Amoxicillin Resistance: An In Vivo Study on the Effects of an Approved Formulation on Antibiotic Resistance in Broiler Chickens
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experimentation Conditions
2.2. Metagenomic Analysis
2.3. Bioinformatic Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| ARG | Antimicrobial resistance gene |
| ESBL | Extended-spectrum beta-lactamase |
| MDR | Multidrug-resistant |
| MGE | Mobile genetic element |
References
- Marshall, B.M.; Levy, S.B. Food Animals and Antimicrobials: Impacts on Human Health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Dhanani, A.S.; Block, G.; Dewar, K.; Forgetta, V.; Topp, E.; Beiko, R.G.; Diarra, M.S. Genomic Comparison of Non-Typhoidal Salmonella enterica serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky Isolates from Broiler Chickens. PLoS ONE 2015, 10, e0128773. [Google Scholar] [CrossRef]
- Rochegüe, T.; Haenni, M.; Mondot, S.; Astruc, C.; Cazeau, G.; Ferry, T.; Madec, J.-Y.; Lupo, A. Impact of Antibiotic Therapies on Resistance Genes Dynamic and Composition of the Animal Gut Microbiota. Animals 2021, 11, 3280. [Google Scholar] [CrossRef]
- Hegde, N.V.; Kariyawasam, S.; DebRoy, C. Comparison of Antimicrobial Resistant Genes in Chicken Gut Microbiome Grown on Organic and Conventional Diet. Vet. Anim. Sci. 2016, 1–2, 9–14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marosevic, D.; Cervinkova, D.; Vlkova, H.; Videnska, P.; Babak, V.; Jaglic, Z. In Vivo Spread of Macrolide-Lincosamide-Streptogramin B (MLSB) Resistance--a Model Study in Chickens. Vet. Microbiol. 2014, 171, 388–396. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, Y.; Guo, L.; Zhang, S.; Wu, R.; Fang, X.; Xu, H.; Nie, Q. Metagenomic Analysis Reveals the Microbiome and Antibiotic Resistance Genes in Indigenous Chinese Yellow-Feathered Chickens. Front. Microbiol. 2022, 13, 930289. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Han, G.G.; Lee, H.-B.; Lee, S.-M.; Kang, S.-K.; Jin, G.-D.; Park, J.; Chae, B.J.; Choi, Y.H.; Kim, E.B.; et al. Prohibition of Antibiotic Growth Promoters Has Affected the Genomic Profiles of Lactobacillus salivarius Inhabiting the Swine Intestine. PLoS ONE 2017, 12, e0186671. [Google Scholar] [CrossRef]
- Paul, S.S.; Rama Rao, S.V.; Hegde, N.; Williams, N.J.; Chatterjee, R.N.; Raju, M.V.L.N.; Reddy, G.N.; Kumar, V.; Phani Kumar, P.S.; Mallick, S.; et al. Effects of Dietary Antimicrobial Growth Promoters on Performance Parameters and Abundance and Diversity of Broiler Chicken Gut Microbiome and Selection of Antibiotic Resistance Genes. Front. Microbiol. 2022, 13, 905050. [Google Scholar] [CrossRef]
- Li, J.; Hao, H.; Dai, M.; Zhang, H.; Ning, J.; Cheng, G.; Shabbir, M.A.B.; Sajid, A.; Yuan, Z. Resistance and Virulence Mechanisms of Escherichia coli Selected by Enrofloxacin in Chicken. Antimicrob. Agents Chemother. 2019, 63, e01824-18. [Google Scholar] [CrossRef]
- Data on Antimicrobial Resistance. Available online: https://ec.europa.eu/commission/presscorner/detail/en/ip_22_6951 (accessed on 16 April 2023).
- Battah, B. Emerging of Bacterial Resistance: An Ongoing Threat during and after the Syrian Crisis. J. Infect. Dev. Ctries. 2021, 15, 179–184. [Google Scholar] [CrossRef]
- Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2022. Available online: https://www.who.int/publications-detail-redirect/9789240062702 (accessed on 19 May 2024).
- Giono-Cerezo, S.; Santos-Preciado, J.I.; Morfín-Otero, M.D.R.; Torres-López, F.J.; Alcántar-Curiel, M.D. Antimicrobial Resistance. Its Importance and Efforts to Control It. GMM 2020, 156, 172–180. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance; HM Government and Wellcome Trust: London, UK, 2014. [Google Scholar]
- Woyda, R.; Oladeinde, A.; Abdo, Z. Chicken Production and Human Clinical Escherichia coli Isolates Differ in Their Carriage of Antimicrobial Resistance and Virulence Factors. Appl. Environ. Microbiol. 2023, 89, e01167-22. [Google Scholar] [CrossRef] [PubMed]
- Gautron, J.; Dombre, C.; Nau, F.; Feidt, C.; Guillier, L. Review: Production Factors Affecting the Quality of Chicken Table Eggs and Egg Products in Europe. Animal 2022, 16, 100425. [Google Scholar] [CrossRef] [PubMed]
- Bahri, S.I.S.; Ariffin, A.S.; Mohtar, S. Critical Review on Food Security in Malaysia for Broiler Industry. Int. J. Acad. Res. Bus. Soc. Sci. 2019, 9, 869–876. [Google Scholar] [CrossRef]
- Maciel-Guerra, A.; Baker, M.; Hu, Y.; Wang, W.; Zhang, X.; Rong, J.; Zhang, Y.; Zhang, J.; Kaler, J.; Renney, D.; et al. Dissecting Microbial Communities and Resistomes for Interconnected Humans, Soil, and Livestock. ISME J. 2023, 17, 21–35. [Google Scholar] [CrossRef]
- FAO. Meat Market Review: Emerging Trends and Outlook; FAO: Rome, Italy, 2020. [Google Scholar]
- Estévez-Moreno, L.X.; Miranda-de La Lama, G.C. Meat Consumption and Consumer Attitudes in México: Can Persistence Lead to Change? Meat Sci. 2022, 193, 108943. [Google Scholar] [CrossRef]
- Devi, S.M.; Balachandar, V.; Lee, S.I.; Kim, I.H. An Outline of Meat Consumption in the Indian Population—A Pilot Review. Korean J. Food Sci. Anim. Resour. 2014, 34, 507–515. [Google Scholar] [CrossRef]
- Essősy, M.; Fodor, I.; Ihnáth, Z.; Karancsi, Z.; Kovács, D.; Szalai, K.V.; Szentmiklósi, D.; Jerzsele, Á. The Possibilities of Antibiotic-Free Broiler-Hen Fattening, with Special Reference to the Use of Pre- and Probiotics. Magyar Állatorvosok Lapja 2020, 142, 397–407. [Google Scholar]
- Farkas, M.; Könyves, L.; Csorba, S.; Farkas, Z.; Józwiák, Á.; Süth, M.; Kovács, L. Biosecurity Situation of Large-Scale Poultry Farms in Hungary According to the Databases of National Food Chain Safety Office Centre for Disease Control and Biosecurity Audit System of Poultry Product Board of Hungary in the Period of 2021–2022. Magyar Állatorvosok Lapja 2024, 146, 723–742. [Google Scholar] [CrossRef]
- Hetényi, N.; Bersényi, A.; Hullár, I. Physiological Effects of Medium-Chain Fatty Acids and Triglycerides, and Their Potential Use in Poultry and Swine Nutrition: A Literature Review. Magyar Állatorvosok Lapja 2024, 146, 651–659. [Google Scholar] [CrossRef]
- Jerzsele, Á.; Somogyi, Z.; Szalai, M.; Kovács, D. Effects of Fermented Wheat Germ Extract on Artificial Salmonella Typhimurium Infection in Broiler Chickens. Magyar Állatorvosok Lapja 2020, 142, 77–85. [Google Scholar]
- Jócsák, G.; Schilling-Tóth, B.; Bartha, T.; Tóth, I.; Ondrašovičová, S.; Kiss, D.S. Metal Nanoparticles—Immersion in the „tiny” World of Medicine. Magyar Állatorvosok Lapja 2025, 147, 115–127. [Google Scholar] [CrossRef]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antiprotozoal and Antifungal Efficiency of Propolis—Part 2. Magyar Állatorvosok Lapja 2022, 144, 691–704. [Google Scholar]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antibacterial Efficiency of Propolis—Part 1. Magyar Állatorvosok Lapja 2022, 144, 285–298. [Google Scholar]
- Kovács, L.; Hejel, P.; Farkas, M.; László, L.; Könyves, K. Study Report on the Effect of a Litter Treatment Product Containing Bacillus licheniformis and Zeolite in Male Fattening Turkey Flock. Magyar Állatorvosok Lapja 2024, 146, 291–305. [Google Scholar] [CrossRef]
- Sebők, C.; Márton, R.A.; Meckei, M.; Neogrády, Z.; Mátis, G. Antimicrobial Peptides as New Tools to Combat Infectious Diseases. Magyar Állatorvosok Lapja 2024, 146, 181–191. [Google Scholar] [CrossRef]
- Petrilla, J.; Mátis, G.; Molnár, A.; Jerzsele, Á.; Pál, L.; Gálfi, P.; Neogrády, Z.; Dublecz, K. In Vitro Investigation of the Antibacterial Efficacy of Butyrate on Various Campylobacter jejuni Strains. MÁL 2021, 143, 57–64. [Google Scholar]
- Jones-Dias, D.; Clemente, L.; Moura, I.B.; Sampaio, D.A.; Albuquerque, T.; Vieira, L.; Manageiro, V.; Caniça, M. Draft Genomic Analysis of an Avian Multidrug Resistant Morganella morganii Isolate Carrying qnrD1. Front. Microbiol. 2016, 7, 1660. [Google Scholar] [CrossRef][Green Version]
- Börjesson, S.; Bengtsson, B.; Jernberg, C.; Englund, S. Spread of Extended-Spectrum Beta-Lactamase Producing Escherichia coli Isolates in Swedish Broilers Mediated by an Incl Plasmid Carrying Bla(CTX-M-1). Acta Vet. Scand. 2013, 55, 3. [Google Scholar] [CrossRef]
- Lambrecht, E.; Van Coillie, E.; Van Meervenne, E.; Boon, N.; Heyndrickx, M.; Van de Wiele, T. Commensal E. coli Rapidly Transfer Antibiotic Resistance Genes to Human Intestinal Microbiota in the Mucosal Simulator of the Human Intestinal Microbial Ecosystem (M-SHIME). Int. J. Food Microbiol. 2019, 311, 108357. [Google Scholar] [CrossRef]
- Murray, M.; Salvatierra, G.; Dávila-Barclay, A.; Ayzanoa, B.; Castillo-Vilcahuaman, C.; Huang, M.; Pajuelo, M.J.; Lescano, A.G.; Cabrera, L.; Calderón, M.; et al. Market Chickens as a Source of Antibiotic-Resistant Escherichia coli in a Peri-Urban Community in Lima, Peru. Front. Microbiol. 2021, 12, 635871. [Google Scholar] [CrossRef]
- Suzuki, K.; Yossapol, M.; Sugiyama, M.; Asai, T. Effects of Antimicrobial Administration on the Prevalence of Antimicrobial-Resistant Escherichia coli in Broiler Flocks. Jpn. J. Infect. Dis. 2019, 72, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.I.; Kuenzli, E.; Campos-Madueno, E.I.; Büdel, T.; Rattanavong, S.; Vongsouvath, M.; Hatz, C.; Endimiani, A. Antimicrobial-Resistant Escherichia coli Strains and Their Plasmids in People, Poultry, and Chicken Meat in Laos. Front. Microbiol. 2021, 12, 708182. [Google Scholar] [CrossRef]
- Timmermans, M.; Wattiau, P.; Denis, O.; Boland, C. Colistin Resistance Genes mcr-1 to mcr-5, Including a Case of Triple Occurrence (mcr-1, -3 and -5), in Escherichia coli Isolates from Faeces of Healthy Pigs, Cattle and Poultry in Belgium, 2012-2016. Int. J. Antimicrob. Agents 2021, 57, 106350. [Google Scholar] [CrossRef]
- Deblais, L.; Kathayat, D.; Helmy, Y.A.; Closs, G.; Rajashekara, G. Translating “Big Data”: Better Understanding of Host-Pathogen Interactions to Control Bacterial Foodborne Pathogens in Poultry. Anim. Health Res. Rev. 2020, 21, 15–35. [Google Scholar] [CrossRef] [PubMed]
- Gundran, R.S.; Cardenio, P.A.; Villanueva, M.A.; Sison, F.B.; Benigno, C.C.; Kreausukon, K.; Pichpol, D.; Punyapornwithaya, V. Prevalence and Distribution of blaCTX-M, blaSHV, blaTEM Genes in Extended- Spectrum β- Lactamase- Producing E. coli Isolates from Broiler Farms in the Philippines. BMC Vet. Res. 2019, 15, 227. [Google Scholar] [CrossRef] [PubMed]
- Persoons, D.; Haesebrouck, F.; Smet, A.; Herman, L.; Heyndrickx, M.; Martel, A.; Catry, B.; Berge, A.C.; Butaye, P.; Dewulf, J. Risk Factors for Ceftiofur Resistance in Escherichia coli from Belgian Broilers. Epidemiol. Infect. 2011, 139, 765–771. [Google Scholar] [CrossRef]
- Afridi, O.K.; Ali, J.; Chang, J.H. Next-Generation Sequencing Based Gut Resistome Profiling of Broiler Chickens Infected with Multidrug-Resistant Escherichia coli. Animals 2020, 10, 2350. [Google Scholar] [CrossRef]
- Tofani, S.; Albini, E.; Blasi, F.; Cucco, L.; Lovito, C.; Maresca, C.; Pesciaroli, M.; Orsini, S.; Scoccia, E.; Pezzotti, G.; et al. Assessing the Load, Virulence and Antibiotic-Resistant Traits of ESBL/ampc E. coli from Broilers Raised on Conventional, Antibiotic-Free, and Organic Farms. Antibiotics 2022, 11, 1484. [Google Scholar] [CrossRef]
- Jiang, H.-X.; Lü, D.-H.; Chen, Z.-L.; Wang, X.-M.; Chen, J.-R.; Liu, Y.-H.; Liao, X.-P.; Liu, J.-H.; Zeng, Z.-L. High Prevalence and Widespread Distribution of Multi-Resistant Escherichia coli Isolates in Pigs and Poultry in China. Vet. J. 2011, 187, 99–103. [Google Scholar] [CrossRef]
- Ding, Y.; Zwe, Y.H.; Chin, S.F.; Kohli, G.S.; Drautz-Moses, D.I.; Givskov, M.; Schlundt, J.; Schuster, S.C.; Yuk, H.-G.; Yang, L. Characterization of a Novel Multidrug Resistance Plasmid pSGB23 Isolated from Salmonella enterica subspecies enterica serovar Saintpaul. Gut Pathog. 2018, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Carvalheira, A.; Casquete, R.; Silva, J.; Teixeira, P. Prevalence and Antimicrobial Susceptibility of Acinetobacter spp. Isolated from Meat. Int. J. Food Microbiol. 2017, 243, 58–63. [Google Scholar] [CrossRef]
- Dos Santos Bersot, L.; Carbonera, N.R.; Rodrigues Valcanaia, C.D.; Viana, C.; Nero, L.A. Multidrug-Resistant and Extended-Spectrum β-Lactamase-Producing Salmonella enterica serotype Heidelberg Is Widespread in a Poultry Processing Facility in Southern Brazil. J. Food Prot. 2021, 84, 2053–2058. [Google Scholar] [CrossRef]
- Hu, Y.; He, Y.; Wang, Y.; Cui, S.; Chen, Q.; Liu, G.; Chen, Q.; Zhou, G.; Yang, B.; Huang, J.; et al. Epidemic condition and molecular subtyping of ciprofloxacin and cefotaxime co-resistant Salmonella Indiana isolated from retail chicken carcasses in six provinces, China. Zhonghua Yu Fang Yi Xue Za Zhi 2015, 49, 716–721. [Google Scholar]
- Aruwa, C.E.; Pillay, C.; Nyaga, M.M.; Sabiu, S. Poultry Gut Health—Microbiome Functions, Environmental Impacts, Microbiome Engineering and Advancements in Characterization Technologies. J. Anim. Sci. Biotechnol. 2021, 12, 119. [Google Scholar] [CrossRef] [PubMed]
- Montoro-Dasi, L.; Villagra, A.; de Toro, M.; Pérez-Gracia, M.T.; Vega, S.; Marin, C. Assessment of Microbiota Modulation in Poultry to Combat Infectious Diseases. Animals 2021, 11, 615. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zhang, Y.; Xiao, K.; Jiang, F.; Wang, H.; Tang, D.; Liu, D.; Liu, B.; Liu, Y.; He, X.; et al. The Chicken Gut Metagenome and the Modulatory Effects of Plant-Derived Benzylisoquinoline Alkaloids. Microbiome 2018, 6, 211. [Google Scholar] [CrossRef]
- Han, G.G.; Kim, E.B.; Lee, J.; Lee, J.-Y.; Jin, G.; Park, J.; Huh, C.-S.; Kwon, I.-K.; Kil, D.Y.; Choi, Y.-J.; et al. Relationship between the Microbiota in Different Sections of the Gastrointestinal Tract, and the Body Weight of Broiler Chickens. SpringerPlus 2016, 5, 911. [Google Scholar] [CrossRef]
- Wise, M.G.; Siragusa, G.R. Quantitative Analysis of the Intestinal Bacterial Community in One- to Three-Week-Old Commercially Reared Broiler Chickens Fed Conventional or Antibiotic-Free Vegetable-Based Diets. J. Appl. Microbiol. 2007, 102, 1138–1149. [Google Scholar] [CrossRef]
- Singh, P.; Karimi, A.; Devendra, K.; Waldroup, P.W.; Cho, K.K.; Kwon, Y.M. Influence of Penicillin on Microbial Diversity of the Cecal Microbiota in Broiler Chickens. Poult. Sci. 2013, 92, 272–276. [Google Scholar] [CrossRef]
- Sahin-Tóth, J.; Kovács, E.; Tóthpál, A.; Juhász, J.; Forró, B.; Bányai, K.; Havril, K.; Horváth, A.; Ghidán, Á.; Dobay, O. Whole Genome Sequencing of Coagulase Positive Staphylococci from a Dog-and-Owner Screening Survey. PLoS ONE 2021, 16, e0245351. [Google Scholar] [CrossRef]
- Modi, A.; Vai, S.; Caramelli, D.; Lari, M. The Illumina Sequencing Protocol and the NovaSeq 6000 System. Methods Mol. Biol. 2021, 2242, 15–42. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 April 2022).
- Krueger, F. Trim Galore. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 14 January 2022).
- Dinghua MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph|Bioinformatics|Oxford Academic. Available online: https://academic.oup.com/bioinformatics/article/31/10/1674/177884 (accessed on 25 April 2022).
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequences (RefSeq): A Curated Non-Redundant Sequence Database of Genomes, Transcripts and Proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020.
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S. Microbiome R Package. Available online: https://github.com/microbiome/microbiome (accessed on 14 January 2022).
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of Mobile Genetic Elements Associated with Antibiotic Resistance in Salmonella enterica Using a Newly Developed Web Tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting Plasmid Sequences in Metagenomic Data Using Genome Signatures. Nucleic Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, W.; Zhang, H.; Wang, J.; Zhang, W.; Gao, J.; Wu, S.; Qi, G. Supplemental Bacillus subtilis DSM 32315 Manipulates Intestinal Structure and Microbial Composition in Broiler Chickens. Sci. Rep. 2018, 8, 15358. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Microbiological Effects of Sublethal Levels of Antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic Resistance Is Ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Y.; Lin, J. Functional Cloning and Characterization of Antibiotic Resistance Genes from the Chicken Gut Microbiome. Appl. Environ. Microbiol. 2012, 78, 3028–3032. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Silbergeld, E.K. Learning from Agriculture: Understanding Low-Dose Antimicrobials as Drivers of Resistome Expansion. Front. Microbiol. 2014, 5, 284. [Google Scholar] [CrossRef]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Torres, C.; Hammami, S. High Prevalence of mcr-1 Encoding Colistin Resistance and First Identification of blaCTX-M-55 in ESBL/CMY-2-Producing Escherichia coli Isolated from Chicken Faeces and Retail Meat in Tunisia. Int. J. Food Microbiol. 2020, 318, 108478. [Google Scholar] [CrossRef]
- Bradford, P.A. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef]
- Gousia, P.; Economou, V.; Bozidis, P.; Papadopoulou, C. Vancomycin-Resistance Phenotypes, Vancomycin-Resistance Genes, and Resistance to Antibiotics of Enterococci Isolated from Food of Animal Origin. Foodborne Pathog. Dis. 2015, 12, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Breidenstein, E.B.M.; Hancock, R.E.W. Creeping Baselines and Adaptive Resistance to Antibiotics. Drug Resist. Updates 2011, 14, 1–21. [Google Scholar] [CrossRef] [PubMed]
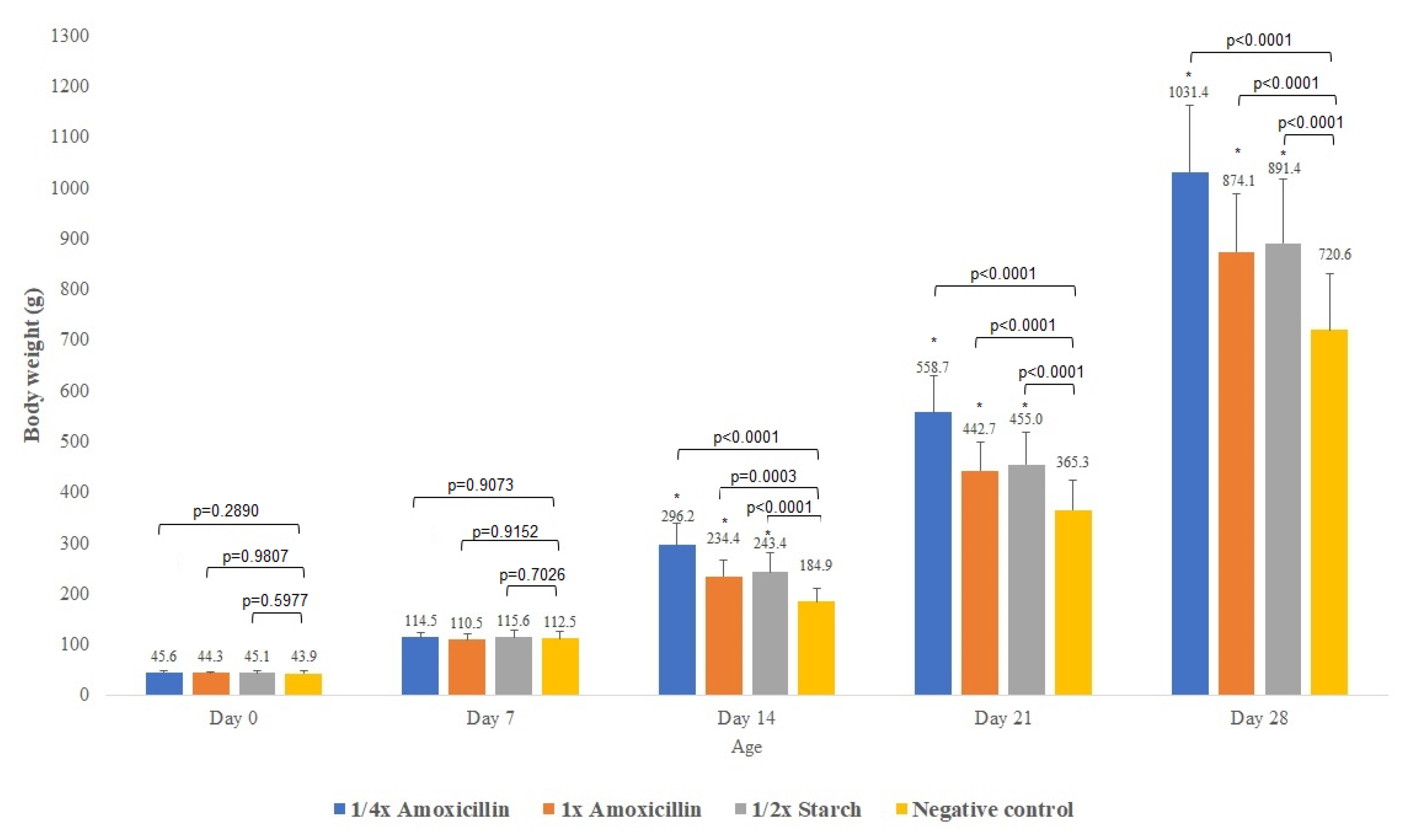

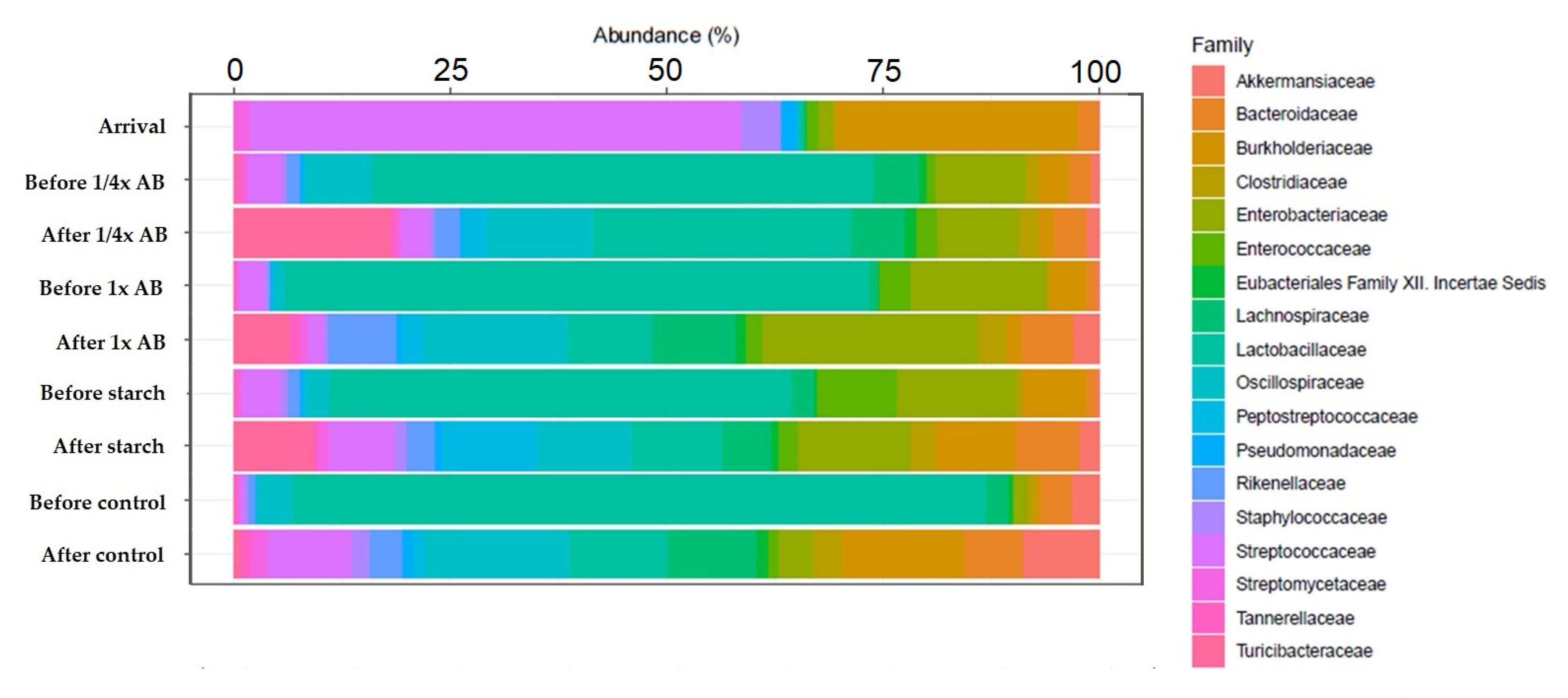
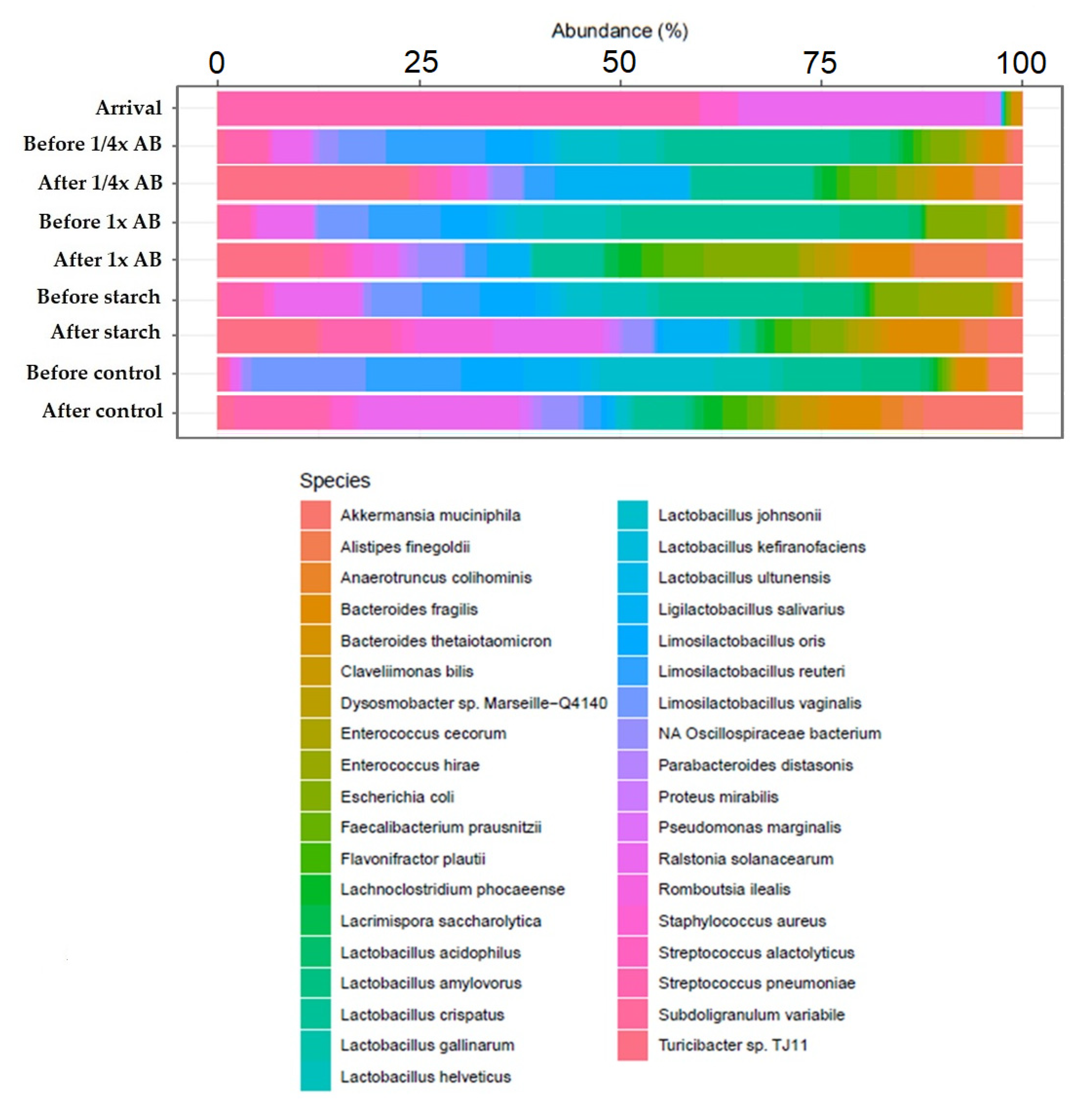
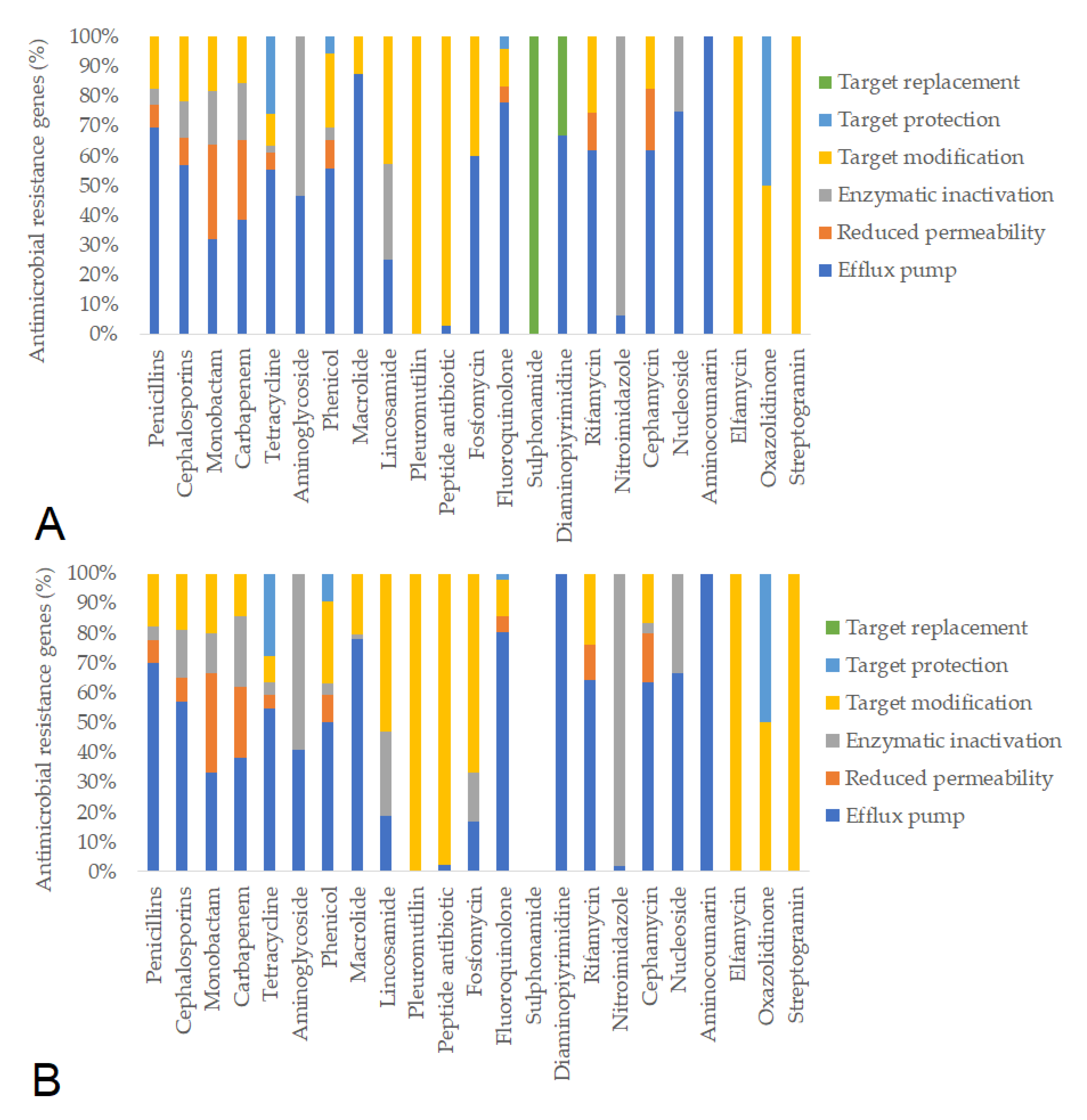

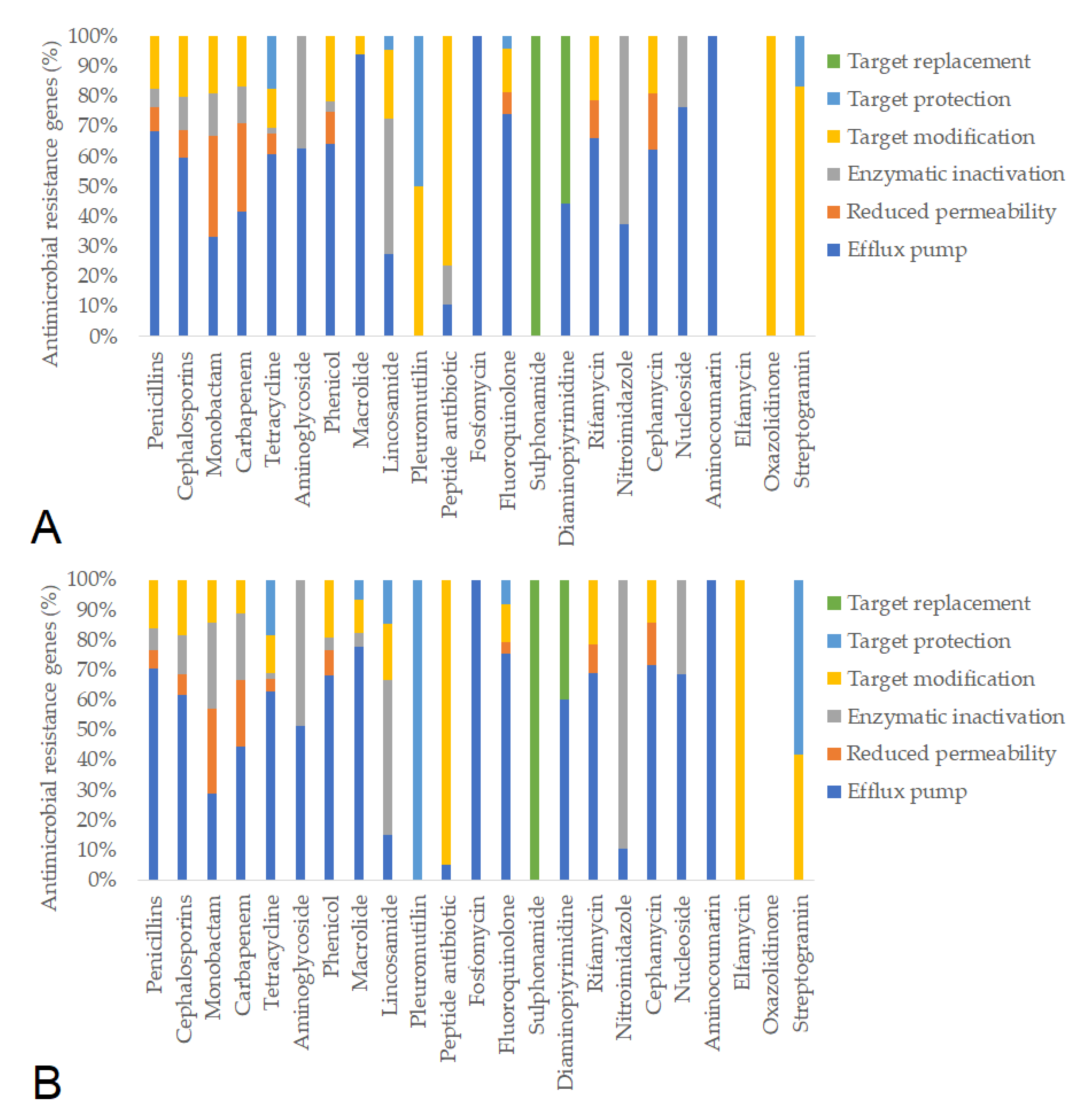

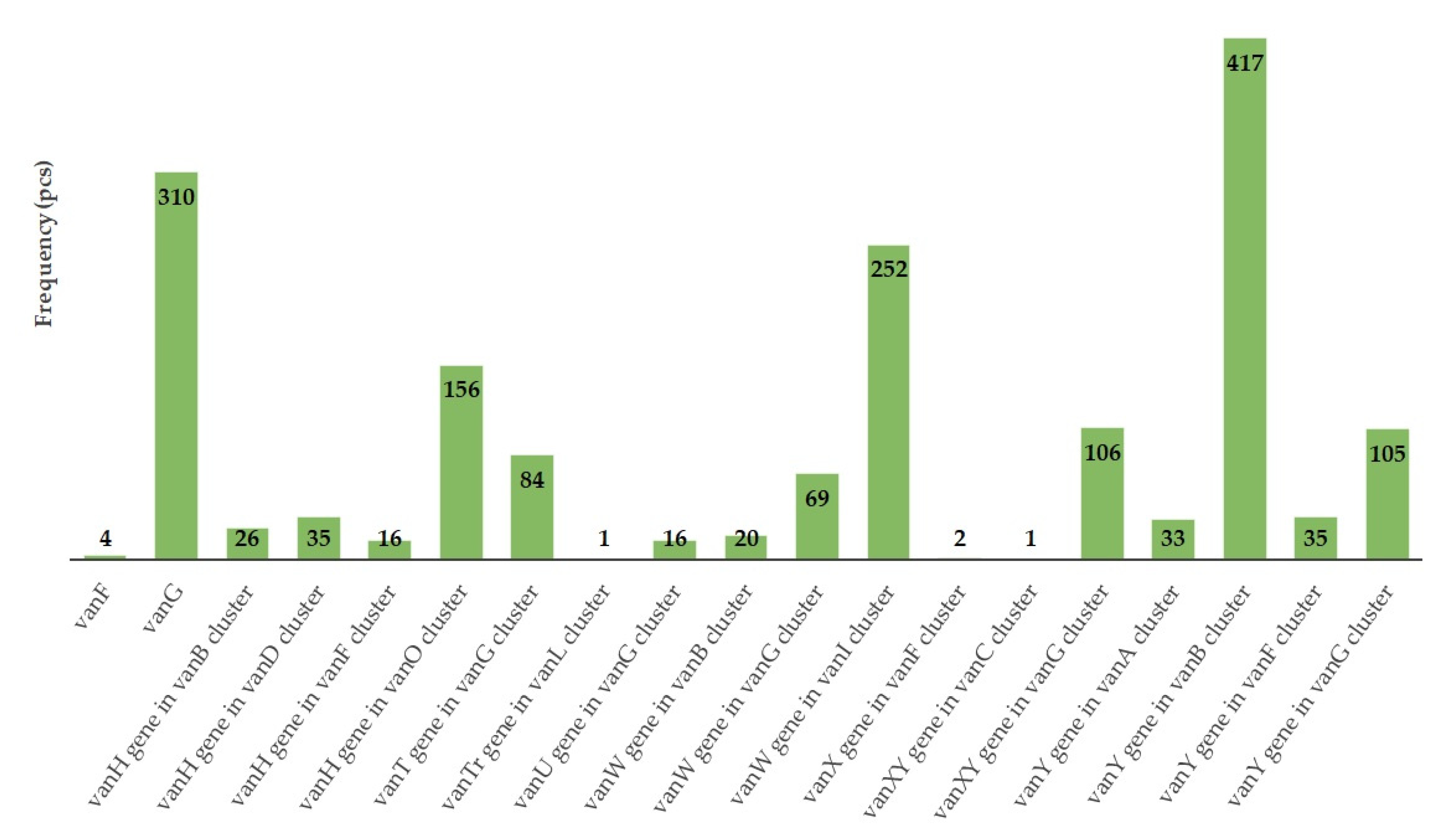
| Day of Treatment | Average BW (kg) | Group Size (n) | Dose 1× (g) | Dose ¼× (g) | Placebo Starch ½× (g) |
|---|---|---|---|---|---|
| 1 | 0.4 | 30 | 0.48 | 0.12 | 0.24 |
| 2 | 0.6 | 30 | 0.72 | 0.18 | 0.36 |
| 3 | 0.6 | 30 | 0.72 | 0.18 | 0.36 |
| 4 | 0.6 | 30 | 0.72 | 0.18 | 0.36 |
| 5 | 0.8 | 30 | 0.96 | 0.24 | 0.48 |
| 6 | 0.8 | 30 | 0.96 | 0.24 | 0.48 |
| 7 | 0.9 | 30 | 1.08 | 0.27 | 0.54 |
| 8 | 0.9 | 30 | 1.08 | 0.27 | 0.54 |
| 9 | 1 | 30 | 1.2 | 0.3 | 0.6 |
| 10 | 1 | 30 | 1.2 | 0.3 | 0.6 |
| Treatment | ¼× Amoxicillin | 1× Amoxicillin | ½× Starch | Negative Control | ||||
|---|---|---|---|---|---|---|---|---|
| Before | After | Before | After | Before | After | Before | After | |
| Plasmid | 105 (±8.9) | 66 (±3.2) | 66 (±6.4) | 64 (±7.8) | 59 (±0.6) | 58 (±5.0) | 110 (±10.0) | 89 (±6.0) |
| Phage | 13 (±1.7) | 13 (±2.0) | 5 (±3.1) | 6 (±3.1) | 12 (±5.6) | 7 (±6.7) | 5 (±3.8) | 17 (±8.6) |
| MGE * | 34 (±1.5) | 19 (±3.1) | 33 (±2.0) | 36 (±6.6) | 31 (±4.2) | 22 (±3.8) | 24 (±9.5) | 27 (±7.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerek, Á.; Szabó, Á.; Jerzsele, Á. Amoxicillin Resistance: An In Vivo Study on the Effects of an Approved Formulation on Antibiotic Resistance in Broiler Chickens. Animals 2025, 15, 1944. https://doi.org/10.3390/ani15131944
Kerek Á, Szabó Á, Jerzsele Á. Amoxicillin Resistance: An In Vivo Study on the Effects of an Approved Formulation on Antibiotic Resistance in Broiler Chickens. Animals. 2025; 15(13):1944. https://doi.org/10.3390/ani15131944
Chicago/Turabian StyleKerek, Ádám, Ábel Szabó, and Ákos Jerzsele. 2025. "Amoxicillin Resistance: An In Vivo Study on the Effects of an Approved Formulation on Antibiotic Resistance in Broiler Chickens" Animals 15, no. 13: 1944. https://doi.org/10.3390/ani15131944
APA StyleKerek, Á., Szabó, Á., & Jerzsele, Á. (2025). Amoxicillin Resistance: An In Vivo Study on the Effects of an Approved Formulation on Antibiotic Resistance in Broiler Chickens. Animals, 15(13), 1944. https://doi.org/10.3390/ani15131944






