The Nariño Cat, the Tigrinas and Their Problematic Systematics and Phylogeography: The Real Story
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Samples
2.2. Mitochondrial DNA
2.3. Statistical Analyses
2.3.1. Genetic Distances
2.3.2. Phylogenetic Trees
2.3.3. Spatial Monmonier’s Algorithm Analysis
2.3.4. BAPS Analyses
3. Results
3.1. Genetic Distances Between the Nariño Cat and Other Tigrina Specimens and Species of the Genus Leopardus
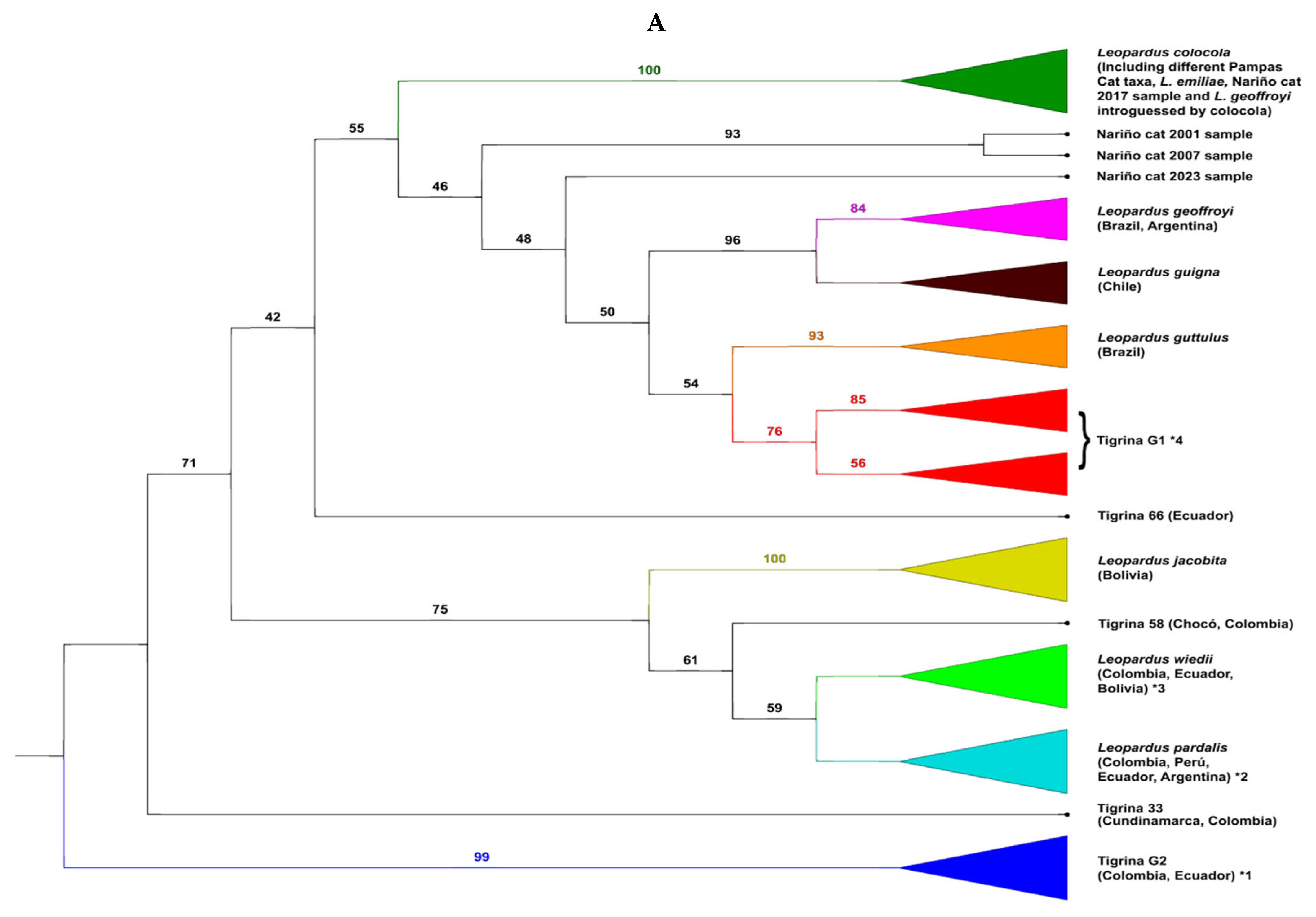
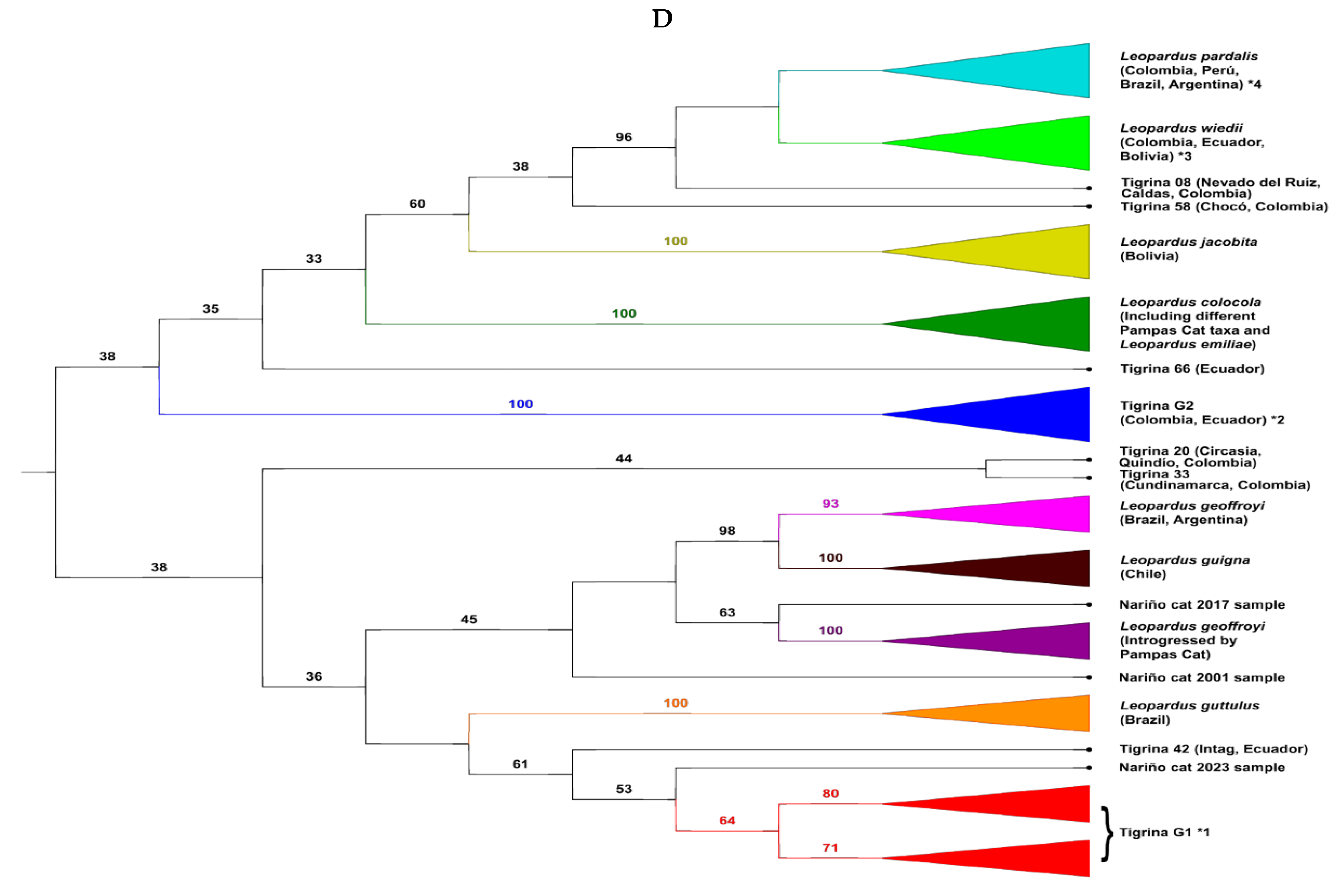
3.2. The Situation of the Different Nariño Cat Samples in the Phylogenetic Tree of the Leopardus Genus
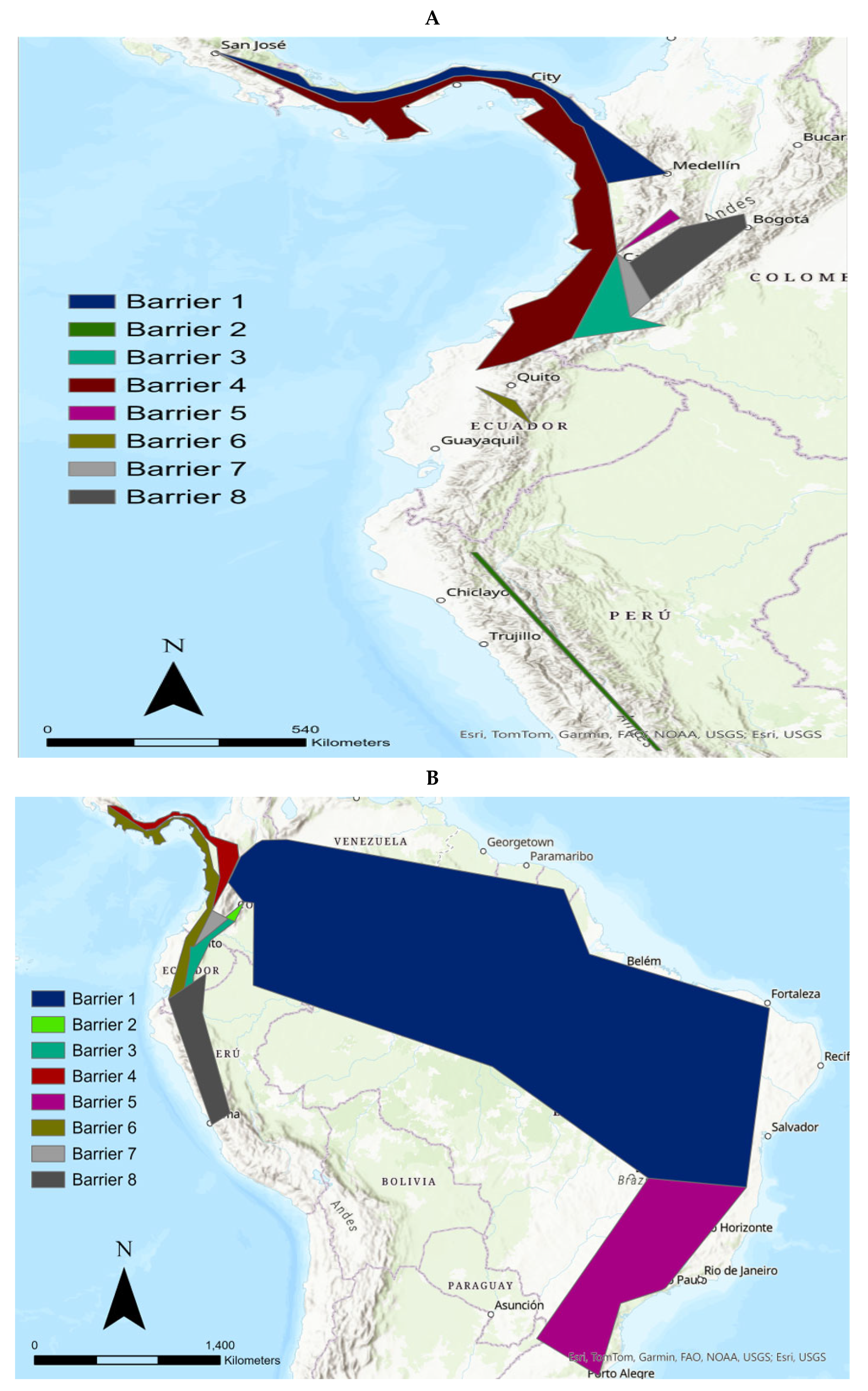
3.3. The Differentiation of the Nariño Cat Using the Monmonier Algorithm (MA) and BAPS
4. Discussion
4.1. Why Is the Phylogeny of the Leopardus Species Complicated?
4.2. The Phylogenetic Situation of the Nariño Cat, of Other Tigrinas, and Why the mtDNA Sequences of the Nariño Cat Were Not Consistent Throughout the Years
4.3. Potential Geographical Barriers Affecting the Tigrinas in the Northern Andean Cordilleras
4.4. The Nariño Cat and Its Scientific Name
4.5. What Conclusions of Astorquiza et al. [23] and Marín-Puerta [24] Regarding the Nariño Cat Were Incorrect?
- (1)
- The authors only compared their DNA sequences of the Nariño cat with two Colombian tigrinas and with two Pampas cats [23], and with the same two Colombian tigrinas, and one Central American tigrina, one L. guttulus, one L. t. emiliae, and one L. colocola [24] (these four last sequences taken from the study of Lescroart et al. [19]). We have shown that the systematic relationships of the Nariño cat (and the other “rare” tigrinas) depends on the number of other specimens of the Leopardus genus used in the comparison. Clearly, the comparison with only two northern Andes tigrinas is not enough. It is relevant to mention the genomic-whole analysis used by Lescroart et al. [19] to try resolving the phylogeny of the Leopardus genus. Nonetheless, they only used two Colombian and one Central American tigrinas (three specimens) to carry out their analyses. Although they correctly discriminated against these northern tigrinas from L. guttulus and L. t. emiliae (as Marín-Puerta [24] did) they concluded that only a group of tigrina is present in northern-western South America and Central America. Herein we showed at the mtND5 gene, analyzing 164 specimens of Leopardus, which included 63 specimens representing the “traditional” tigrina, that the 37 northern Andean and Central American tigrinas were divided, at least, into six different groups (without counting the Nariño cat). The largest group (Tigrina G1) was represented by 16 specimens (the two Colombian tigrinas reported by Astorquiza et al. [23] and Lescroart et al. [19] and the Central American tigrinas were inside this group). The second group (Tigrina G2) was composed of nine tigrinas while the third group (eight specimens) was composed of tigrinas introgressed/hybridized with the margay and the ocelot. The other four tigrinas made up isolated branches. For mitogenomes, we analyzed 102 specimens of the Leopardus genus, including 42 specimens of tigrina. The 34 northern Andean and Central American tigrinas were divided, at least (without counting the Nariño cat), into eight different groups. The largest group (Tigrina G1) was represented by 13 specimens (including the tigrinas from Astorquiza et al. [23] and Lescroart et al. [19] and the Central American tigrinas). The second group (Tigrina G2) was composed of eight tigrinas. The third group was integrated by five tigrinas introgressed/hybridized by the margay and ocelot and later appeared as five isolated branches formed by one or two “rare” tigrina(s).
- (2)
- Astorquiza et al. [23] commented that the three fragments of the mtDNA they studied (16S rRNA, COX2-ATP8, and ND4-ND5) totaled 845 bp. This contrasts with the 16,756 bp we studied for the mitogenome dataset. Of these three molecular markers they studied, the mtND5 gene was the best discriminator among species of the Leopardus genus. The primers that they employed for ND4-ND5 only amplified 145 bp, whereas we obtained nearly 1800 bp (although we used 315 bp of this gene to carry out the comparisons with the sequences deposited in GenBank). Thus, their study of the Nariño cat not only used far fewer samples, but they also studied a much smaller portion of mitochondrial DNA compared to the study by Ruiz-García et al. [8]. In fact, some of the mitochondrial genes used by Astorquiza et al. [23] (such as 16S rRNA and ATP8) did not highly differentiate the Nariño cat from Tigrina G1. However, other mt genes such as ND5, ND6, Cyt-b, or COX3 differentiated well both small spotted cats. Marín-Puerta [24] used the same mt sequences generated by Astorquiza et al. [23]. Therefore, his molecular data lacks novelty and has exactly the same problems as those shown by Astorquiza et al. [23].
- (3)
- Astorquiza et al. [23] textually affirmed that the Nariño cat showed 100% identity with sequences of the two Colombian tigrinas that they studied, although its identity with L. colocola sequences ranged from 88 to 98%. An analysis of the authors’ Supplementary Tables demonstrates that their affirmation is incorrect. Furthermore, these supplementary Tables contain mistakes. For example, Supplementary Table S3, shows that the genetic distance for COX2-ATP8 between the Nariño cat and the two tigrinas was 0. Nonetheless, the authors reported a genetic distance of 0.27% between the two Colombian tigrinas. If the sequences of the two tigrinas were not identical, then one of both tigrinas could not be identical to the sequence of the Nariño cat. Thus, it is materially impossible for the Nariño cat to be simultaneously identical to both tigrinas. Clearly, the authors made some mistakes in their numerical estimations. Second, Supplementary Table S4 for ND4-ND5 showed that the Nariño cat was differentiated from one of the tigrinas by 0.47%. Supplementary Table S5 indicates that the Nariño cat was differentiated from one of the tigrinas by 0.16% and from the other by 0.08%. These genetic differentiation values were very small, but they do not support a 100% genetic identity between the Nariño cat and the two Colombian tigrinas as the authors claimed. Marín-Puerta [24] also repeated that the Nariño cat and the two Colombian tigrinas used by Astorquiza et al. [23] and by himself had a 100% of genetic identity. However, in his ML tree showed that the Nariño cat was in a different branch in reference to the two Colombian tigrinas. Thus, it is materially impossible that the Nariño cat had a 100% of genetic identity with these two Colombian tigrinas.
- (4)
- Another refutable affirmation of Astorquiza et al. [23] and Marín-Puerta [24] concerns the morphotype of the Nariño cat. They used the argument that the Nariño cat skin was classified by Nascimento and Feijó [5] within morphogroup I. Effectively, the Nariño cat by the disposition of its rosettes is more related to morphogroup I than to morphogroup II (L. t. emiliae) or to morphogroup III (L. guttulus). But this statement is misleading. Nascimento and Feijó [5] also showed, for instance (Figure 10, p. 246), that a skin of margay (NMNH388255, Rio Cunucunuma, Belen, Venezuela) was extremely like the skin of a tigrina from morphogroup I (USNM374861, El Manteco, Bolivar, Venezuela). Therefore, based on Nascimiento and Feijó [5], this margay skin could be classified in Morphogroup I, being a margay a different species from a tigrina. That is, Leopardus species other than tigrinas could also be classified in morphogroup I using the reasoning of Nascimento and Feijó [5]. The fact that the Nariño cat by the disposition and color of its rosettes belongs to morphogroup I does not mean it is not strongly different in other coat characteristics to other tigrinas from morphogroup I. Indeed, if anyone looks at Figure 1 from Astorquiza et al. [23], they can easily observe that the Nariño cat skin (Figure 1e) is clearly different from the Pampas cat skin (Figure 1a–c), but it is just as easy to observe that the Nariño cat skin is totally distinguishable from the other tigrina skin analyzed by these authors (Figure 1d). It is certain that the Nariño cat skin has a similar size to other tigrina skins, and no skull (or other bones) are associated with this skin to be compared with material of other tigrina and this limited quantitative morphological comparisons weaken the case for the Nariño cat’s distinctiveness. However, an indisputable fact is that the overall physical appearance of the skin of the Nariño cat is totally distinguishable from any other skin of the different taxa recognized within the Leopardus genus. On the other hand, Marín-Puerta [24] added an alleged new proof against the skin differentiation of the Nariño cat. He commented that the intense reddish coat color of the Nariño cat’s skin is an artifact of bad preservation (he forgets the texture of the coat, the intense black hood of the head and the middle area of the back, the non-existence of the white ventral fur of the traditional tigrilla, etc.) and that this skin was exposed to smoke or prolonged exposure to intense heat, considering the unusual coloration of the hindfeet bones. This is a very unbelievable argument for several reasons. First, the preservation of this skin was excellent until at least 2017 and it was not subjected to chemical or physical treatments that would have mistreated it because, if it had been, it would have been difficult for us to obtain DNA of such good quality as to obtain its complete mitogenome. Second, to say that it was smoked or that it was subjected to very high temperatures is totally incomprehensible since this skin comes from a very cold place in the Andean Páramo at 3100 masl and where the technique of smoking hunted animals is not usually used, unlike what happens in the Amazon. When an animal smokes, the blood is destroyed and the reddish color in the bones of the legs is a symptom that the blood was preserved because it was exposed to very low temperatures that preserved well its morphological characteristics as well as its DNA (i.e., from the Peruvian mummies found at high altitudes). In fact, other skins from other tigrina obtained in the IAVH museum and that were treated with chemical procedures or heat did not offer DNA of good quality or in high quantities as the Nariño cat skin did.
- (5)
- Astorquiza et al. [23] claimed that the Nariño cat sequences they generated would be deposited in GenBank upon submission of their paper. However, to date these sequences have not been submitted to GenBank and, therefore, it is impossible to compare them with our Nariño cat’s sequences. Likely, Marín-Puerta [24] claimed that the genetic data used in Ruíz-García et al. [8] has not been previously published and lacks traceability, making the study impossible to replicate. This statement is not entirely true. The senior author of the current article (MR-G) has uploaded to GenBank around 460 sequences of specimens of the Leopardus genus, whilst the senior author or director of [23,24] (HR-CH) is collaterally associated with seven sequences of specimens of the same genus. On the other hand, Marín-Puerta [24] has not either uploaded in GenBank the sequences of the Nariño cat that he worked in his thesis. Perhaps, they may not be able to replicate the present study because they do not have as large numbers of northern Andean tigrina samples and with geographic origins as diversified as what we show in this study.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macdonald, D.W.; Loveridge, A.J. Biology and Conservation of Wild Felids; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Sunquist, F.; Sunquist, M. The Wild Cat Book: Everything You Ever Wanted to Know About Cats; The University of Chicago Press: Chicago, IL, USA, 2014. [Google Scholar]
- Bellani, G.G. Felines of the World: Discovery in Taxonomic Classification and History; Academic Press: London, UK, 2020. [Google Scholar]
- Kitchener, A.C.; Breitenmoser-Würsten, C.; Eizirik, E.; Gentry, A.; Werdelin, L.; Wilting, A.; Yamaguchi, N.; Abramov, A.V.; Christiansen, P.; Driscoll, C.; et al. A revised taxonomy of the Felidae. The final report of the Cat Classification Task Force of the IUCN/SSC Cat Specialist group. Cat News Spec. Issue 2017, 11, 1–80. [Google Scholar]
- Nascimento, F.O.; Feijó, A. Taxonomic revision of the tigrina Leopardus tigrinus (Schreber, 1775) species group (Carnivora, Felidae). Papéis Avulsos Zool. 2017, 57, 231–264. [Google Scholar] [CrossRef]
- Nascimento, F.O.; Cheng, J.; Feijó, A. Taxonomic revision of the pampas cat Leopardus colocola complex (Carnivora: Felidae): An integrative approach. Zool. J. Linn. Soc. 2021, 191, 575–611. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Pinedo-Castro, M.; Shostell, J.M. Small spotted bodies with multiple specific mitochondrial DNAs: Existence of diverse and differentiated tigrina lineages or species (Leopardus spp.: Felidae, Mammalia) throughout Latin America. Mitochondrial DNA Part A 2018, 29, 993–1014. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Pinedo-Castro, M.; Shostell, J.M. Morphological and genetics support for a hitherto undescribed spotted cat species (Genus Leopardus; Felidae, Carnivora) from the Southern Colombian Andes. Genes 2023, 14, 1266. [Google Scholar] [CrossRef]
- García-Perea, R. The Pampas cat group (genus Lynchailurus Severtzov, 1858) (Carnivora: Felidae), a systematic and biogeographic review. Am. Mus. Novitat. 1994, 3096, 35. [Google Scholar]
- Cossíos, D.; Lucherini, M.; Ruiz-García, M.; Angers, B. Influence of ancient glacial periods on the Andean fauna: The case of the pampas cat (Leopardus colocolo). BMC Evol. Biol. 2009, 9, 68. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Cossíos, D.; Lucherini, M.; Yáñez, J.; Pinedo-Castro, M.; Angers, B. Population genetics and spatial structure in two Andean cats (the Pampas cat, Leopardus pajeros and the Andean Mountain cat, L. jacobita) by means of nuclear and mitochondrial markers and some notes on skull biometrics. In Molecular Population Genetics, Evolutionary Biology and Biological Conservation of Neotropical Carnivores; Ruiz-García, M., Shostell, J., Eds.; Nova Science Publishers Inc.: New York, NY, USA, 2013; pp. 187–244. [Google Scholar]
- Cabrera, A. Catálogo de los mamíferos de América del Sur. I (Metatheria, Unguiculata, Carnıvora). Rev. Del. Mus. Argent. Cienc. Nat. “Bernardo Rivadavia” 1958, 4, 1–307. [Google Scholar]
- Cabrera, A. Los félidos vivientes de la Republica de Argentina. Rev. Del. Mus. Argent. Cienc. Nat. 1961, 6, 161–247. [Google Scholar]
- Wilson, D.E.; Reeder, D.A.M. Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2005; pp. 1–142. [Google Scholar]
- Trigo, T.C.; Freitas, T.R.O.; Kunzler, G.; Cardoso, L.; Silva, J.C.R.; Johnson, W.E.; O’Brien, S.J.; Bonatto, S.L.; Eizirik, E. Inter-species hybridization among Neotropical cats of the genus Leopardus, and evidence for an introgressive hybrid zone between L. geoffroyi and L. tigrinus in southern Brazil. Mol. Ecol. 2008, 17, 4317–4333. [Google Scholar] [CrossRef]
- Trigo, T.C.; Schneider, A.; Oliveira, T.G.; Lehugeur, L.M.; Silveira, L.; Freitas, T.R.; Eizirik, E. Molecular data reveal complex hybridization and a cryptic species of Neotropical wild cat. Curr. Biol. 2013, 23, 2528–2533. [Google Scholar] [CrossRef] [PubMed]
- Trigo, T.C.; Tirelli, F.P.; de Freitas, T.R.O.; Eizirik, E. Comparative assessment of genetic and morphological variation at an extensive hybrid zone between two wild cats in Southern Brazil. PLoS ONE 2014, 9, e108469. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Davis, B.W.; Eizirik, E.; Murphy, W.J. Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res. 2016, 26, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lescroart, J.; Bonilla-Sánchez, A.; Napolitano, C.; Buitrago-Torres, D.L.; Ramírez-Chaves, H.E.; Pulido-Santacruz, P.; Murphy, W.J.; Svardal, H.; Eizirik, E. Extensive Phylogenomic Discordance and the Complex Evolutionary History of the Neotropical Cat Genus Leopardus. Mol. Biol. Evol. 2023, 40, msad255. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Lescroart, J.; Figueiró, H.V.; Torres-Florez, J.P.; Villela, P.M.S.; Coutinho, L.L.; Freitas, P.D.; Johnson, W.E.; Antunes, A.; Galetti, P.M., Jr.; et al. Genomic signatures of divergent ecological strategies in a recent radiation of neotropical wild cats. Mol. Biol. Evol. 2022, 39, msac117. [Google Scholar] [CrossRef]
- Soraggi, S.; Wiuf, C.; Albrechtsen, A. Powerful inference with the D-statistic on low-coverage whole-genome data. G3 Genes Genomes Genet. 2018, 8, 551–566. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Payán, C.E.; Hernández-Camacho, J.I. Possible records of Lynchailurus in southwestern Colombia. Cat News 2003, 38, 35–36. [Google Scholar]
- Astorquiza, J.M.; Noguera-Urbano, E.A.; Cabrera-Ojeda, C.; Cepeda-Quilindo, B.; González-Maya, J.F.; Eizirik, E.; Bonilla-Sánchez, A.; Buitrago, D.L.; Pulido-Santacruz, P.; Ramírez-Chaves, H.E. Distribution of the northern pampas cat, Leopardus garleppi, in northern South America, confirmation of its presence in Colombia and genetic analysis of a controversial record from the country. Mammalia 2023, 87, 606–614. [Google Scholar] [CrossRef]
- Marín-Puerta, S. Leopardus narinensis (Ruiz-García, Pinedo-Castro y Shostell, 2023) es un Sinónimo Menor de Leopardus Pardinoides (Gray 1867): La Necesidad de Buenas Prácticas en las Propuestas Nomenclaturales y Taxonómicas. Bachelor’s Thesis, University of Caldas, Manizales, Colombia, 2024. [Google Scholar]
- Johnson, W.E.; O’Brien, S.J. Phylogenetic reconstruction of the Felidae using 16S rRNA and NADH-5 mitochondrial genes. J. Mol. Evol. 1997, 44, S98–S116. [Google Scholar] [CrossRef]
- Culver, M.; Johnson, W.E.; Pecon-Slattery, J.; O’Brien, S.J. Genomic ancestry of the American puma (Puma concolor). J. Hered. 2000, 91, 186–197. [Google Scholar] [CrossRef]
- Napolitano, C.; Johnson, W.E.; Sanderson, J.; O’Brien, S.J.; Hoelzel, A.R.; Freer, R.; Dunstone, N.; Ritland, K.; Ritland, C.E.; Poulin, E. Phylogeography and population history of Leopardus guigna, the smallest American felid. Conserv. Genet. 2014, 15, 631–653. [Google Scholar] [CrossRef]
- Johnson, W.E.; Culver, M.; Iriarte, J.A.; Eizirik, E.; Seymour, K.L.; O’Brien, S.J. Tracking the evolution of the elusive Andean Mountain cat (Oreailurus jacobita) from mitochondrial DNA. J. Hered. 1998, 89, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, O.; Hebler, J.; Poinar, H.N.; Paabo, S.; Vigilant, L. Unreliable mtDNA data due to nuclear insertions: A cautionary tale from analysis of humans and other great apes. Mol. Ecol. 2004, 13, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Raaum, R.L.; Sterner, K.N.; Noviello, C.M.; Stewart, C.B.; Disotell, T.R. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: Concordance with fossil and nuclear DNA evidence. J. Hum. Evol. 2005, 48, 237–257. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Larkin, A. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Bradley, R.D.; Baker, R.J. A test of the genetic species concept: Cytochrome-b sequences and mammals. J. Mammal. 2001, 82, 960–973. [Google Scholar] [CrossRef]
- Kartavtsev, Y. Divergence at Cyt-b and Co-1 mtDNA genes on different taxonomic levels and genetics of speciation in animals. Mitochondrial DNA 2011, 22, 55–65. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; de Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. Ser. B 2003, 270 (Suppl. S1), S96–S99. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Kakusan, T.A. Aminosan: Two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Resour. 2011, 11, 914–921. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Intraspecific gene genealogies: Trees grafting into networks. Trends Ecol. Evol. 2001, 16, 37–45. [Google Scholar] [CrossRef]
- Lanave, C.; Preparata, G.; Saccone, C.; Serio, G. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 1984, 20, 86–93. [Google Scholar] [CrossRef]
- Tavare, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 1986, 17, 57–86. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Monmonier, M.S. Maximum-difference barriers: An alternative numerical regionalization method. Geogr. Anal. 2010, 5, 245–261. [Google Scholar] [CrossRef]
- Miller, M.P. Alleles In space (AIS): Computer software for the joint analysis of interindividual spatial and genetic information. J. Hered. 2005, 96, 722–724. [Google Scholar] [CrossRef]
- Dupanloup, I.; Schneider, S.; Excoffier, L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 2002, 11, 2571–2581. [Google Scholar] [CrossRef]
- Manel, S.; Schwartz, M.L.; Luikart, G.; Taberlet, P. Landscape genetics: Combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Manni, F.; Guerard, E.; Heyer, E. Geographic patterns of (genetic, morphologic, linguistic) variation: How barriers can be detected by using Monmonier’s algorithm. Hum. Biol. 2004, 76, 173–190. [Google Scholar] [CrossRef]
- Watson, D.F. Contouring: A Guide to the Analysis and Display of Spatial Data; Pergamon Press: New York, NY, USA, 1992. [Google Scholar]
- Brouns, G.; De Wulf, A.; Constales, D. Delaunay triangulation algorithms useful for multibeam echosounding. J. Surv. Eng. 2003, 129, 79–84. [Google Scholar] [CrossRef]
- Corander, J.; Marttinen, P.; Sirén, J.; Tang, J. BAPS: Bayesian Analysis of Population Structure; Department of Mathematics, ÅboAkademi University: Helsinki, Finland, 2013; Available online: http://www.abo.fi/mnf/mate/jc/smack_index_eng.html (accessed on 18 December 2023).
- ESRI. ArcGIS Pro 3.2.2 Desktop; Environmental Systems Research Institute: Redlands, CA, USA, 2023. [Google Scholar]
- Azurduy, H. Nota sobre el primer espécimen de museo para Leopardus tigrinus en Bolivia. Kempffiana 2005, 1, 47–50. [Google Scholar]
- Johnson, W.E.; Eizirik, E.; Pecon-Slattery, J.; Murphy, W.J.; Antunes, A.; Teeling, E.; O’Brien, S.J. The late Miocene radiation of modern Felidae: A genetic assessment. Science 2006, 311, 73–77. [Google Scholar] [CrossRef]
- Li, G.; Figueiró, H.V.; Eizirik, E.; Murphy, W.J. Recombination-aware phylogenomics reveals the structured genomic landscape of hybridizing cat species. Mol Biol. Evol. 2019, 36, 2111–2126. [Google Scholar] [CrossRef]
- Van der Hammen, T. La paleoecología de Suramérica Tropical. Cuarenta años de investigación de la historia del medio ambiente y de la vegetación. In Historia, Ecología y Vegetación; Corporación Colombiana para la Amazonía-Araracuara: Bogotá, Colombia, 1992; pp. 1–411. [Google Scholar]
- Gregory-Wodzicki, K.M. Uplift history of the central and northern Andes: A review. Geol. Soc. Amer. Bull. 2000, 112, 1091–1105. [Google Scholar] [CrossRef]
- Hibbins, M.S.; Hahn, M.W. Phylogenomic approaches to detecting and characterizing introgression. Genetics 2022, 220, iyab173. [Google Scholar] [CrossRef]
- Figueiró, H.V.; Li, G.; Trindade, F.J.; Assis, J.; Pais, F.; Fernandes, G.; Santos, S.H.D.; Hughes, G.M.; Komissarov, A.; Antunes, A.; et al. Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci. Adv. 2017, 3, e1700299. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Jaramillo, M.F.; Cáceres-Martínez, C.H.; Shostell, J.M. The Phylogeographic Structure of the mountain coati (Nasuella olivacea; Procyonidae, Carnivora), and phylogenetic relationships with the other coati species (Nasua nasua and Nasua narica) as inferred by mitochondrial DNA. Mammal. Biol. 2020, 100, 521–548. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Jaramillo, M.F.; López, J.B.; Rivillas, Y.; Leguizamón, N.; Bello, A.; Shostell, J.M. Mitochondrial and karyotypic evidence reveals a lack of support for the genus Nasuella (Procyonidae, Carnivora). J. Vertebrate Biol. 2022, 71, 21040. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Jaramillo, M.F.; Shostell, J.M. How Many Taxa or Groups are within Nasua nasua and Nasuella olivacea (Procyonidae, Carnivora)? The Mitochondrial Reconstruction of the Complex Evolutionary History of the Coatis throughout the Neotropics and Some Insights into the Systematics of the Genus Bassarycion. J. Phylogenetics Evol. Biol. 2022, 10, 206. [Google Scholar]
- Slon, V.; Mafessoni, F.; Vernot, B.; de Filippo, C.; Grote, S.; Viola, B.; Hajdinjak, M.; Peyrégne, S.; Nagel, S.; Brown, S.; et al. The genome of the offspring of a Neanderthal mother and a Denisovan father. Nat. Res. Lett. 2018, 561, 113–116. [Google Scholar] [CrossRef]
- Wurster-Hill, D.H.; Centerwall, W.R. The interrelationships of chromosome banding patterns in Procyonids, Viverrids and Felids. Cytogenet. Genome Res. 1982, 34, 178–192. [Google Scholar] [CrossRef]
- Davis, B.W.; Raudsepp, T.; Pearks Wilkerson, A.J.; Agarwala, R.; Schäffer, A.A.; Houck, M.; Chowdhary, B.P.; Murphy, W.J. A high-resolution cat radiation hybrid and integrated FISH mapping resource for phylogenomic studies across Felidae. Genomics 2009, 93, 299–304. [Google Scholar] [CrossRef]
- Cho, Y.S.; Hu, L.; Hou, H.L.; Lee, H.; Xu, J.H.; Kwon, S.; Oh, S.; Kim, H.M.; Jho, S.; Kim, S.; et al. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat. Commun. 2013, 4, 2433. [Google Scholar] [CrossRef]
- Harrison, R.G.; Larson, E.L. Hybridization, introgression, and the nature of species boundaries. J. Hered. 2014, 105, 795–809. [Google Scholar]
- Gillespie, R.G.; Bennett, G.M.; De Meester, L.; Feder, J.L.; Fleischer, R.C.; Harmon, L.J.; Hendry, A.P.; Knope, M.L.; Mallet, J.; Martin, C.; et al. Comparing adaptive radiations across space, time, and taxa. J. Hered. 2020, 111, 1–20. [Google Scholar] [CrossRef]
- de Oliveira, T.G.; Fox-Rosales, L.A.; Ramírez-Fernández, J.D.; Cepeda-Duque, J.C.; Zug, R.; Sanchez-Lalinde, C.; Oliveira, M.J.R.; Marinho, P.H.D.; Bonilla-Sánchez, A.; Marques, M.C.; et al. Ecological modeling, biogeography, and phenotypic analyses setting the tiger cats’ hyperdimensional niches reveal a new species. Sci. Rep. 2024, 14, 2395. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.E. The Bogota cat (Felis pardinoides, Gray). Ann. Mag. Nat. Hist. Ser. 1874, 13, 475. [Google Scholar] [CrossRef][Green Version]
- Thomas, O. Notes on Neotropical mammals of the genera Felis, Hapale, Oryzomys, Akodon, and Ctenomys, with descriptions of new species. Ann. Mag. Nat. Hist. Ser. 7 1903, 12, 234–243. [Google Scholar] [CrossRef][Green Version]
- Hernández-Camacho, J.; Hurtado, A.; Ortiz, R.; Walschburger, T. Unidades biogeográficas de Colombia. In La Diversidad Biológica de Iberoamérica; Halffter, I.G., Ed.; Acta Zoológica Mexicana, Instituto de Ecología, A.C.: Mexico City, Mexico, 1992; pp. 105–151. [Google Scholar]
- Haffer, J. Speciation in Amazonian forest birds. Science 1969, 165, 131–137. [Google Scholar] [CrossRef]
- Prance, G.T. Phytogeographica support for the theory of Pleistocene Forest refuges in the Amazon basin, based on evidence from distribution patterns in Chrysobalanaceae, Dichapetalaceae and Lecythidacea. Acta Amaz. 1973, 3, 5–28. [Google Scholar] [CrossRef]
- Prance, G.T. The origin and evolution of the Amazon flora. Interciencia 1978, 3, 207–222. [Google Scholar]
- Brown, K.S. Paleoecological and regional patterns of evolution in neotropical forest butterflies. In Biological Diversification in the Tropics; Prance, G.T., Ed.; Columbia University Press: New York, NY, USA, 1982; pp. 255–308. [Google Scholar]
- Hernández Camacho, J.; Walschburger, T.; Ortiz-Quijano, R.; Hurtado-Guerra, A. Origen y distribución de la biota suramericana y colombiana. In La Diversidad Biológica de Iberoamérica; Halffter, I.G., Ed.; Acta Zoológica Mexicana, Instituto de Ecología, A.C.: Mexico City, Mexico, 1992; pp. 55–104. [Google Scholar]
- Hernández Camacho, J.; Hurtado-Guerra, A.; Ortiz-Quijano, R.; Walschburger, T. Centros de endemismos en Colombia. In La Diversidad Biológica de Iberoamérica; Halffter, I.G., Ed.; Acta Zoológica Mexicana, Instituto de Ecología, A.C.: Mexico City, Mexico, 1992; pp. 175–190. [Google Scholar]
- Cossíos, D.; Walker, R.S.; Lucherini, M.; Ruiz-García, M.; Angers, B. Population structure and conservation of a high-altitude specialist, the Andean cat Leopardus jacobita. Endanger. Species Res. 2012, 16, 283–294. [Google Scholar] [CrossRef]
- Duellman, W. Origin, Evolution and Dispersal of the Andean Herpetofauna; Ann Meet Herepetol League: Lawrence, KS, USA, 1977; pp. 1–7. [Google Scholar]
- Ruiz-García, M.; Arias-Vásquez, J.Y.; Castellanos, A.; Kolter, L.; Shostell, J.M. Molecular evolution (mitochondrial and nuclear microsatellites markers) in the Andean bear (Tremarctos ornatus; Ursidae, Carnivora): How many ESUs are there? In Conservation Genomics of Mammals; Ortega, J., Maldonado, J.E., Eds.; Springer: New York, NY, USA, 2020; pp. 165–193. [Google Scholar]
- Kull, C.; Grosjean, M.; Veit, H. Modeling modern and Late Pleistocene glacio-climatological conditions in the north Chilean Andes (29–30 degrees S). Clim. Chang. 2002, 52, 359–381. [Google Scholar] [CrossRef]
- Marin, J.C.; Casey, C.S.; Kadwell, M.; Yaya, K.; Hoces, D.; Olazabal, J.; Rosadio, R.; Rodriguez, J.; Spotorno, A.; Bruford, M.W.; et al. Mitochondrial phylogeography and demographic history of the Vicuña: Implications for conservation. Heredity 2007, 99, 70–80. [Google Scholar] [CrossRef]
- Yensen, E.; Tarifa, T. Galictis cuja. Mam. Spec. 2003, 728, 1–8. [Google Scholar] [CrossRef]
- Sheffield, S.R.; Thomas, H.H. Mustela frenata. Mam. Spec. 1997, 570, 1–9. [Google Scholar] [CrossRef]
- Imbrie, L.J.; Hays, J.D.; Martinson, D.G.; McIntyre, A. The orbital theory of Pleistocene climate: Support from a revised chronology of the marine δ18O record. In Milankovitch and Climate; Berger, A., Imbrie, J., Hays, J.D., Kukla, G., Saltzman, B., Eds.; D Reidel: Dordrecht, The Netherlands; London, UK, 1984; pp. 269–305. [Google Scholar]
- Gottelli, D.; Marino, J.; Sillero-Zubiri, C.; Funk, S.M. The effect of the last glacial age on speciation and population structure of the endangered Ethiopian wolf (Canis simensis). Mol. Ecol. 2004, 13, 2275–2286. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. Population structure and estimates of gene flow in the homosporous fern Polystichum munitum. Evolution 1987, 41, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.G.; Cosson, J.F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef]
- Sersic, A.N.; Cosacov, A.; Cocucci, A.A.; Johnson, L.A.; Pozner, R.; Avila, L.J.; Sites, J.W.; Morando, M. Emerging phylogeographical patterns of plants and terrestrial vertebrates from Patagonia. Biol. J. Linn. Soc. 2011, 103, 475–494. [Google Scholar] [CrossRef]
- Schreber, J.C.D. Die Säugethiere in Abbildungen nach der Natur mit Beschreibungen; v. 1; Wolfgang: Erlangen, Germany, 1775; 2(14), pl. 99, 100; 2(15): pl. 101, 101B, 105, 106; 2(16): pl. 108. [Google Scholar]
- Schreber, J.C.D. Die Säugethiere in Abbildungen nach der Natur mit Beschreibungen; v. 2; Wolfgang: Erlangen, Germany, 1777; p. 13, pl. 110B; v. 3, pt. 22, pp. 384, 387, 392; v. 3, pt. 23, pp. 393, 394; 396, 397; 407; v. 3, pt. 24, pp. 412, 414; v. 3, pt. 25, pl. 104B, 109B. [Google Scholar]
- Buffon, G. L’Histoire Naturelle, Générale et Particulière, avec la Description du Cabinet du Roi; L’Imprimerie Royale: Paris, France, 1765; Tome XIII (Quadrupèdes X); pp. 1–368. [Google Scholar]
- Gray, J.E. Notes on certain species of cats in the collection of the British Museum. Proc. Zool. Soc. Lond. 1867, 14, 394–405. [Google Scholar]
- Hensel, R.F. Beitrage zur Kenntniss der Saugethiere Sud-Brasiliens; Abhandlungen der Königlich Preussischen Akademie der Wissenschaft: Berlin, Germany, 1872; pp. 1–130. [Google Scholar]
- Allen, J.A. New South American mammals. Bull. Amer. Mus. Nat. Hist. 1915, 34, 625–634. [Google Scholar]
- Allen, J.A. Notes on the synonymy and nomenclature of the smaller spotted cats of Tropical America. Bull. Amer. Mus. Nat. Hist. 1919, 41, 341–419. [Google Scholar]
- Cabrera, A.; Yepes, J. Mamíferos Sud-Americanos (Vida, Costumbres y Descripción); Compañía Argentina de Editores, Soc. de Resp. Ltda: Buenos Aires, Argentina, 1940; pp. 1–344. [Google Scholar]
- Weigel, I. Das Fellmuster der wildlebenden Katzenarten und der Hauskatze in vergleichender und stammesgeschichtlicher Hinsicht. Saügetier. Mitt. 1961, 9, 1–120. [Google Scholar]
- Pinedo-Castro, M.; Ruiz-García, M. Filogeografía del margay (Leopardus wiedii; Felidae, Carnivora): Determinación de posibles subespecies mediante marcadores mitocondriales. Mastozool. Neotrop. 2020, 27, 103–125. [Google Scholar] [CrossRef]
- Schwartz, J.H. Pseudopotto martini: A new genus and species of extant lorisiform primate. In Anthropological Papers of the AMNH; American Museum of Natural History: New York, NY, USA, 1996; Volume 78, pp. 1–14. [Google Scholar]
- Stump, D.P. Taxonomy of the Genus Perodicticus. Ph.D. Thesis, University of Pittburgh, Pittsburgh, PA, USA, 2005; pp. 1–199. [Google Scholar]
- Pääbo, S. Neanderthal Man: In Search of Lost Genomes; Basic Books: New York, NY, USA, 2014. [Google Scholar]
- White, M.J.D. Modes of Speciation; W.H. Freeman: New York, NY, USA, 1978; pp. 1–455. [Google Scholar]
- Li, X.; Jayaprasad, S.; Einarsdottir, E.; Cooper, S.J.B.; Suh, A.; Kawakawi, T.; Palacios-Gimenez, O.M. Chromosome-level genoma assembly of the morabine grasshopper Vandiemenella viatica 19. Sci. Data 2024, 11, 997. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Jaramillo, M.F.; Sánchez-Castillo, S.; Castillo, M.I.; Pinto, C.M.; Shostell, J.M. Effects of Sample Size in the Determination of the True Number of Haplogroups or ESUs Within a Species with Phylogeographic and Conservation Purposes: The Case of Cebus albifrons in Ecuador, and the Kinkajous and Coatis Throughout Latin America. In Molecular Ecology and Conservation Genetics of Neotropical Mammals; Nardelli, M., Túnez., J.I., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2021; pp. 101–148. [Google Scholar]





| A | |||||||
| Comparisons | Nariño Cat 2001 | Nariño Cat 2007 | Nariño Cat 2017 | Nariño Cat 2023 | |||
| Different temporal samples of the Nariño cat | With 2007 sample: 0.5 ± 0.5; with 2017 sample: 5.1 ± 1.4; with 2023 sample: 2.0 ± 0.8 | With 2017 sample: 5.9 ± 0.5; with 2023 sample: 2.0 ± 0.8 | With 2023 sample: 5.6 ± 1.5 | ------------------- | |||
| Tigrina from Circasia, Quindío, Colombia | 6.0 ± 1.5 | 5.2 ± 1.4 | 9.4 ± 2.1 | 3.8 ± 1.2 | |||
| Tigrina run over on the northern highway in the outskirts of Bogotá, Colombia | 4.4 ± 1.3 | 3.7 ± 1.2 | 7.6 ± 1.8 | 2.5 ± 0.9 | |||
| Tigrina run over in the road Bogotá- La Calera, Colombia | 4.4 ± 1.3 | 3.7 ± 1.2 | 7.6 ± 1.8 | 2.3 ± 0.9 | |||
| Tigrina from Astorquiza et al. [23], Caldas, Colombia | 7.1 ± 1.7 | 6.3 ± 1.6 | 10.7 ± 2.3 | 4.9 ± 1.4 | |||
| Tigrina from Astorquiza et al. [23], Antioquia, Colombia | 6.7 ± 1.7 | 5.9 ± 1.5 | 10.2 ± 2.2 | 4.5 ± 1.3 | |||
| Tigrina from the Nariño department, Colombia, but physically different to the Nariño cat | 5.6 ± 1.5 | 4.8 ± 1.4 | 8.9 ± 2.0 | 3.4 ± 1.1 | |||
| Tigrina from Intag, Imbabura, Ecuador | 6.3 ± 1.6 | 5.5 ± 1.5 | 9.8 ± 2.2 | 4.1 ± 1.2 | |||
| Two tigrinas from Costa Rica, Central America | 7.5 ± 1.5 | 6.7 ± 1.7 | 10.2 ± 2.2 | 6.0 ± 1.6 | |||
| Leopardus colocola braccatus | 5.5 ± 1.5 | 6.3 ± 1.6 | 2.3 ± 0.9 | 6.0 ± 1.6 | |||
| Leopardus colocola steinbachi | 5.2 ± 1.4 | 5.9 ± 1.5 | 2.0 ± 0.8 | 5.6 ± 1.5 | |||
| Leopardus guigna | 7.5 ± 1.8 | 7.5 ± 1.8 | 10.2 ± 2.2 | 6.0 ± 1.6 | |||
| Two Leopardus geoffroyi | 7.5 ± 1.8 and 9.2 ± 2.0 | 7.5 ± 1.9 and 8.8 ± 2.0 | 9.7 ± 2.1 | 6.0 ± 1.6 and 7.2 ± 1.8 | |||
| B | |||||||
| Comparisons | Nariño cat 2001 | Nariño cat 2017 | Nariño cat 2023 | ||||
| Different temporal samples of the Nariño cat | With 2017 sample: 5.4 ± 0.009; with 2023 sample: 4.4 ± 0.008 | With 2023 sample: 2.3 ± 0.005 | ------------------- | ||||
| Tigrina from Circasia, Quindío, Colombia | 5.2 ± 0.009 | 5.7 ± 0.009 | 3.9 ± 0.007 | ||||
| Tigrina run over on the northern highway in the outskirts of Bogotá, Colombia | 5.0 ± 0.008 | 2.7 ± 0.006 | 1.1 ± 0.004 | ||||
| Tigrina run over in the road Bogotá- La Calera, Colombia | 5.0 ± 0.008 | 2.7 ± 0.006 | 1.1 ± 0.004 | ||||
| Tigrina from Astorquiza et al. [23], Caldas, Colombia | 6.2 ± 0.010 | 3.7 ± 0.007 | 2.0 ± 0.005 | ||||
| Tigrina from Astorquiza et al. [23], Antioquia, Colombia | 6.2 ± 0.010 | 3.7 ± 0.007 | 1.8 ± 0.005 | ||||
| Tigrina from the Nariño department, Colombia, but physically different to the Nariño cat | 6.2 ± 0.010 | 4.0 ± 0.007 | 2.3 ± 0.005 | ||||
| Tigrina from Intag, Imbabura, Ecuador | 5.5 ± 0.009 | 5.8 ± 0.009 | 4.0 ± 0.007 | ||||
| Two tigrinas from Costa Rica, Central America | 6.2 ± 0.010 | 4.3 ± 0.008 | 3.1 ± 0.006 | ||||
| Leopardus colocola braccatus | 6.4 ± 0.010 | 4.1 ± 0.008 | 5.9 ± 0.009 | ||||
| Leopardus colocola steinbachi | 6.1 ± 0.009 | 4.1 ± 0.008 | 5.9 ± 0.009 | ||||
| Leopardus guigna | 6.4 ± 0.010 | 5.9 ± 0.009 | 4.9 ± 0.008 | ||||
| Leopardus geoffroyi | 6.2 ± 0.010 | 5.6 ± 0.009 | 5.1 ± 0.009 | ||||
| Comparisons | Tigrina from Circasia, Quindío, Colombia | Tigrina Run over on the Northern Highway in the Outskirts of Bogotá, Colombia | Tigrina Run over in the Road Bogotá- La Calera, Colombia | Tigrina from Astorquiza et al. [23], Caldas, Colombia | Tigrina from Astorquiza et al. [23], Antioquia, Colombia | Tigrina from the Nariño Department, Colombia, but Physically Different to the Nariño Cat | Tigrina from Intag, Imbabura, Ecuador | Two tigrinas from Costa Rica, Central America |
|---|---|---|---|---|---|---|---|---|
| Tigrina from Circasia, Quindío, Colombia | - | 2.7 ± 0.006 | 2.7 ± 0.006 | 3.2 ± 0.007 | 3.2 ± 0.007 | 3.2 ± 0.007 | 3.6 ± 0.007 | 4.0 ± 0.007 |
| Tigrina run over on the northern highway in the outskirts of Bogotá, Colombia | 1.3 ± 0.7 | - | 0.0 ± 0.0 | 0.9 ± 0.003 | 0.9 ± 0.003 | 1.2 ± 0.004 | 2.8 ± 0.006 | 1.9 ± 0.005 |
| Tigrina run over in the road Bogotá- La Calera, Colombia | 1.3 ± 0.7 | 0.0 ± 0.0 | - | 0.9 ± 0.003 | 0.9 ± 0.003 | 1.2 ± 0.004 | 2.8 ± 0.006 | 1.9 ± 0.005 |
| Tigrina from Astorquiza et al. [23], Caldas, Colombia | 2.3 ± 0.9 | 2.3 ± 0.9 | 2.3 ± 0.9 | - | 0.22 ± 0.002 | 1.7 ± 0.005 | 3.3 ± 0.007 | 1.7 ± 0.005 |
| Tigrina from Astorquiza et al. [23], Antioquia, Colombia | 2.0 ± 0.8 | 2.0 ± 0.8 | 2.0 ± 0.8 | 0.3 ± 0.3 | - | 1.7 ± 0.005 | 3.3 ± 0.007 | 1.7 ± 0.005 |
| Tigrina from the Nariño department, Colombia, but physically different to the Nariño cat | 1.0 ± 0.6 | 1.0 ± 0.6 | 1.0 ± 0.6 | 2.0 ± 0.8 | 1.6 ± 0.7 | - | 2.5 ± 0.006 | 2.2 ± 0.005 |
| Tigrina from Intag, Imbabura, Ecuador | 1.6 ± 0.7 | 1.6 ± 0.7 | 1.6 ± 0.7 | 2.7 ± 1.0 | 2.3 ± 0.9 | 0.6 ± 0.5 | - | 3.9 ± 0.007 |
| Two tigrinas from Costa Rica, Central America | 3.4 ± 1.1 | 3.4 ± 1.1 | 3.4 ± 1.1 | 2.3 ± 0.7 | 2.0 ± 0.8 | 3.0 ± 1.1 | 3.7 ± 1.2 | - |
| Compared with Consensus Sequence of the Nariño Cat | mtND5 Gene | Complete Mitogenomes |
|---|---|---|
| Tigrina G1 | 3.2 ± 1.0 | 1.3 ± 0.005 |
| Leopardus colocola braccatus | 2.6 ± 0.8 | 4.2 ± 0.006 |
| Leopardus colocola steinbachi | 2.9 ± 0.9 | 4.1 ± 0.005 |
| Leopardus colocola garleppi | 3.4 ± 1.0 | -------------- |
| Leopardus guigna | 6.1 ± 1.4 | 4.2 ± 0.008 |
| Leopardus geoffroyi | 6.2 ± 1.3 | 3.8 ± 0.007 |
| Leopardus geoffroyi introgressed by L. colocola in Bolivia and in Argentina | 3.2 ± 0.9 | 2.8 ± 0.004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-García, M.; Vega, J.; Pinedo-Castro, M.; Shostell, J.M. The Nariño Cat, the Tigrinas and Their Problematic Systematics and Phylogeography: The Real Story. Animals 2025, 15, 1891. https://doi.org/10.3390/ani15131891
Ruiz-García M, Vega J, Pinedo-Castro M, Shostell JM. The Nariño Cat, the Tigrinas and Their Problematic Systematics and Phylogeography: The Real Story. Animals. 2025; 15(13):1891. https://doi.org/10.3390/ani15131891
Chicago/Turabian StyleRuiz-García, Manuel, Javier Vega, Myreya Pinedo-Castro, and Joseph Mark Shostell. 2025. "The Nariño Cat, the Tigrinas and Their Problematic Systematics and Phylogeography: The Real Story" Animals 15, no. 13: 1891. https://doi.org/10.3390/ani15131891
APA StyleRuiz-García, M., Vega, J., Pinedo-Castro, M., & Shostell, J. M. (2025). The Nariño Cat, the Tigrinas and Their Problematic Systematics and Phylogeography: The Real Story. Animals, 15(13), 1891. https://doi.org/10.3390/ani15131891





