Genome-Wide Association Analysis and Genomic Selection for Growth Traits in Grass Carp (Ctenopharyngodon idella)
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Ethics Statement
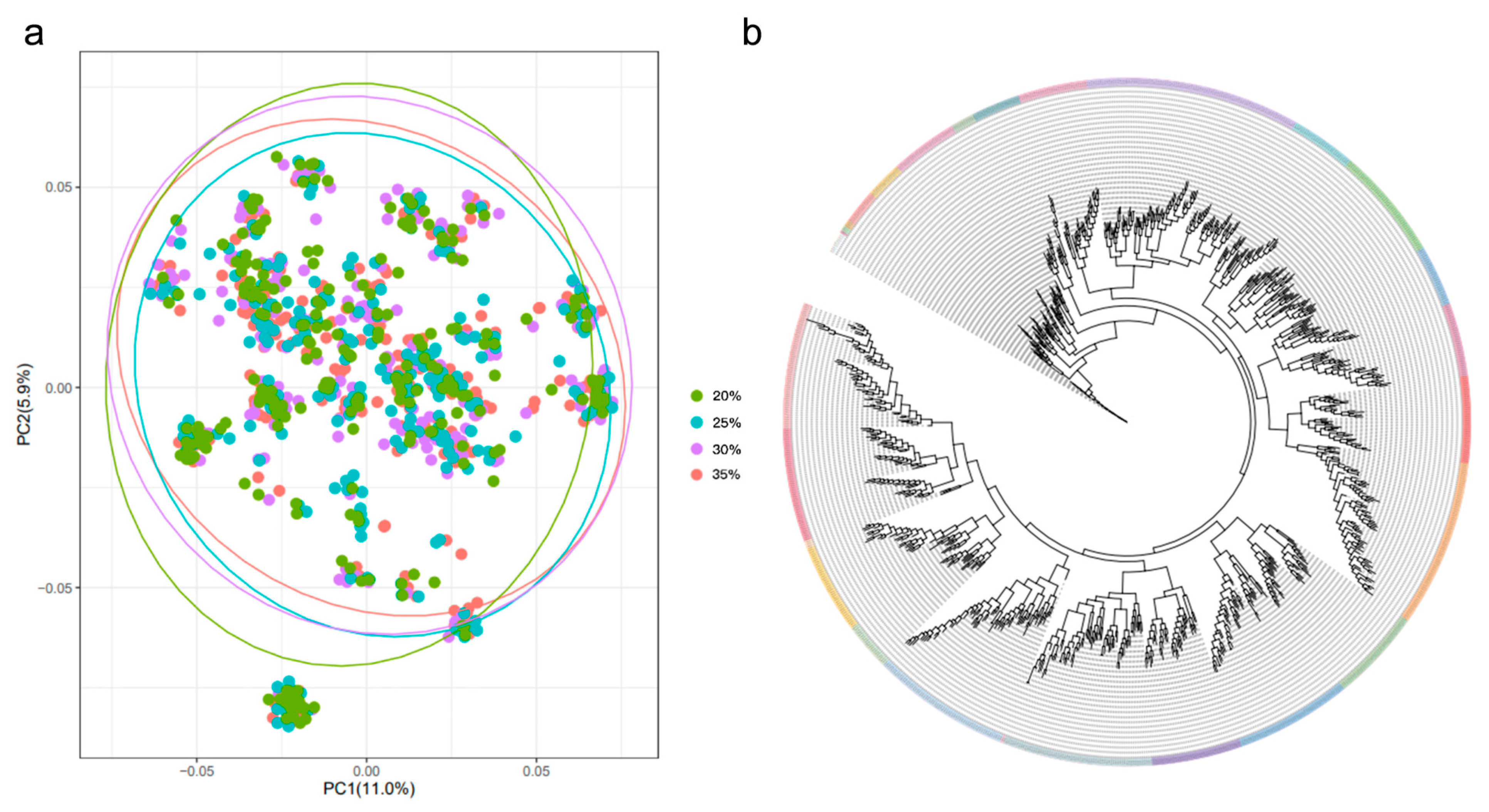
2.2. Sample Preparation for GWAS
2.3. SNP Array Genotyping
2.4. Genome-Wide Association Study
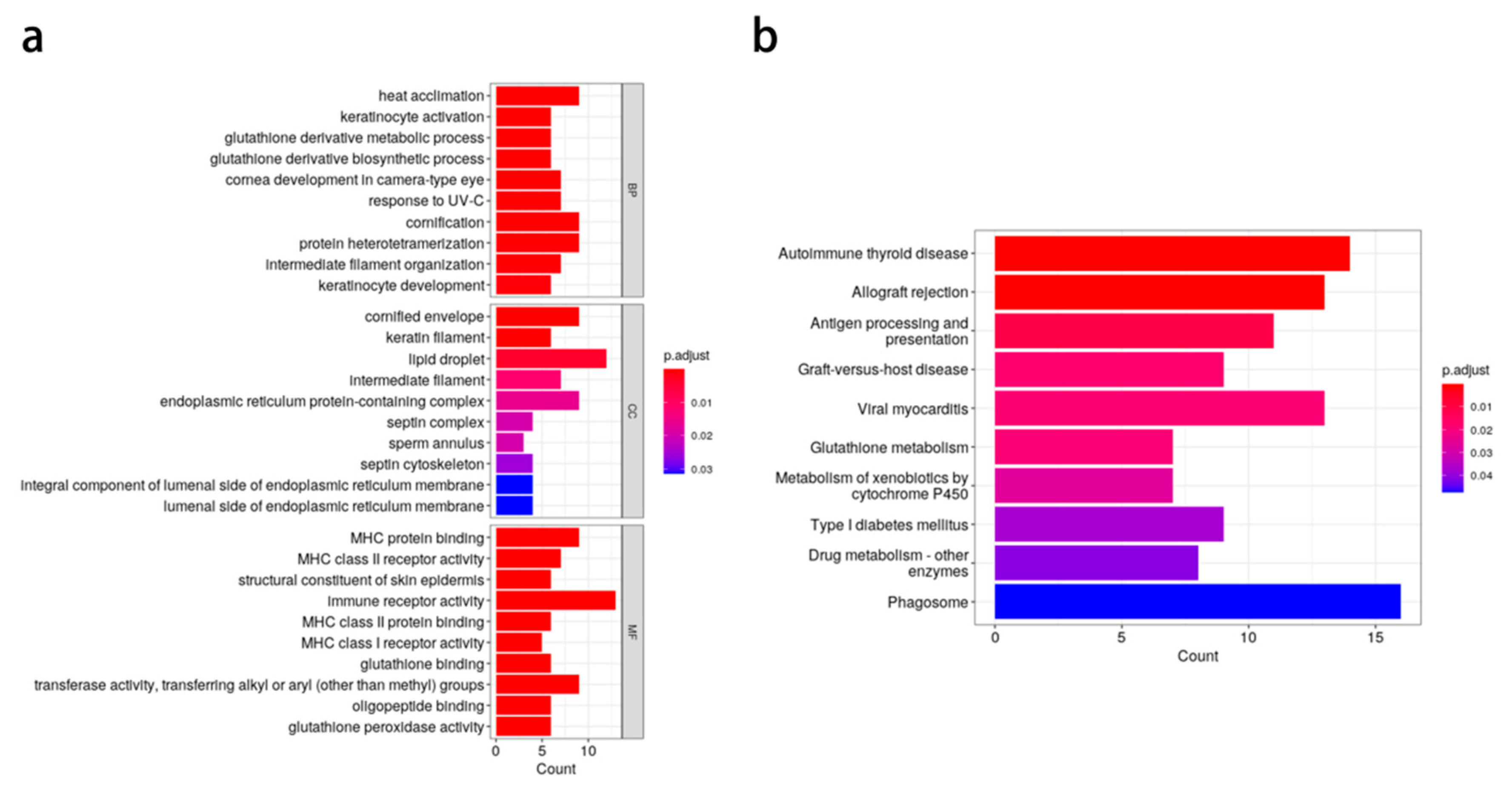
2.5. Candidate Gene Acquisition and Functional Annotation
3. Results
3.1. Statistics of Growth Traits
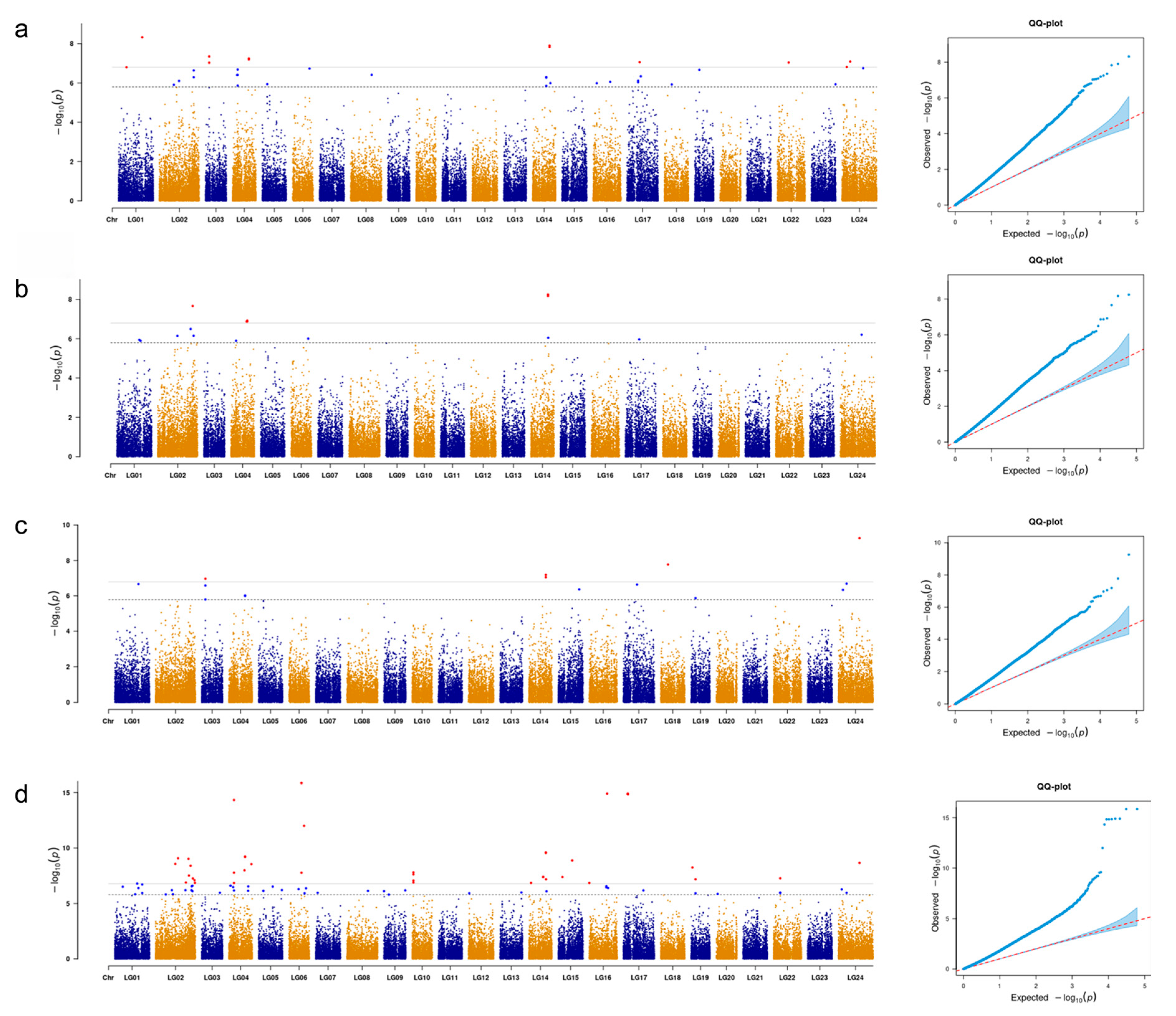
3.2. Genome-Wide Association Analysis
3.3. Candidate Gene Prediction
3.4. Genomic Prediction Within Population
4. Discussion
4.1. Genetic Architecture of Protein-Dependent Growth Traits
4.2. Advancing Precision Breeding Through Genomic Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, C.; Tang, H.; Jiang, Y.; Lu, H.; Chen, Q.; Gui, L.; Qiu, J.; Xu, X.; Li, J.; Shen, Y. Growth Performance and Selection Signatures Revealed by Whole-Genome Resequencing in Genetically Selected Grass Carp (Ctenopharyngodon idella). Aquaculture 2024, 587, 740885. [Google Scholar] [CrossRef]
- Jin, Y.; Tian, L.; Xie, S.; Guo, D.; Yang, H.; Liang, G.; Liu, Y. Interactions between Dietary Protein Levels, Growth Performance, Feed Utilization, Gene Expression and Metabolic Products in Juvenile Grass Carp (Ctenopharyngodon idella). Aquaculture 2015, 437, 75–83. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The Advantages and Limitations of Trait Analysis with GWAS: A Review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Sodeland, M.; Gaarder, M.; Moen, T.; Thomassen, M.; Kjøglum, S.; Kent, M.; Lien, S. Genome-Wide Association Testing Reveals Quantitative Trait Loci for Fillet Texture and Fat Content in Atlantic Salmon. Aquaculture 2013, 408–409, 169–174. [Google Scholar] [CrossRef]
- Jin, Y.; Zhou, T.; Geng, X.; Liu, S.; Chen, A.; Yao, J.; Jiang, C.; Tan, S.; Su, B.; Liu, Z. A Genome-Wide Association Study of Heat Stress-Associated SNPs in Catfish. Anim. Genet. 2017, 48, 233–236. [Google Scholar] [CrossRef]
- Reis Neto, R.V.; Yoshida, G.M.; Lhorente, J.P.; Yáñez, J.M. Genome-Wide Association Analysis for Body Weight Identifies Candidate Genes Related to Development and Metabolism in Rainbow Trout (Oncorhynchus mykiss). Mol. Genet. Genom. 2019, 294, 563–571. [Google Scholar] [CrossRef]
- Castillo-Juárez, H.; Campos-Montes, G.R.; Caballero-Zamora, A.; Montaldo, H.H. Genetic Improvement of Pacific White Shrimp [Penaeus (Litopenaeus) Vannamei]: Perspectives for Genomic Selection. Front. Genet. 2015, 6, 93. [Google Scholar] [CrossRef]
- D’Agaro, E.; Favaro, A.; Matiussi, S.; Gibertoni, P.P.; Esposito, S. Genomic Selection in Salmonids: New Discoveries and Future Perspectives. Aquacult. Int. 2021, 29, 2259–2289. [Google Scholar] [CrossRef]
- Su, S.; Raouf, B.; He, X.; Cai, N.; Li, X.; Yu, J.; Li, J.; Yu, F.; Wang, M.; Tang, Y. Genome Wide Analysis for Growth at Two Growth Stages in A New Fast-Growing Common Carp Strain (Cyprinus carpio L.). Sci. Rep. 2020, 10, 7259. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, J.-Y.; Kim, N. GMStool: GWAS-Based Marker Selection Tool for Genomic Prediction from Genomic Data. Sci. Rep. 2020, 10, 19653. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, H.-C.; Wang, Y.-Y.; Huang, H.-Z. Genome-Wide Association Study Reveals Growth-Related SNPs and Candidate Genes in Mandarin Fish (Siniperca chuatsi). Aquaculture 2022, 550, 737879. [Google Scholar] [CrossRef]
- Bonferroni, C. Teoria Statistica Delle Classi e Calcolo Delle Probabilita. In Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commericiali di Firenze; Primarily Firenze University Press (FUP): Firenze, Italy, 1936; Volume 8, pp. 3–62. [Google Scholar]
- Guo, J.; Zhang, M.; Wang, S.; Xu, X.; Shen, Y.; Li, J. A High-Density Genetic Linkage Map and QTL Mapping for Growth Related Traits in Grass Carp (Ctenopharyngodon idella). Aquaculture 2022, 552, 738041. [Google Scholar] [CrossRef]
- Zhou, S.; Yang, L.; Li, J.; Shen, Y. Genome-Wide Association Study for Growth Traits in Black Carp (Mylopharyngodon piceus). Aquaculture 2025, 595, 741582. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, L.; Wu, X.; Li, B.; Huang, W.; Weng, Z.; Lin, Z.; Song, L.; Guo, Y.; Meng, Z.; et al. Identification of Candidate Growth-Related SNPs and Genes Using GWAS in Brown-Marbled Grouper (Epinephelus fuscoguttatus). Mar. Biotechnol. 2020, 22, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-Y.; Luo, L.-F.; Wang, Z.-Y.; Yu, Y.; Nie, C.-H.; Guo, X.-Z.; Gao, Z.-X. Identification of Novel SNPs and Candidate Genes Significantly Affecting Growth in Grass Carp (Ctenopharyngodon idella) through GWAS Analysis. Aquaculture 2024, 591, 741129. [Google Scholar] [CrossRef]
- Chen, G.; Zhou, Y.; Yu, X.; Wang, J.; Luo, W.; Pang, M.; Tong, J. Genome-Wide Association Study Reveals SNPs and Candidate Genes Related to Growth and Body Shape in Bighead Carp (Hypophthalmichthys nobilis). Mar. Biotechnol. 2022, 24, 1138–1147. [Google Scholar] [CrossRef]
- Ghosal, A.; Lambrecht, N.; Subramanya, S.B.; Kapadia, R.; Said, H.M. Conditional Knockout of the Slc5a6 Gene in Mouse Intestine Impairs Biotin Absorption. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G64–G71. [Google Scholar] [CrossRef]
- Wang, J.; Yu, X.; Chen, G.; Zhang, Y.; Tong, J. Molecular Characterization of Slc5a6a and Its Association with Growth and Body Conformation in Bighead Carp (Hypophthalmichthys nobilis). Aquac. Rep. 2022, 27, 101394. [Google Scholar] [CrossRef]
- Lee, E.-R.; Kim, J.-Y.; Kang, Y.-J.; Ahn, J.-Y.; Kim, J.-H.; Kim, B.-W.; Choi, H.-Y.; Jeong, M.-Y.; Cho, S.-G. Interplay between PI3K/Akt and MAPK Signaling Pathways in DNA-Damaging Drug-Induced Apoptosis. Biochim. Biophys. Acta 2006, 1763, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Pimm, J.; McQuillin, A.; Thirumalai, S.; Lawrence, J.; Quested, D.; Bass, N.; Lamb, G.; Moorey, H.; Datta, S.R.; Kalsi, G.; et al. The Epsin 4 Gene on Chromosome 5q, Which Encodes the Clathrin-Associated Protein Enthoprotin, Is Involved in the Genetic Susceptibility to Schizophrenia. Am. J. Human Genet. 2005, 76, 902–907. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guipponi, M.; Toh, M.-Y.; Tan, J.; Park, D.; Hanson, K.; Ballana, E.; Kwong, D.; Cannon, P.Z.F.; Wu, Q.; Gout, A.; et al. An Integrated Genetic and Functional Analysis of the Role of Type II Transmembrane Serine Proteases (TMPRSSs) in Hearing Loss. Human Mutat. 2008, 29, 130–141. [Google Scholar] [CrossRef]
- Hao, Y.; Jia, X.; Yuan, L.; Liu, Y.; Gui, L.; Shen, Y.; Li, J.; Xu, X. Genome-Wide Association Study Reveals Growth-Related SNPs and Candidate Genes in Grass Carp (Ctenopharyngodon idella). Aquaculture 2023, 577, 739979. [Google Scholar] [CrossRef]
- Azodi, C.B.; Bolger, E.; McCarren, A.; Roantree, M.; de los Campos, G.; Shiu, S.-H. Benchmarking Parametric and Machine Learning Models for Genomic Prediction of Complex Traits. G3 Genes Genomes Genet. 2019, 9, 3691–3702. [Google Scholar] [CrossRef]
- Tsai, H.-Y.; Hamilton, A.; Tinch, A.E.; Guy, D.R.; Gharbi, K.; Stear, M.J.; Matika, O.; Bishop, S.C.; Houston, R.D. Genome Wide Association and Genomic Prediction for Growth Traits in Juvenile Farmed Atlantic Salmon Using a High Density SNP Array. BMC Genom. 2015, 16, 969. [Google Scholar] [CrossRef]
- Lu, S.; Liu, Y.; Yu, X.; Li, Y.; Yang, Y.; Wei, M.; Zhou, Q.; Wang, J.; Zhang, Y.; Zheng, W.; et al. Prediction of Genomic Breeding Values Based on Pre-Selected SNPs Using ssGBLUP, WssGBLUP and BayesB for Edwardsiellosis Resistance in Japanese Flounder. Genet. Sel. Evol. 2020, 52, 49. [Google Scholar] [CrossRef]
- Correa, K.; Bangera, R.; Figueroa, R.; Lhorente, J.P.; Yáñez, J.M. The Use of Genomic Information Increases the Accuracy of Breeding Value Predictions for Sea Louse (Caligus rogercresseyi) Resistance in Atlantic Salmon (Salmo salar). Genet. Sel. Evol. 2017, 49, 15. [Google Scholar] [CrossRef]
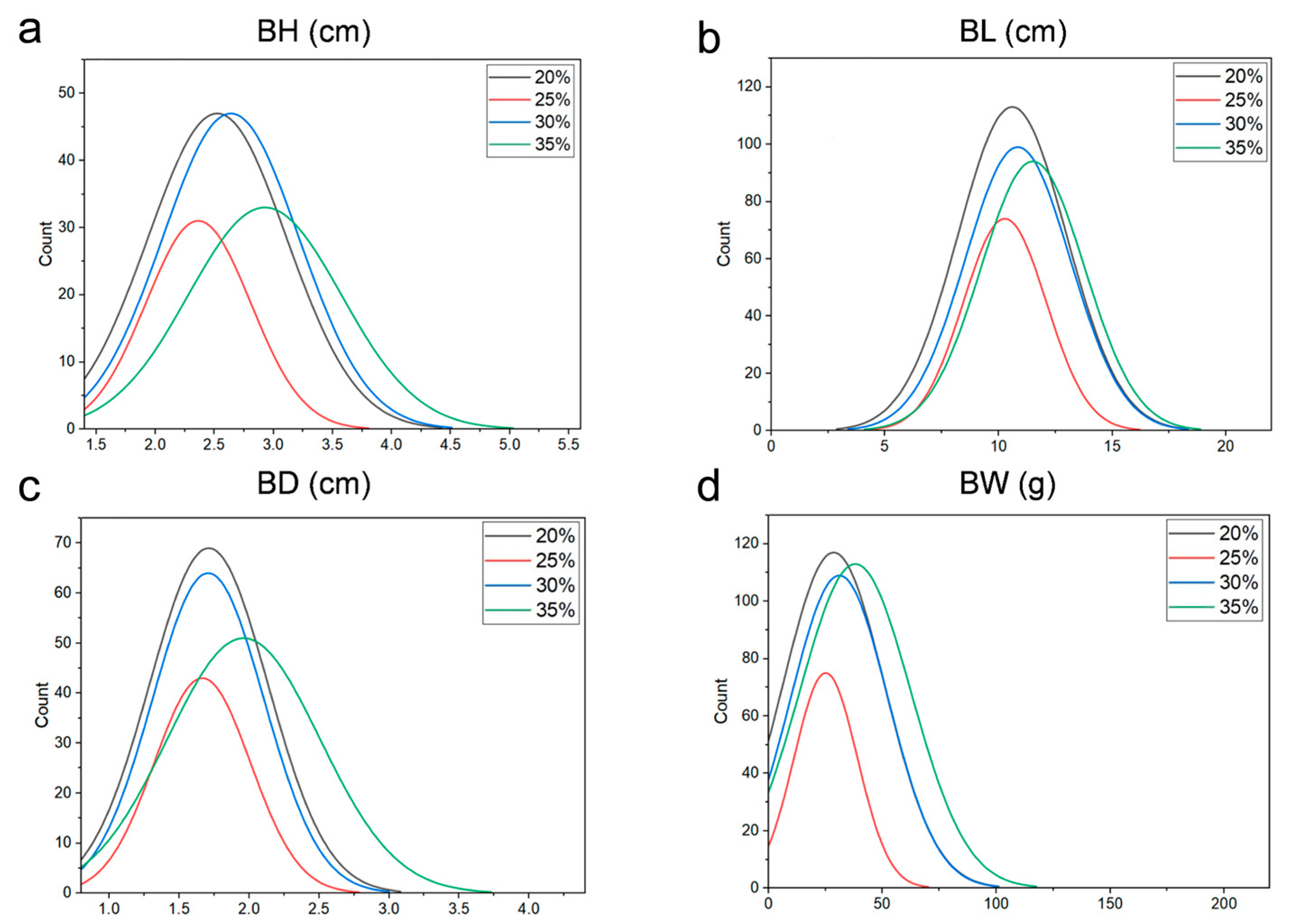
| 20% Protein Feed | 25% Protein Feed | 30% Protein Feed | 35% Protein Feed | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BL (cm) | BH (cm) | BD (cm) | BW (g) | BL (cm) | BH (cm) | BD (cm) | BW (g) | BL (cm) | BH (cm) | BD (cm) | BW (g) | BL (cm) | BH (cm) | BW (cm) | BD (g) | |
| average value | 10.60 | 2.52 | 1.71 | 28.60 | 10.27 | 2.36 | 1.67 | 25.11 | 18.84 | 2.64 | 1.71 | 31.10 | 11.51 | 2.93 | 1.96 | 30.08 |
| max value | 19.61 | 4.61 | 3.24 | 153.94 | 15.67 | 3.64 | 2.74 | 73.7 | 19.23 | 4.64 | 3.41 | 141.65 | 21.40 | 5.37 | 4.24 | 207.97 |
| min value | 7.11 | 1.61 | 1.00 | 7.04 | 6.19 | 1.70 | 1.08 | 8.68 | 1.41 | 1.70 | 1.06 | 9.40 | 7.37 | 1.76 | 1.09 | 9.17 |
| Phenotype | GWAS | Method | Select Markers | RRB (Train/Val./Test) | RF (Train/Val./Test) | DNN (Train/Val./Test) | CNN (Train/Val./Test) |
|---|---|---|---|---|---|---|---|
| BH | glm | RRB | 2138 | 0.96/0.86/0.51 | 0.99/0.45/0.57 | 0.00/0.14/0.00 | 0.85/0.47/0.39 |
| RRB_BTS | 2195 | 0.96/0.86/0.51 | 0.99/0.47/0.60 | 0.08/0.27/0.44 | 0.85/0.53/0.53 | ||
| mlm | RRB | 1529 | 0.95/0.87/0.65 | 0.99/0.52/0.63 | 0.02/0.10/0.27 | 0.87/0.64/0.67 | |
| RRB_BTS | 1572 | 0.95/0.87/0.65 | 0.99/0.54/0.63 | 0.07/0.06/0.10 | 0.87/0.61/0.65 | ||
| emmax | RRB | 1678 | 0.95/0.88/0.70 | 0.99/0.50/0.58 | 0.05/0.06/0.00 | 0.86/0.63/0.61 | |
| RRB_BTS | 1701 | 0.95/0.87/0.70 | 0.99/0.51/0.58 | 0.00/0.12/−0.26 | 0.85/0.67/0.55 | ||
| emmaxQ | RRB | 1579 | 0.94/0.86/0.71 | 0.99/0.50/0.55 | 0.12/0.25/0.11 | 0.87/0.64/0.59 | |
| RRB_BTS | 1636 | 0.94/0.86/0.71 | 0.99/0.49/0.57 | 0.00/0.10/−0.12 | 0.87/0.68/0.66 | ||
| BL | glm | RRB | 2094 | 0.95/0.85/0.50 | 0.98/0.45/0.65 | 0.90/0.57/0.35 | 0.88/0.57/0.52 |
| RRB_BTS | 2154 | 0.94/0.85/0.51 | 0.98/0.48/0.66 | 0.86/0.56/0.41 | 0.92/0.54/0.42 | ||
| mlm | RRB | 1478 | 0.94/0.87/0.71 | 0.99/0.54/0.70 | 0.89/0.70/0.59 | 0.93/0.66/0.66 | |
| RRB_BTS | 1515 | 0.94/0.87/0.71 | 0.99/0.50/0.71 | 0.87/0.69/0.71 | 0.93/0.66/0.64 | ||
| emmax | RRB | 1758 | 0.950.88/0.72 | 0.99/0.48/0.66 | 0.88/0.69/0.59 | 0.92/0.60/0.55 | |
| RRB_BTS | 1780 | 0.95/0.87/0.71 | 0.99/0.47/0.66 | 0.89/0.71/0.65 | 0.91/0.65/0.61 | ||
| emmaxQ | RRB | 1687 | 0.94/0.87/0.72 | 0.99/0.46/0.68 | 0.89/0.70/0.60 | 0.92/0.62/0.68 | |
| RRB_BTS | 1719 | 0.94/0.87/0.72 | 0.99/0.45/0.69 | 0.90/0.66/0.64 | 0.91/0.62/0.56 | ||
| BD | glm | RRB | 2070 | 0.96/0.86/0.34 | 0.98/0.49/0.49 | 0.15/0.29/0.21 | 0.72/0.47/0.30 |
| RRB_BTS | 2134 | 0.96/0.85/0.34 | 0.98/0.50/0.48 | 0.12/0.15/−0.01 | 0.78/0.46/0.35 | ||
| mlm | RRB | 1640 | 0.94/0.84/0.59 | 0.99/0.53/0.55 | 0.09/0.11/0.15 | 0.79/0.58/0.39 | |
| RRB_BTS | 1649 | 0.94/0.83/0.60 | 0.99/0.53/0.56 | −0.01/0.10/0.17 | 0.80/0.58/0.53 | ||
| emmax | RRB | 1880 | 0.95/0.85/0.62 | 0.98/0.47/0.50 | 0.10/0.18/−0.05 | 0.82/0.59/0.55 | |
| RRB_BTS | 1880 | 0.95/0.85/0.62 | 0.98/0.47/0.50 | 0.10/0.18/−0.04 | 0.82/0.59/0.55 | ||
| emmaxQ | RRB | 1903 | 0.94/0.85/0.60 | 0.99/0.48/0.51 | 0.03/0.18/0.18 | 0.80/0.62/0.57 | |
| RRB_BTS | 1949 | 0.94/0.85/0.60 | 0.99/0.49/0.51 | 0.03/0.10/0.35 | 0.84/0.57/0.49 | ||
| BW | glm | RRB | NA | NA | NA | NA | NA |
| RRB_BTS | NA | NA | NA | NA | NA | ||
| mlm | RRB | NA | NA | NA | NA | NA | |
| RRB_BTS | NA | NA | NA | NA | NA | ||
| emmax | RRB | 1524 | 0.94/0.86/0.74 | 0.98/0.48/0.70 | 0.93/0.78/0.72 | 0.89/0.68/0.64 | |
| RRB_BTS | 1554 | 0.94/0.86/0.74 | 0.98/0.48/0.70 | 0.94/0.77/0.73 | 0.91/0.67/0.56 | ||
| emmaxQ | RRB | 1533 | 0.94/0.86/0.76 | 0.98/0.47/0.71 | 0.94/0.78/0.79 | 0.89/0.68/0.68 | |
| RRB_BTS | 1571 | 0.94/0.86/0.76 | 0.98/0.48/0.71 | 0.94/0.77/0.76 | 0.93/0.68/0.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Yu, Q.; Lv, W.; Sheng, T.; Gui, L.; Qiu, J.; Xu, X.; Li, J. Genome-Wide Association Analysis and Genomic Selection for Growth Traits in Grass Carp (Ctenopharyngodon idella). Animals 2025, 15, 1888. https://doi.org/10.3390/ani15131888
Chen Y, Yu Q, Lv W, Sheng T, Gui L, Qiu J, Xu X, Li J. Genome-Wide Association Analysis and Genomic Selection for Growth Traits in Grass Carp (Ctenopharyngodon idella). Animals. 2025; 15(13):1888. https://doi.org/10.3390/ani15131888
Chicago/Turabian StyleChen, Yuxuan, Qiaozhen Yu, Wenyao Lv, Tao Sheng, Lang Gui, Junqiang Qiu, Xiaoyan Xu, and Jiale Li. 2025. "Genome-Wide Association Analysis and Genomic Selection for Growth Traits in Grass Carp (Ctenopharyngodon idella)" Animals 15, no. 13: 1888. https://doi.org/10.3390/ani15131888
APA StyleChen, Y., Yu, Q., Lv, W., Sheng, T., Gui, L., Qiu, J., Xu, X., & Li, J. (2025). Genome-Wide Association Analysis and Genomic Selection for Growth Traits in Grass Carp (Ctenopharyngodon idella). Animals, 15(13), 1888. https://doi.org/10.3390/ani15131888





