrossRef] [PubMed]
- Temerario, L.; Monaco, D.; Mastrorocco, A.; Martino, N.A.; Cseh, S.; Lacalandra, G.M.; Ciani, E.; Dell’Aquila, M.E. New Strategies for Conservation of Gentile Di Puglia Sheep Breed, an Autochthonous Capital of Millennial Tradition in Southern Italy. Animals 2023, 13, 2371. [Google Scholar] [CrossRef] [PubMed]
- Torres-Rovira, L.; Succu, S.; Pasciu, V.; Manca, M.E.; Gonzalez-Bulnes, A.; Leoni, G.G.; Pennino, M.G.; Spezzigu, A.; Gallus, M.; Dattena, M.; et al. Postnatal Pituitary and Follicular Activation: A Revisited Hypothesis in a Sheep Model. Reproduction 2016, 151, 215–225. [Google Scholar] [CrossRef]
- Kochhar, H.; Wu, B.; Morris, L.; Buckrell, B.; Pollard, J.; Basrur, P.; King, W. Maturation Status, Protein Synthesis and Developmental Competence of Oocytes Derived from Lambs and Ewes. Reprod. Domest. Anim. 2002, 37, 19–25. [Google Scholar] [CrossRef]
- Ledda, S.; Bogliolo, L.; Leoni, G.; Naitana, S. Production and Lambing Rate of Blastocysts Derived from In Vitro Matured Oocytes after Gonadotropin Treatment of Prepubertal Ewes. J. Anim. Sci. 1999, 77, 2234. [Google Scholar] [CrossRef]
- Palmerini, M.G.; Nottola, S.A.; Leoni, G.G.; Succu, S.; Borshi, X.; Berlinguer, F.; Naitana, S.; Bekmukhambetov, Y.; Macchiarelli, G. In Vitromaturation Is Slowed in Prepubertal Lamb Oocytes: Ultrastructural Evidences. Reprod. Biol. Endocrinol. 2014, 12, 115. [Google Scholar] [CrossRef]
- Bebbere, D.; Ariu, F.; Bogliolo, L.; Masala, L.; Murrone, O.; Fattorini, M.; Falchi, L.; Ledda, S. Expression of Maternally Derived KHDC3, NLRP5, OOEP and TLE6 Is Associated with Oocyte Developmental Competence in the Ovine Species. BMC Dev. Biol. 2014, 14, 40. [Google Scholar] [CrossRef]
- Leoni, G.G.; Bebbere, D.; Succu, S.; Berlinguer, F.; Mossa, F.; Galioto, M.; Bogliolo, L.; Ledda, S.; Naitana, S. Relations between Relative MRNA Abundance and Developmental Competence of Ovine Oocytes. Mol. Reprod. Dev. 2007, 74, 249–257. [Google Scholar] [CrossRef]
- Leoni, G.G.; Palmerini, M.G.; Satta, V.; Succu, S.; Pasciu, V.; Zinellu, A.; Carru, C.; Macchiarelli, G.; Nottola, S.A.; Naitana, S.; et al. Differences in the Kinetic of the First Meiotic Division and in Active Mitochondrial Distribution between Prepubertal and Adult Oocytes Mirror Differences in Their Developmental Competence in a Sheep Model. PLoS ONE 2015, 10, e0124911. [Google Scholar] [CrossRef]
- Masala, L.; Ariu, F.; Bogliolo, L.; Bellu, E.; Ledda, S.; Bebbere, D. Delay in Maternal Transcript Degradation in Ovine Embryos Derived from Low Competence Oocytes. Mol. Reprod. Dev. 2018, 85, 427–439. [Google Scholar] [CrossRef]
- Ptak, G. Developmental and Functional Evidence of Nuclear Immaturity in Prepubertal Oocytes. Hum. Reprod. 2006, 21, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Ren, P.; Liu, K.; Qiu, C.; Fan, L.; Li, J.; Hou, J. Transcriptomic Comparison of Ovarian Granulosa Cells between Adult Sheep and Prepubertal Lambs. BMC Genom. 2022, 23, 151. [Google Scholar] [CrossRef] [PubMed]
- Khatir, H.; Lonergan, P.; Carolan, C.; Mermillod, P. Prepubertal Bovine Oocyte: A Negative Model for Studying Oocyte Developmental Competence. Mol. Reprod. Dev. 1996, 45, 231–239. [Google Scholar] [CrossRef]
- Tutt, D.A.R.; Guven-Ates, G.; Kwong, W.Y.; Simmons, R.; Sang, F.; Silvestri, G.; Canedo-Ribeiro, C.; Handyside, A.H.; Labrecque, R.; Sirard, M.-A.; et al. Developmental, Cytogenetic and Epigenetic Consequences of Removing Complex Proteins and Adding Melatonin during In Vitro Maturation of Bovine Oocytes. Front. Endocrinol. 2023, 14, 1280847. [Google Scholar] [CrossRef]
- Al-Mutary, M.; Al-Ghadi, M.; Al-Himaidi, A.; Iwamoto, D.; Al-Anazi, Y.; Ammari, A.; Ahmad, J.; Al-Khedhairy, A. Using RT-PCR and Glutathione Level to Study the Effect of Follicular Fluid on In Vitro Maturation and Gene Expression of Sheep Oocytes. Saudi J. Biol. Sci. 2019, 26, 1216–1222. [Google Scholar] [CrossRef]
- Azari-Dolatabad, N.; Raes, A.; Pavani, K.C.; Asaadi, A.; Angel-Velez, D.; Van Damme, P.; Leroy, J.L.M.R.; Van Soom, A.; Pascottini, O.B. Follicular Fluid during Individual Oocyte Maturation Enhances Cumulus Expansion and Improves Embryo Development and Quality in a Dose-Specific Manner. Theriogenology 2021, 166, 38–45. [Google Scholar] [CrossRef]
- Chung, S.O.; Choi, Y.H.; Kim, M.K.; Cho, W.K. The Effects of Follicular Fluid on In Vitro Maturation of Bovine Follicular Oocytes. Yonsei Med. J. 1974, 15, 147–155. [Google Scholar] [CrossRef]
- Kim, K.; Mitsumizo, N.; Fujita, K.; Utsumi, K. The Effects of Follicular Fluid on In Vitro Maturation, Oocyte Fertilization and the Development of Bovine Embryos. Theriogenology 1996, 45, 787–799. [Google Scholar] [CrossRef]
- Sun, F.J.; Holm, P.; Irvine, B.; Seamark, R.F. Effect of Sheep and Human Follicular Fluid on the Maturation of Sheep Oocytes In Vitro. Theriogenology 1994, 41, 981–988. [Google Scholar] [CrossRef]
- Tian, H.; Liu, K.; Zhang, Y.; Qi, Q.; Wang, C.; Guan, H.; Yan, F.; Hou, J. Adult Follicular Fluid Supplementation During In Vitro Maturation Improves the Developmental Competence of Prepubertal Lamb Oocytes. Theriogenology 2019, 130, 157–162. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, L.; Xu, A.; NIMA, D.; Dong, Z. Effects of Stem Cell Factor in Follicular Fluid and Granulosa Cells on Oocyte Maturity and Clinical Pregnancy. Medicine 2023, 102, e36749. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhao, C.; Chen, B.; Yu, X.; Zhou, Y.; Ni, D.; Zhang, X.; Zhang, J.; Ling, X.; Zhang, Z.; et al. Follicular Fluid C3a-Peptide Promotes Oocyte Maturation through F-Actin Aggregation. BMC Biol. 2023, 21, 285. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, M.A.; Hunter, A.G. Inhibitory Effect of Bovine Follicular Fluid on In Vitro Maturation of Bovine Oocytes. J. Dairy Sci. 1993, 76, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Dostál, J.; Pavlok, A. Isolation and Characterization of Maturation Inhibiting Compound in Bovine Follicular Fluid. Reprod. Nutr. Dev. 1996, 36, 681–690. [Google Scholar]
- Pawlak, P.; Warzych, E.; Cieslak, A.; Malyszka, N.; Maciejewska, E.; Madeja, Z.E.; Lechniak, D. The Consequences of Porcine IVM Medium Supplementation with Follicular Fluid Become Reflected in Embryo Quality, Yield and Gene Expression Patterns. Sci. Rep. 2018, 8, 15306. [Google Scholar] [CrossRef]
- Huang, X.; Hong, L.; Wu, Y.; Chen, M.; Kong, P.; Ruan, J.; Teng, X.; Wei, Z. Raman Spectrum of Follicular Fluid: A Potential Biomarker for Oocyte Developmental Competence in Polycystic Ovary Syndrome. Front. Cell Dev. Biol. 2021, 9, 777224. [Google Scholar] [CrossRef]
- Huang, Y.; Tu, M.; Qian, Y.; Ma, J.; Chen, L.; Liu, Y.; Wu, Y.; Chen, K.; Liu, J.; Ying, Y.; et al. Age-Dependent Metabolomic Profile of the Follicular Fluids from Women Undergoing Assisted Reproductive Technology Treatment. Front. Endocrinol. 2022, 13, 818888. [Google Scholar] [CrossRef]
- Ji, J.; Zhu, X.; Zhang, Y.; Shui, L.; Bai, S.; Huang, L.; Wang, H.; Fan, S.; Zhang, Z.; Luo, L.; et al. A Proteomic Analysis of Human Follicular Fluid: Proteomic Profile Associated with Embryo Quality. Reprod. Sci. 2024, 31, 199–211. [Google Scholar] [CrossRef]
- Lazzarino, G.; Pallisco, R.; Bilotta, G.; Listorti, I.; Mangione, R.; Saab, M.; Caruso, G.; Amorini, A.; Brundo, M.; Lazzarino, G.; et al. Altered Follicular Fluid Metabolic Pattern Correlates with Female Infertility and Outcome Measures of In Vitro Fertilization. Int. J. Mol. Sci. 2021, 22, 8735. [Google Scholar] [CrossRef]
- Ledee, N.; Lombroso, R.; Lombardelli, L.; Selva, J.; Dubanchet, S.; Chaouat, G.; Frankenne, F.; Foidart, J.M.; Maggi, E.; Romagnani, S.; et al. Cytokines and Chemokines in Follicular Fluids and Potential of the Corresponding Embryo: The Role of Granulocyte Colony-Stimulating Factor. Hum. Reprod. 2008, 23, 2001–2009. [Google Scholar] [CrossRef]
- Revelli, A.; Piane, L.D.; Casano, S.; Molinari, E.; Massobrio, M.; Rinaudo, P. Follicular Fluid Content and Oocyte Quality: From Single Biochemical Markers to Metabolomics. Reprod. Biol. Endocrinol. 2009, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, D.; Roura, M.; Pérez-Trujillo, M.; Soto-Heras, S.; Paramio, M.-T. Fatty Acids and Metabolomic Composition of Follicular Fluid Collected from Environments Associated with Good and Poor Oocyte Competence in Goats. Int. J. Mol. Sci. 2022, 23, 4141. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhang, L.; Qi, Q.; Li, J.; Yan, F.; Hou, J. Growth Hormone Treatment Improves the Development of Follicles and Oocytes in Prepubertal Lambs. J. Ovarian Res. 2023, 16, 132. [Google Scholar] [CrossRef]
- Wu, Y.; Lin, J.; Han, B.; Wang, L.; Chen, Y.; Liu, M.; Huang, J. Proteomic Profiling of Follicle Fluids after Superstimulation in One-month-old Lambs. Reprod. Domest. Anim. 2018, 53, 186–194. [Google Scholar] [CrossRef]
- Mastrorocco, A.; Martino, N.A.; Marzano, G.; Lacalandra, G.M.; Ciani, E.; Roelen, B.A.J.; Dell’Aquila, M.E.; Minervini, F. The Mycotoxin Beauvericin Induces Oocyte Mitochondrial Dysfunction and Affects Embryo Development in the Juvenile Sheep. Mol. Reprod. Dev. 2019, 86, 1430–1443. [Google Scholar] [CrossRef]
- Marzano, M.; Fosso, B.; Manzari, C.; Grieco, F.; Intranuovo, M.; Cozzi, G.; Mulè, G.; Scioscia, G.; Valiente, G.; Tullo, A.; et al. Complexity and Dynamics of the Winemaking Bacterial Communities in Berries, Musts, and Wines from Apulian Grape Cultivars through Time and Space. PLoS ONE 2016, 11, e0157383. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author Correction: Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. SILVA: A Comprehensive Online Resource for Quality Checked and Aligned Ribosomal RNA Sequence Data Compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Parallelization of the MAFFT Multiple Sequence Alignment Program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple Statistical Identification and Removal of Contaminant Sequences in Marker-Gene and Metagenomics Data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.B.; Simpson, G.; Solymos, P.; Stevenes, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0-2. 2012. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 21 November 2022).
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Angeletti, S. Matrix Assisted Laser Desorption Time of Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Microbiology. J. Microbiol. Methods 2017, 138, 20–29. [Google Scholar] [CrossRef]
- Santos, M.V.D.O.; Queiroz Neta, L.B.d.; Borges, A.A.; Pereira, A.F. Influence of Commercially Available Follicle Stimulating Hormone on the In Vitro Maturation of Bovine Oocytes. Semin. Cienc. Agrar. 2017, 38, 1393. [Google Scholar] [CrossRef]
- Martino, N.A.; Marzano, G.; Mangiacotti, M.; Miedico, O.; Sardanelli, A.M.; Gnoni, A.; Lacalandra, G.M.; Chiaravalle, A.E.; Ciani, E.; Bogliolo, L.; et al. Exposure to Cadmium During In Vitro Maturation at Environmental Nanomolar Levels Impairs Oocyte Fertilization Through Oxidative Damage: A Large Animal Model Study. Reprod. Toxicol. 2017, 69, 132–145. [Google Scholar] [CrossRef]
- Martino, N.A.; Ariu, F.; Bebbere, D.; Uranio, M.F.; Chirico, A.; Marzano, G.; Sardanelli, A.M.; Cardinali, A.; Minervini, F.; Bogliolo, L.; et al. Supplementation with Nanomolar Concentrations of Verbascoside During In Vitro Maturation Improves Embryo Development by Protecting the Oocyte against Oxidative Stress: A Large Animal Model Study. Reprod. Toxicol. 2016, 65, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Dell’Aquila, M.E.; Asif, S.; Temerario, L.; Mastrorocco, A.; Marzano, G.; Martino, N.A.; Lacalandra, G.M.; Roelen, B.A.; Carluccio, A.; Robbe, D.; et al. Ochratoxin A Affects Oocyte Maturation and Subsequent Embryo Developmental Dynamics in the Juvenile Sheep Model. Mycotoxin Res. 2021, 37, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Somoskoi, B.; Martino, N.A.; Cardone, R.A.; Lacalandra, G.M.; Dell’Aquila, M.E.; Cseh, S. Different Chromatin and Energy/Redox Responses of Mouse Morulae and Blastocysts to Slow Freezing and Vitrification. Reprod. Biol. Endocrinol. 2015, 13, 22. [Google Scholar] [CrossRef] [PubMed]
- Ariu, F.; Bogliolo, L.; Leoni, G.; Falchi, L.; Bebbere, D.; Nieddu, S.M.; Zedda, M.T.; Pau, S.; Ledda, S. Effect of Caffeine Treatment Before Vitrification on MPF and MAPK Activity and Spontaneous Parthenogenetic Activation of In Vitro Matured Ovine Oocytes. Cryo Lett. 2014, 35, 530–536. [Google Scholar]
- Manders, E.M.M.; Verbeek, F.J.; Aten, J.A. Measurement of Co-localization of Objects in Dual-colour Confocal Images. J. Microsc. 1993, 169, 375–382. [Google Scholar] [CrossRef]
- Han, G.; Vaishnava, S. Microbial Underdogs: Exploring the Significance of Low-Abundance Commensals in Host-Microbe Interactions. Exp. Mol. Med. 2023, 55, 2498–2507. [Google Scholar] [CrossRef]
- de Cena, J.A.; Zhang, J.; Deng, D.; Damé-Teixeira, N.; Do, T. Low-Abundant Microorganisms: The Human Microbiome’s Dark Matter, a Scoping Review. Front. Cell. Infect. Microbiol. 2021, 11, 689197. [Google Scholar] [CrossRef]
- Li, W.I.; Brackett, B.G.; Halper, J. Culture Supernatant of Lactobacillus acidophilus Stimulates Proliferation of Embryonic Cells. Exp. Biol. Med. 2005, 230, 494–500. [Google Scholar] [CrossRef]
- Ghaffarilaleh, V.; Fouladi-Nashta, A.; Paramio, M.-T. Effect of α-Linolenic Acid on Oocyte Maturation and Embryo Development of Prepubertal Sheep Oocytes. Theriogenology 2014, 82, 686–696. [Google Scholar] [CrossRef]
- Reader, K.L.; Cox, N.R.; Stanton, J.-A.L.; Juengel, J.L. Effects of Acetyl-L-Carnitine on Lamb Oocyte Blastocyst Rate, Ultrastructure, and Mitochondrial DNA Copy Number. Theriogenology 2015, 83, 1484–1492. [Google Scholar] [CrossRef]
- Tian, H.; Qi, Q.; Yan, F.; Wang, C.; Hou, F.; Ren, W.; Zhang, L.; Hou, J. Enhancing the Developmental Competence of Prepubertal Lamb Oocytes by Supplementing the In Vitro Maturation Medium with Sericin and the Fibroblast Growth Factor 2—Leukemia Inhibitory Factor—Insulin-like Growth Factor 1 Combination. Theriogenology 2021, 159, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiang, X.; Wu, Y.; Lin, J.; Zhang, L.; Yang, N.; Huang, J. Effect of Milrinone on the Developmental Competence of Growing Lamb Oocytes Identified with Brilliant Cresyl Blue. Theriogenology 2016, 86, 2020–2027. [Google Scholar] [CrossRef]
- da Silveira, J.C.; Andrade, G.M.; Del Collado, M.; Sampaio, R.V.; Sangalli, J.R.; Silva, L.A.; Pinaffi, F.V.L.; Jardim, I.B.; Cesar, M.C.; Nogueira, M.F.G.; et al. Supplementation with Small-Extracellular Vesicles from Ovarian Follicular Fluid During In Vitro Production Modulates Bovine Embryo Development. PLoS ONE 2017, 12, e0179451. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.; Brown, L.; Maryam, M.; Vij, R.; Smith, D.F.Q.; Burnet, M.C.; Kyle, J.E.; Heyman, H.M.; Ramirez, J.; Prados-Rosales, R.; et al. Listeria Monocytogenes Virulence Factors, Including Listeriolysin O, Are Secreted in Biologically Active Extracellular Vesicles. J. Biol. Chem. 2019, 294, 1202–1217. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Jahromi, L.P.; Fuhrmann, G. Bacterial Extracellular Vesicles: Understanding Biology Promotes Applications as Nanopharmaceuticals. Adv. Drug Deliv. Rev. 2021, 173, 125–140. [Google Scholar] [CrossRef]
- Bromfield, J.J.; Sheldon, I.M. Lipopolysaccharide Initiates Inflammation in Bovine Granulosa Cells via the TLR4 Pathway and Perturbs Oocyte Meiotic Progression In Vitro. Endocrinology 2011, 152, 5029–5040. [Google Scholar] [CrossRef]
- Zhao, S.-J.; Pang, Y.-W.; Zhao, X.-M.; Du, W.-H.; Hao, H.-S.; Zhu, H.-B. Effects of Lipopolysaccharide on Maturation of Bovine Oocyte In Vitro and Its Possible Mechanisms. Oncotarget 2017, 8, 4656–4667. [Google Scholar] [CrossRef]
- Magata, F.; Shimizu, T. Effect of Lipopolysaccharide on Developmental Competence of Oocytes. Reprod. Toxicol. 2017, 71, 1–7. [Google Scholar] [CrossRef]
- Yan, K.; Cui, K.; Nie, J.; Zhang, H.; Sui, L.; Zhang, H.; Yang, X.; Xu, C.-L.; Liang, X. Mogroside V Protects Porcine Oocytes from Lipopolysaccharide-Induced Meiotic Defects. Front. Cell Dev. Biol. 2021, 9, 639691. [Google Scholar] [CrossRef]
- Rasekhi, M.; Mohammadi-Sangcheshmeh, A.; Daliri, M.; Bakhtiarizadeh, M.; Shariati, V.; Rahimi, M.; Hajarizadeh, A.; Nazari, S.A.; Ross, P.J.; Tvrdá, E. Transcriptional Profile of Ovine Oocytes Matured under Lipopolysaccharide Treatment In Vitro. Theriogenology 2020, 157, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Yoo, D.K.; Lee, S.-H. Effect of Lipopolysaccharide (LPS) Exposure on the Reproductive Organs of Immature Female Rats. Dev. Reprod. 2016, 20, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, L.; Charitos, I.A.; Colella, M.; Palmirotta, R.; Jirillo, E. Blood Microbiota and Its Products: Mechanisms of Interference with Host Cells and Clinical Outcomes. Hematol. Rep. 2024, 16, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, K.; Ando, C.; Tashiro, K.; Kuhara, S.; Okamura, S.; Nakano, S.; Takagi, Y.; Miki, T.; Nakashima, Y.; Hirakawa, H. Polymerase Chain Reaction Detection of Bacterial 16S RRNA Gene in Human Blood. Microbiol. Immunol. 2008, 52, 375–382. [Google Scholar] [CrossRef]
- Gordon, I. Maturing the Oocyte. In Laboratory Production of Cattle Embryos; CABI Publishing: Wallingford, UK, 2003; pp. 112–157. [Google Scholar]
- Chandra, V. I In Vitro i Strategies to Enhance Oocyte Developmental Competence. Front. Biosci. 2020, 12, 543. [Google Scholar] [CrossRef]
- Watson, A.J. Oocyte Cytoplasmic Maturation: A Key Mediator of Oocyte and Embryo Developmental Competence. J. Anim. Sci. 2007, 85, E1–E3. [Google Scholar] [CrossRef]
- Asaf, S.; Leitner, G.; Furman, O.; Lavon, Y.; Kalo, D.; Wolfenson, D.; Roth, Z. Effects of Escherichia coli- and Staphylococcus aureus-Induced Mastitis in Lactating Cows on Oocyte Developmental Competence. Reproduction 2014, 147, 33–43. [Google Scholar] [CrossRef]
- Alvarado Rincón, J.A.; Gindri, P.C.; Mion, B.; de Ávila, F.G.; Barbosa, A.A.; Maffi, A.S.; Pradieé, J.; Mondadori, R.G.; Corrêa, M.N.; Ligia Margareth Cantarelli, P.; et al. Early Embryonic Development of Bovine Oocytes Challenged with LPS In Vitro or In Vivo. Reproduction 2019, 158, 453–463. [Google Scholar] [CrossRef]
| Condition # | Medium | Atmosphere | Temperature |
|---|---|---|---|
| 1 | Blood agar | Aerobically | 30 °C |
| 2 | Blood agar | Aerobically | 37 °C |
| 3 | Blood agar | Anaerobically | 30 °C |
| 4 | Blood agar | Candle jar (O2 reduced/ CO2 enhanced atmosphere) | 37 °C |
| 5 | MacConkey agar | Aerobically | 30 °C |
| 6 | MacConkey agar | Aerobically | 37 °C |
| FF1 | FF2 | FF3 | |
|---|---|---|---|
| Phylum: Proteobacteria | 25.88 | 99.90 | 99.89 |
| Genus: | |||
| Escherichia-Shigella | 25.875 | 99.507 | 99.858 |
| Burkholderia-Caballeronia-Paraburkholderia | 0.000 | 0.371 | 0.016 |
| Oligella | 0.000 | 0.013 | 0.003 |
| Pseudomonas | 0.003 | 0.004 | 0.001 |
| Achromobacter | 0.000 | 0.002 | 0.003 |
| Uncultured | 0.000 | 0.001 | 0.003 |
| Enhydrobacter | 0.000 | 0.000 | 0.000 |
| Brevundimonas | 0.001 | 0.000 | 0.000 |
| Uncultured | 0.000 | 0.001 | 0.000 |
| Methylobacterium-Methylorubrum | 0.000 | 0.000 | 0.001 |
| Pseudoxanthomonas | 0.000 | 0.000 | 0.000 |
| FF1 | FF2 | FF3 | |
| Phylum: Firmicutes | 74.114 | 0.069 | 0.081 |
| Genus | |||
| Streptococcus | 74.080 | 0.033 | 0.023 |
| Enterococcus | 0.019 | 0.009 | 0.013 |
| Staphylococcus | 0.015 | 0.023 | 0.035 |
| Lactobacillus | 0.000 | 0.001 | 0.002 |
| Faecalibacterium | 0.000 | 0.0004 | 0.003 |
| Blautia | 0.000 | 0.000 | 0.002 |
| Fusicatenibacter | 0.000 | 0.001 | 0.000 |
| Roseburia | 0.000 | 0.000 | 0.002 |
| Aerococcus | 0.000 | 0.001 | 0.000 |
| Christensenellaceae_R-7_group | 0.000 | 0.000 | 0.000 |
| Coprococcus | 0.000 | 0.000 | 0.0005 |
| Leuconostoc | 0.000 | 0.000 | 0.001 |
| FF1 | FF2 | FF3 | |
| Phylum | |||
| Bacteroidota | 0.000 | 0.004 | 0.011 |
| Genus | |||
| Bacteroides | 0.000 | 0.002 | 0.003 |
| Cloacibacterium | 0.000 | 0.000 | 0.006 |
| Prevotella | 0.000 | 0.000 | 0.001 |
| Empedobacter | 0.000 | 0.002 | 0.000 |
| Alistipes | 0.000 | 0.0004 | 0.000 |
| Parabacteroides | 0.000 | 0.000 | 0.000 |
| Chryseobacterium | 0.000 | 0.000 | 0.001 |
| FF1 | FF2 | FF3 | |
| Phylum | |||
| Actinobacteriota | 0.006 | 0.020 | 0.016 |
| Genus | |||
| Corynebacterium | 0.000 | 0.003 | 0.004 |
| Dietzia | 0.000 | 0.002 | 0.000 |
| Cutibacterium | 0.004 | 0.011 | 0.008 |
| Kocuria | 0.002 | 0.001 | 0.000 |
| Georgenia | 0.000 | 0.000 | 0.004 |
| Lawsonella | 0.000 | 0.001 | 0.000 |
| Nocardioides | 0.000 | 0.001 | 0.000 |
| FFs | Bacterial Species (Strain) | MALDI-TOF MS ID Log Score |
|---|---|---|
| FF-1 | Streptococcus infantarius subsp. Infantarius | 2.19 |
| Escherichia coli (lactose ‘+’, non-hemolytic) | 2.12 | |
| Escherichia coli (lactose ‘−’, non-hemolytic) | 2.48 | |
| FF-2 | Escherichia coli (lactose ‘+’, non-hemolytic) | 2.26 |
| Burkholderia cepacia | 2.19 | |
| FF-3 | Escherichia coli (lactose ‘+’, α-hemolytic) | 2.44 |
| Well Supplement | N° of Evaluated Oocytes (Replicates) | Nuclear Chromatin Configurations Number (%) | |||||
|---|---|---|---|---|---|---|---|
| GV | GVBD | MI-TI | MII | Abnormal | Activated | ||
| CTRL | 120 (5) | 12 (10) | 10 (8) | 7 (6) | 80 (67) | 7 (6) | 4 (3) |
| vehCTRL | 93 (4) | 8 (9) | 7 (8) a | 6 (6) | 64 (69) | 6 (6) a | 2 (2) |
| FF-S1 | 70 (3) | 8 (11) | 4 (6) | 4 (6) | 45 (64) | 9 (13) | 0 (0) |
| FF-S2 | 72 (3) | 12 (17) | 13 (18) b | 4 (6) | 39 (54) | 3 (4) | 1 (1) |
| FF-S3 | 72 (3) | 5 (7) | 8 (11) | 7 (10) | 40 (55) | 12 (17) b | 0 (0) |
| Well Supplement | N° of Analyzed MII Oocytes | Mitochondria Distribution Pattern Number (%) | ||
|---|---|---|---|---|
| Perinuclear and Subplasmalemmal | Small Aggregates | Abnormal | ||
| CTRL | 80 | 42 (53) | 37 (46) | 1 (1) |
| vehCTRL | 64 | 26 (41) | 38 (59) | 0 (0) |
| FF-S1 | 45 | 20 (44) | 24 (53) | 1 (2) |
| FF-S2 | 39 | 16 (41) | 23 (59) | 0 (0) |
| FF-S3 | 40 | 19 (48) | 20 (50) | 1 (2) |
|
Well Supple-ment (Replicates) | N° of Insemi-nated Oocytes | Embryo Development to N (%)/Inseminated | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 2 | Day 7 | ||||||||||
|
2–4 Cell | 4–8 Cell | 8–16 Cell |
Total Cleaved/ Insemi-nated |
2–4 Cell |
4–8 Cell | 8–16 Cell | M + B |
Total Cleaved/ Insemi-nated | M + B/Cleaved | ||
| CTRL (7) | 168 | 19 (11) | 22 (13) | 12 (7) | 53/168 (32) x | 19 (11) | 17 (10) | 20 (12) | 4 (2) | 60/168 (36) z | 4/60 (7) |
| vehCTRL (7) | 161 | 14 (9) a | 15 (9) | 5 (3) | 34/161 (21) y | 9 (6) a | 11 (7) a | 12 (7) | 4 (2) | 36/161 (22) a w | 4/36 (11) |
| FF-S1 (4) | 89 | 8 (9) | 13 (15) | 1 (1) | 22/89 (25) | 9 (10) | 12 (13) | 5 (6) | 3 (3) | 29/89 (33) | 3/29 (10) |
| FF-S2 (6) | 133 | 20 (15) | 17 (13) | 1 (1) | 38/133 (29) | 12 (9) | 22 (17) c | 9 (7) | 5 (4) | 48/133 (36) c | 5/48 (10) |
| FF-S3 (5) | 91 | 21 (23) c | 7 (8) | 0 (0) | 28/91 (31) | 13 (14) b | 15 (16) b | 4 (4) | 2 (2) | 34/91 (37) b | 2/34 (6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mrenoshki, S.; Temerario, L.; Mastrorocco, A.; Visci, G.; Notario, E.; Marzano, M.; Martino, N.A.; Mrenoshki, D.; Lacalandra, G.M.; Pesole, G.; et al. Taxonomic Profile of Cultivable Microbiota from Adult Sheep Follicular Fluid and Its Effects on In Vitro Development of Prepubertal Lamb Oocytes. Animals 2025, 15, 1951. https://doi.org/10.3390/ani15131951
Mrenoshki S, Temerario L, Mastrorocco A, Visci G, Notario E, Marzano M, Martino NA, Mrenoshki D, Lacalandra GM, Pesole G, et al. Taxonomic Profile of Cultivable Microbiota from Adult Sheep Follicular Fluid and Its Effects on In Vitro Development of Prepubertal Lamb Oocytes. Animals. 2025; 15(13):1951. https://doi.org/10.3390/ani15131951
Chicago/Turabian StyleMrenoshki, Slavcho, Letizia Temerario, Antonella Mastrorocco, Grazia Visci, Elisabetta Notario, Marinella Marzano, Nicola Antonio Martino, Daniela Mrenoshki, Giovanni Michele Lacalandra, Graziano Pesole, and et al. 2025. "Taxonomic Profile of Cultivable Microbiota from Adult Sheep Follicular Fluid and Its Effects on In Vitro Development of Prepubertal Lamb Oocytes" Animals 15, no. 13: 1951. https://doi.org/10.3390/ani15131951
APA StyleMrenoshki, S., Temerario, L., Mastrorocco, A., Visci, G., Notario, E., Marzano, M., Martino, N. A., Mrenoshki, D., Lacalandra, G. M., Pesole, G., & Dell’Aquila, M. E. (2025). Taxonomic Profile of Cultivable Microbiota from Adult Sheep Follicular Fluid and Its Effects on In Vitro Development of Prepubertal Lamb Oocytes. Animals, 15(13), 1951. https://doi.org/10.3390/ani15131951







