Simple Summary
Increasing the yield of healthy piglets per litter is economically important for pig production, yet constrained by biological factors affecting sow reproductive capacity. This study investigated genetic solutions using the Large White breed, known for its high reproductive performance. We recorded nine litter traits across 2096 sows, including total births and piglet weights. Advanced DNA analysis identified 153,782 genetic variations linked to these traits. A new breeding method improved sow productivity predictions by 6–13% compared to conventional approaches. Six genomic regions influencing litter size were discovered, notably the GPR12 as a pivotal gene for litter size. These findings advance genomic strategies for improving reproductive efficiency in swine.
Abstract
(1) Background: Litter size traits are critical for pig breeding efficiency but pose challenges due to low heritability and sex-limited influences. This study aimed to elucidate the genetic architecture and identify candidate genes for these traits in Large White pigs using genomic selection (GS) and genome-wide association analyses (GWAS). (2) Methods: This study utilized phenotypic data from nine litter size traits in Large White sows. Genotyping-by-sequencing (GBS) was performed to obtain genotype data, retaining 153,782 high-quality SNPs after quality control. Genetic evaluation was conducted using single-step genomic best linear unbiased prediction (ssGBLUP), with genetic parameters (heritability and genetic correlations) estimated via an animal model (repeatability model). To assess prediction accuracy, 10-fold cross-validation was employed to compare traditional BLUP with ssGBLUP. Furthermore, a single-step genome-wide association study (ssGWAS) integrated genomic information and pedigree-based relationship matrices to screen for significant SNPs associated with litter size traits across the genome. Functional analysis of key candidate genes was subsequently conducted based on ssGWAS results. (3) Results: Heritabilities for litter traits ranged from 0.01 to 0.06. ssGBLUP improved genomic prediction accuracy by 6.38–13.33% over BLUP. Six genomic windows explaining 1.07–1.77% of genetic variance were identified via ssGWAS, highlighting GPR12 on SSC11 as a key candidate gene linked to oocyte development. (4) Conclusions: This study demonstrates the efficacy of ssGBLUP for low-heritability traits and identifies GPR12 as a pivotal gene for litter size. Prioritizing NHB and LBWT in breeding programs could enhance genetic gains while mitigating adverse effects on piglet health. These findings advance genomic strategies for improving reproductive efficiency in swine.
1. Introduction
Litter size is a crucial breeding trait for pig production efficiency and profitability. Improving the total number of births and the number of healthy births in pigs has long been a primary goal for breeders. However, genetic improvement of litter size is challenging due to its low heritability and the significant impact of sex-limited factors [1,2,3,4]. Additionally, increasing the total number of piglets born has significantly increased this trait but also heightened piglet mortality [5]. Furthermore, an increase in the number of piglets often results in more piglets with low birth weights, which is associated with various long-term negative effects [6,7]. Genomic selection (GS) is a vital and effective strategy for determining litter size traits, production traits, and other economic traits, particularly for traits with low heritability or sex-limited influence such as litter size [8,9].
In GS, incorporating prior information, such as candidate genes or quantitative trait loci (QTLs) affecting traits, can enhance GS accuracy [10,11,12]. Genome-wide association studies (GWAS), a common method, have allowed researchers to discover QTLs associated with traits. Notable examples include the identification of ESR1, FSH, EPOR, NR4A1, GNB2L1, and PRLR variants associated with litter size traits in pigs [13]. To date, approximately 57,041 QTLs are associated with different traits in pigs, and more than 7568 QTLs are associated with reproduction traits in pigs (https://www.animalgenome.org/cgi-bin/QTLdb/SS/index; accessed on 5 April 2025) [13]. A general method for conducting GWAS involves fitting a univariate linear mixed model for marker association tests with a single phenotype or fitting a Bayesian sparse linear mixed model to estimating variance in phenotypes explained by single nucleotide polymorphism (SNP) heritability [14,15,16]. When ungenotyped animals have more phenotypic information, the common method for GWAS becomes difficult to perform, or there will be a loss of accuracy [17,18]. Compared to common methods, single-step GWAS (ssGWAS) can better integrate phenotypes, pedigrees, and genotyping data [19,20]. Additionally, ssGWAS can reduce noise, highlight the most significant peaks, and be adapted to complex ordinal models, such as repeatability models [18].
In many scenarios, whole-genome sequencing remains challenging, costly, and time-consuming in agricultural breeding. However, by simplifying genome sequencing, the cost of genotyping animals can be greatly reduced, and imputation-based strategies can significantly increase the density of genetic markers [21]. High-density genetic markers can effectively identify linkage disequilibrium (LD) in large populations and address issues related to genetic diversity and spatial structure [22]. Additionally, for non-model species without reference genomes, genotyping-by-sequencing (GBS) technology is well suited for revealing genetic diversity, genotyping, and genetic structure [23].
Large White pigs, one of the most commonly used pig breeds, are often employed as dam lines in modern commercial settings due to their advantages of large litters, high milk production, and strong maternal instincts. Therefore, the objective of this work was to perform high-density GS and ssGWAS using GBS technology in the Large White population to improve the accuracy of estimated breeding values and map the genomic regions associated with litter size traits.
2. Materials and Methods
2.1. Animals and Traits
The animals used in this study were sourced from 10 swine breeding herds in southern China, owned and managed by Wens Foodstuff Group Co., Ltd. (Yunfu, Guangdong, China). Data were collected from these herds between December 2010 and December 2020. The total number of litter sizes (total number born, TNB) observed was 170,027, which was available from 62,208 Large White sows. The study considered TNB, number of piglets born alive (NBA), number of healthy births (NHB), rate of NHB (rNHB), number of weak births (NWB), number of deformed fetuses (NDF), number of stillborn fetuses (NSB), mummified pigs (MUMM), and litter weight at birth (LBWT). Litters were removed from the data if they contained at most 3 piglets in the TNB, whereas litters of 22 piglets or larger were all considered “22”. Records of reproductive performance of 8 pairs or more were all considered “8”, and only 22,139 sows had a single observation. TNB is the number of all piglets born at the same birth, which equals the sum of NBA, NSB, and MUMM. NBA equals the sum of NHB, NWB, and NDF for all surviving piglets. NHB means that the birth weight is not less than 1.0 kg; in contrast, NWB means that the body weight is less than 1.0 kg, but NSB and NWB have no genetic defects. rNHB is the ratio of NHB to TNB. NDF refers to piglets with genetic defects, such as limb defects or reproductive system disorders. NSB refers to the number of dead piglets at birth or during pregnancy. In MUMM, a pig fetus dies or degenerates after the 35th day of pregnancy without abortion. LBWT is the total weight of all NBA within 24 h of birth.
2.2. Genotyping and Genotype Quality Control
The extraction of genomic DNA from ear tissue and the genotyping of whole-genome SNPs by the GBS technique have been explained in detail in previous studies. In detail, genomic DNA was digested with EcoRI/MspI restriction enzymes, ligated to barcoded adapters, and PCR-amplified. After Agilent 2100 fragment size selection, raw reads were quality-filtered (base quality ≥ 20; length ≥ 85 bp) and aligned to Sscrofa11.1 using TASSEL5.0, with genotype imputation via Beagle5.1 [24]. Quality control of the genotypes was performed using Plink (version 1.9 beta) [25], and the following criteria were used: SNPs with a minor allele frequency less than 0.01, SNPs that deviated significantly from Hardy–Weinberg equilibrium (HWE) (p < 10−6), SNPs with an individual call rate lower than 0.95, SNPs with a call rate lower than 0.99, and SNPs where the number of individuals in both homozygous genotypes (i.e., AA and BB) was less than 30 were excluded. After quality control, a total of 153,782 SNPs on autosomal chromosomes of Sus scrofa (SSCs) in 2096 pigs were available for further analysis.
2.3. Estimation of Genetic Parameters and Prediction Accuracy of GS
Genetic correlations and heritabilities between the nine litter size traits were estimated. The statistical model for describing the phenotypic data was as follows:
where Y is the vector of phenotypic records, b is the vector of the fixed discrete effect of farrowing farm-year-month and parity, a is the vector of additive genotypic values (genomic estimated breeding value, GEBV), t is the vector of permanent environmental effects, e is the vector of residuals, and X, Z, and W are the known design matrices for b, a, and t, respectively. A univariate animal model and a bivariate animal model were used to calculate heritability and genetic correlation, respectively. Genetic parameters (heritabilities and correlations) were estimated using the BLUPF90 software suite (version 2.60). Genetic correlation magnitudes were classified as follows: very high—|r| ≥ 0.80; high—0.40 ≤ |r| < 0.80; low—0.2 < |r| < 0.40; very low—|r| < 0.2.
In this study, 10-fold cross validation was used to estimate the prediction performance of best linear unbiased prediction (BLUP) and single-step genomic BLUP (ssGBLUP) [26]. Each dataset was randomly divided into ten parts, nine of which were used as training datasets, and the remainder were used for validation. The prediction accuracy of the GEBV was as follows:
where is the expected accuracy of predicting genetic values [27,28], PEV is the predictor error variance, and is the additive genetic variance.
2.4. ssGWAS
The ssGWAS method is an improvement on the inverse of a numerator relationship matrix A−1, with H−1 replacing A−1 [29]:
where is the inverse of a genomic relationship matrix and where is a numerator relationship matrix for genotyped animals. The weighted genomic relationship matrix (G*) was as follows [30]:
where Z is a matrix of gene content adjusted for allele frequencies, D is a weight matrix for SNPs (initially D = I), and is a variance ratio renormalizing constant. According to VanRaden et al. [31]. The calculated SNP effects and weights for the GWAS were as follows [19]:
- (1)
- First, let D = I.
- (2)
- Calculate G.
- (3)
- The GEBV was calculated via ssGBLUP.
- (4)
- The GEBV is converted to calculate the SNP effects () as follows:
- (5)
- The SNP weights are calculated as follows:
- (6)
- The SNP weights are normalized to keep the total genetic variance constant.
- (7)
- Loop to 2.
A previous study of this population demonstrated that LD (measured as r2 = 0.2) extends to approximately 0.3 Mb [24]. To ensure our window size captured haplotypic blocks reflecting the population’s LD structure, we adopted 0.3 Mb as the genomic window for ssGWAS. The proportion of genetic variance explained by the i-th window of successive SNPs was as follows [18]:
where is the genetic value of the i-th window that consists of a region of successive SNPs located within 0.3 Mb, is the total genetic variance, is a vector genotype of the j-th SNP for all animals, is the effect of the j-th SNP within the i-th window, and m is the number of SNPs within the i-th window. Overlapping windows were considered for variance calculations. Therefore, the top SNP was defined as contributing approximately equally to the 0.3 Mb-adjacent SNP window. The BLUPF90 family of programs was used to perform these analyses [32,33,34,35].
2.5. Identification of Candidate Genes and Analysis of Functional Enrichment
The candidate QTLs associated with litter size traits in the Large White population were defined as genomic windows that explained more than 1% of the genetic variance, and genes that were less than 0.3 Mb away from the potential SNPs were selected and identified as candidate genes via the Ensembl database (https://www.ensembl.org/; accessed on 5 April 2025) [36]. To better understand the biological processes and pathways associated with these candidate genes, Metascape [37] (https://metascape.org/; accessed on 5 April 2025) was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. Metascape is based on the Homo sapiens (human) database and uses the default parameters. GO terms and KEGG pathways for which Fisher’s exact p value was less than 0.01 were considered significant.
3. Results
3.1. Descriptive Statistics for the Litter Size Traits
Statistics on the number of animals with or without records and genotypes in the Large White population are shown in Table 1. A total of 2096 pigs were genotyped, and 2036 of these also had phenotypic data recorded. Additionally, extensive genealogical databases were utilized in the study. Table 2 presents the descriptive statistics of the observed phenotypes. The coefficient of variation for the litter size traits ranged from 19% to 212%, indicating substantial phenotypic variation in the litter size traits within the Large White population. The kurtosis of TNB, NBA, NHB, and rNHB was close to zero, suggesting that these traits are suitable for animal models. In contrast, NDF, NSB, and MUMM may not be very suitable for animal models.

Table 1.
Statistics on the number of animals with or without records and genotypes in the Large White population.

Table 2.
Descriptive statistics for litter size traits a in the Large White population.
3.2. Heritabilities and Repeatabilities of the Litter Size Traits
The estimated variance components (additive genetic, permanent environmental, residual, and phenotypic variance), heritabilities, and repeatability of the litter size traits, along with their standard errors in the Large White population from the BLUP and ssGBLUP models, are shown in Table 3. No significant differences were observed between the two models. Heritability estimates for the litter size traits ranged from 0.01 to 0.06, indicating low heritability traits. Among the traits, TNB had the highest heritability (0.06), while NDF and MUMM had the lowest heritabilities (0.01). The repeatability estimates for the litter size traits ranged from 0.02 to 0.14. TNB and LBWT had the highest repeatability (0.14) and NDF had the lowest repeatability (0.02).

Table 3.
Estimated variance components and heritabilities for litter size a traits in the Large White population.
3.3. Genetic Correlations and Phenotypic Correlations Between the Litter Size Traits
The estimated genetic correlations and phenotypic correlations (lower and upper triangular sections, respectively), along with their standard errors in parentheses, between the nine litter size traits in the Large White population are shown in Table 4. High positive genetic correlations were estimated between TNB and other traits, except for rNHB. A very high positive genetic correlation of 0.90 was estimated between TNB and NBA. Negative correlations were estimated between rNHB and most traits (such as TNB, NBA, NWB, NDF, NSB, and MUMM), with a very high negative correlation of −0.78 between rNHB and NSB. Large differences were not observed between the genetic correlations and phenotypic correlations.

Table 4.
Genetic correlations (below the diagonal) and phenotypic correlations (above the diagonal) for litter size traits a in the Large White population.
3.4. Prediction Accuracies of (G) EBV with BLUP and ssGBLUP in Litter Size Traits
The expected accuracies of predicting genetic values (Rg) across litter size traits for EBV and GEBV selection in the Large White population are shown in Table 5. The expected accuracies of BLUP ranged from 0.47 to 0.64, while the expected accuracies of ssGBLUP ranged from 0.50 to 0.70. The expected accuracies indicated that ssGBLUP had greater accuracy in genomic selection compared to traditional BLUP, with an increase ranging from 6.38% to 13.33%. Additionally, the expected accuracies of both BLUP and ssGBLUP showed very little variation (~0.01).

Table 5.
Accuracy of (G)EBV in the validation group using the BLUP and ssGBLUP methods.
3.5. Summary of the ssGWAS Results for Litter Size Traits
The ssGWAS results were represented by the proportion of genetic variance explained by 0.3 Mb windows for the litter size traits (Figure 1). The genomic windows that explained more than 1.0% of the additive genetic variance in the litter size traits are shown in Table 6 and Tables S1–S5, along with the candidate genes near these genomic windows. In total, six genomic windows explaining between 1.07% and 1.77% of the additive genetic variance associated with litter size traits were identified. Additionally, two, one, one, one, and one genomic windows for TNB, NBA, NWB, NDF, and MUMM, respectively, were identified. No genomic windows were discovered for NHB, rNHB, NSB, and LBWT. In total, 26 candidate genes associated with litter size traits were identified, with the same candidate genes identified for TNB and NWB.
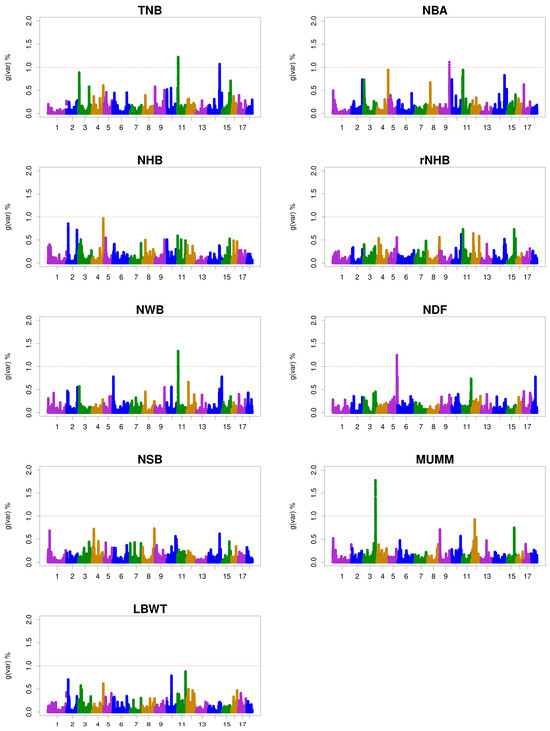
Figure 1.
Manhattan plots for the percentage of genetic variance by 0.3 Mb window for litter size traits. gVar (%) represents the proportion of genetic variance explained by the 0.3 Mb window. The horizontal coordinate represents the chromosomes of pigs: total number born (TNB), number of piglets born alive (NBA), number of healthy births (NHB), rate of NHB (rNHB), number of weak births (NWB), number of deform fetus (NDF), number of stillborn (NSB), mummified pigs (MUMM), litter weight at birth (LBWT), and gVar (%) = the proportion of the genetic variance explained by the 0.3 Mb-adjacent SNP window.

Table 6.
Details of genomic regions and genes associated with litter size traits a in the Large White population.
3.6. Functional Enrichment Analysis
Clusters with enriched terms related to litter size traits are shown in Table 7. The results of the GO analysis revealed that these genes were enriched in six functional categories, including signaling by receptor tyrosine kinases, cell morphogenesis involved in differentiation, cardiac muscle tissue development, negative regulation of cell differentiation, regulation of GTPase activity, and the RHO GTPase cycle (p < 0.01).

Table 7.
The significant pathways for the positional candidate genes for litter size traits in Large White pigs.
4. Discussion
4.1. Genetic Parameter Statistics
Heritability estimates for the litter size traits ranged from 0.01 to 0.06. Imboonta reported that the heritability of TNB in Thai Landrace pigs in the first four parities ranged from 0.02 to 0.11 [38]. Lundgren reported that the heritability of TNB in Landrace sows was 0.09 [39]. Camargo reported that the additive genetic variance and heritability of NBA in Landrace pigs were 0.90 and 0.09, respectively [40]. Damgaard reported that the heritability of NBA was 0.12 [41]. Lopez reported that heritability estimates for TNB ranged from 0.072 to 0.102, 0.090 to 0.099, and 0.109 to 0.121; from 0.087 to 0.110, 0.088 to 0.100, and 0.099 to 0.107 for NBA; and from 0.027 to 0.031, 0.050 to 0.053, and 0.073 to 0.081 for NSB in the Duroc, Landrace, and Yorkshire breeds, respectively [42]. Ogawa reported that the estimated heritabilities of TNB, NBA, NSB, and LBWT were 0.12, 0.12, 0.08, and 0.18, respectively, in Landrace pigs and 0.12, 0.10, 0.08, and 0.18, respectively, in Large White pigs [2]. These reports revealed low heritabilities in litter size traits. Compared to previous reports, our study found the lowest additive genetic variance and heritability.
Consistent with prior reports [43,44,45], TNB and NBA exhibit a very high genetic cor-relation (r ≥ 0.90), indicating similar selection effects. However, TNB-based selection is suboptimal due to its positive associations with NWB, NDF, NSB, and MUMM [46]. Alternatively, NHB and LBWT emerge as superior target traits given their contrasting correlation patterns: strong positive links with desirable traits (TNB/NBA/LBWT: NHB = 0.76/0.87/0.87; LBWT = 0.63/0.73/0.87); minimal associations with adverse outcomes (NWB/NDF/NSB/MUMM: NHB = 0.10/0.23/−0.11/0.11; LBWT = −0.04/0.23/−0.07/0.17). This supports Ogawa’s conclusion that LBWT optimizes NBA genetic improvement [2].
4.2. Genomic Prediction Accuracy
Genomic selection is widely regarded as a successful tool for genetic improvement in various livestock and plant species [47,48]. The single-step genomic best linear unbiased prediction (ssGBLUP) method has been extensively researched and applied [49]. In the present study, the accuracies of GEBV with ssGBLUP improved by 6.38% to 13.33% over the accuracies of the BLUP model for litter size traits. Teissier reported that the improvement between ssGBLUP and BLUP ranged from 5% to 7% [50]. However, the proportion of genetic variance explained by markers depends on the accuracy of GS. The size and family structure of the reference population may influence the accuracy of GS [51]. Additionally, identifying linkage disequilibrium depends on high-density genetic markers [52]. Thus, increasing the density of genetic markers may improve the accuracy of GS. Asora reported that accuracy increased as the number of markers and training sizes increased [53]. In this study, a high density of genetic markers and a large reference population significantly increased the accuracy of GS. In the future, we will continue to increase the size of the reference population and aim to improve or maintain the accuracy of GS.
4.3. Candidate Regions and Genes
In total, six genomic windows were identified. Among these significant windows, the region located at SSC11: 4.33–4.50 Mb explained 1.22% and 1.33% of the genetic variances for TNB and NWB, respectively. This region also explained relatively high genetic variance for NBA and rNHB (0.94%, 0.73%), with the top SNP located at SSC11: 4,423,721 bp. These findings indicate that these traits are highly genetically correlated. The gene adjacent to the top SNP (SSC11: 4,423,721 bp) was G protein-coupled receptor 12 (GPR12). Hinckley reported that GPR12 was detected in the oocytes of Xenopus laevis, mice, and rats. Overexpression of GPR12 in Xenopus laevis oocytes prevented progesterone-induced meiotic resumption. In reorganization systems, GPR12 and GPR3 appear to stimulate Gs activities in a ligand-independent fashion, suggesting that both GPR3 and GPR12 play key roles in controlling cAMP levels in the oocyte and in meiotic arrest [54]. Brown reported that GPR12 may be involved in physiological processes such as the maintenance of oocyte meiotic arrest and brain development, as well as pathological conditions such as metastatic cancer [55]. Zhang reported that GPR12 suppresses esophageal migration and promotes apoptosis in cancer and hypopharyngeal cancer [56]. Additionally, we determined the LD pattern of the SNPs in the region of SSC11: 4.0–4.8 Mb, which indicates a high level of LD between the top and nearby SNPs (Figure 2, left). These findings suggest potential selection signatures for litter size traits in the Large White population. Thus, we speculate that GPR12 is a strong candidate gene for determining litter size traits because of its key role in the mechanisms of oocyte development.
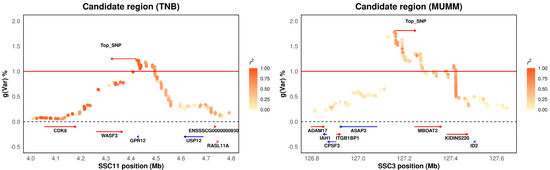
Figure 2.
Candidate region plots illustrating the two major candidate regions on SSC11 and SSC3. The results are displayed for the total number born (TNB) at approximately 4.0–4.8 Mb on SSC11 (left) and for mummified pigs (MUMM) at 126.8–127.8 Mb on SSC3 (right). Various levels of linkage disequilibrium between the primary SNP and surrounding SNPs are represented by distinct colors. gVar (%) denotes the proportion of the genetic variance explained by the 0.3 Mb-adjacent SNP window.
The second most important genomic window was located at SSC3: 127.1–127.4 Mb, explaining 1.77% of the genetic variance in MUMM. The genes adjacent to the top SNP (SSC3: 127,157,661) were ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 (ASAP2), and membrane-bound O-acyltransferase domain containing 2 (MBOAT2). Fujii reported that the novel driver gene ASAP2 is a potential druggable target in pancreatic cancer [57]. Tekola-Ayele reported that ASAP2 is associated with birth weight in humans and that the link between birth weight and placental DNA methylation is the opposite of what was previously reported in cord blood [58]. Tebani reported that ASAP2 is associated with malignancies in different tissues and organs [59]. Zhou reported that circ-MBOAT2 modulates tumor development and glutamine catabolism via the miR-433-3p/GOT1 axis in pancreatic cancer, suggesting circ-MBOAT2 may be a therapeutic target for pancreatic cancer [60]. We also determined the LD pattern of the top SNPs in the region around SSC3: 126.8–127.8 Mb (Figure 2, right). We did not discover a high level of LD between the top and nearby SNPs. Therefore, while ASAP2 and MBOAT2 are strong candidate genes for determining litter size traits, further study is needed.
We also discovered other candidate genes associated with litter size traits. For example, treatment interventions targeting CDK8 may have clinical benefits for beta-catenin-induced malignancies [61]. The WASF3 gene promotes the invasion and metastasis of breast cancer cells that have undergone epithelial-to-mesenchymal transition, making it a potential target for inhibiting breast cancer cell infiltration and metastasis [62]. USP12 acts as a regulator of neuron protein homeostasis and mHTT-mediated neurodegeneration [63]. The expression of RASL11A in primary colorectal tumors is lower than in normal mucosa [64]. Loss-of-function variants of biallelic WDR11 are associated with intellectual disability and microcephaly [65]. FGFR2 has been identified as a driver of intrahepatic disease [66]. RABGAP1 is expressed mainly in follicular, granular, and epidermal cells [67]. High expression of CACYBP in hepatocellular carcinoma patients with poor prognosis is required for hepatocellular carcinoma cell growth both in vitro and in vivo [68]. The MRPS-14 mutation can cause perinatal hypertrophic cardiomyopathy, lactate toxicity, growth retardation, deformities, and neurological involvement in neonates [69]. Loss of TNR results in nonprogressive neurodevelopmental disorders, with spasticity and transient visual impairment [70]. TMTC2 variants are associated with familial bipolar sensorineural hearing loss and auditory neuropathy spectrum disorders [71]. The KIDINS220 gene mutation, mediated by TrkA, plays a role in human brain ventriculomegaly [72]. These genes are associated with various human diseases, given the potential roles of the GO terms related to cell morphogenesis involved in differentiation. The GO enrichment results revealed that many genes are involved in cell morphogenesis and cardiac muscle tissue development.
5. Conclusions
In this study, the genetic parameters were estimated, and GS and ssGWAS were performed for the litter size traits of the Large White population. NHB and LBWT may be target traits, and their use may be beneficial for the genetic improvement of NBA. The accuracies of GEBV with ssGBLUP improved by 6.38% to 13.33% over the accuracies of the BLUP model in terms of litter size traits. ssGWAS results for these traits revealed a series of candidate genes, and GPR12 is a strong candidate gene for determining litter size traits. These findings reveal the complexity of the genetic mechanisms underlying litter size traits and provide guidance for genetic improvement through genetic selection in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani15121724/s1. Table S1: The explained genetic variance of SNPs within significant windows for TNB; Table S2: The explained genetic variance of SNPs within significant windows for NBA; Table S3: The explained genetic variance of SNPs within significant windows for NWB; Table S4: The explained genetic variance of SNPs within significant windows for NDF; Table S5: The explained genetic variance of SNPs within significant windows for MUMM.
Author Contributions
Conceptualization, C.T. and Z.W.; methodology, C.T.; software, Y.H.; validation, C.T.; formal analysis, Y.H. and Y.Z.; investigation, D.W., X.H. and C.T.; resources, J.Y. and C.T.; data curation, Y.H., D.W., X.H., Y.Z., J.Y. and C.T.; writing—original draft preparation, Y.H.; writing—review and editing, Y.H. and C.T.; visualization, C.T.; supervision, C.T. and Z.W.; project administration, C.T. and Z.W.; funding acquisition, Z.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Guangdong Province, China (Grant No. 2018B030313011).
Institutional Review Board Statement
All procedures related to the animals in this study were performed in strict accordance with the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of the People’s Republic of China (Approval Number: 2006-398). All the experimental designs for the animals were approved by the ethics committee of South China Agricultural University (SCAU, Guangzhou, China). The experimental animals were not anesthetized or euthanized for the purpose of this study. Wens Foodstuff Group Co., Ltd. (Guangdong, China) confirmed that the sample collection of this study was approved. There was no use of human participants, data, or tissues.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genotype data were deposited into the European Variation Archive database under accession number PRJEB79359 and are available at the following URL: https://www.ebi.ac.uk/eva/?eva-study=PRJEB79359. The phenotypic data are not publicly accessible, as the populations consist of the nucleus herd of Wens Foodstuff Group Co., Ltd. However, this data is available from the corresponding author upon reasonable request.
Acknowledgments
Many thanks to all the staff at the pig core breeding farms of Wens Foodstuff Group Co., Ltd. (Yunfu, Guangdong, China) for record collection.
Conflicts of Interest
Authors Xiaoyan He, Dan Wu, and Yuxing Zhang were employed by Wens Foodstuff Group Co., Ltd. This company provided the pigs used in this study from a swine breeding herd that it owns and manages. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Hanenberg, E.H.A.T.; Knol, E.F.; Merks, J.W.M. Estimates of Genetic Parameters for Reproduction Traits at Different Parities in Dutch Landrace Pigs. Livest. Prod. Sci. 2001, 69, 179–186. [Google Scholar] [CrossRef]
- Ogawa, S.; Konta, A.; Kimata, M.; Ishii, K.; Uemoto, Y.; Satoh, M. Estimation of Genetic Parameters for Farrowing Traits in Purebred Landrace and Large White Pigs. Anim. Sci. J. 2019, 90, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Tan, C.; Hu, X.; Wang, A.; Wu, Z. Genetic Parameters for Reproductive Traits at Different Parities in Large White Pigs. J. Anim. Sci. 2018, 96, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gan, M.; Yang, X.; Zhu, P.; Luo, Y.; Liu, B.; Zhu, K.; Cheng, W.; Chen, L.; Zhao, Y.; et al. Estimation of genetic parameters of pig reproductive traits. Front. Vet. Sci. 2023, 10, 1172287. [Google Scholar] [CrossRef]
- Högberg, A.; Rydhmer, L. A Genetic Study of Piglet Growth and Survival. Acta Agric. Scand. Sect. Anim. Sci. 2000, 50, 300–303. [Google Scholar] [CrossRef]
- Feldpausch, J.A.; Jourquin, J.; Bergstrom, J.R.; Bargen, J.L.; Bokenkroger, C.D.; Davis, D.L.; Gonzalez, J.M.; Nelssen, J.L.; Puls, C.L.; Trout, W.E.; et al. Birth Weight Threshold for Identifying Piglets at Risk for Preweaning Mortality. Transl. Anim. Sci. 2019, 3, 633–640. [Google Scholar] [CrossRef]
- Stange, K.; Miersch, C.; Sponder, G.; Röntgen, M. Low birth weight influences the postnatal abundance and characteristics of satellite cell subpopulations in pigs. Sci. Rep. 2020, 10, 6149. [Google Scholar] [CrossRef]
- Knol, E.F.; Mathur, P.K. Genomics-Based Selection for Reproduction and Adaptation in Pigs. Biosci. Proc. 2019. [Google Scholar] [CrossRef]
- Garrick, D.J. The Role of Genomics in Pig Improvement. Anim. Prod. Sci. 2017, 57, 2360. [Google Scholar] [CrossRef]
- Zhang, Z.; Ober, U.; Erbe, M.; Zhang, H.; Gao, N.; He, J.; Li, J.; Simianer, H. Improving the Accuracy of Whole Genome Prediction for Complex Traits Using the Results of Genome Wide Association Studies. PLoS ONE 2014, 9, e93017. [Google Scholar] [CrossRef]
- Zhu, D.; Zhao, Y.; Zhang, R.; Wu, H.; Cai, G.; Wu, Z.; Wang, Y.; Hu, X. Genomic prediction based on selective linkage disequilibrium pruning of low-coverage whole-genome sequence variants in a pure Duroc population. Genet. Sel. Evol. 2023, 55, 72. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Geng, H.; Yang, B.; Yin, Z.; Liu, Y. Integrating QTL and expression QTL of PigGTEx to improve the accuracy of genomic prediction for small population in Yorkshire pigs. Anim. Genet. 2025, 56, e70001. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-L.; Carissa; Park, A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef]
- Hoffman, G.E. Correcting for Population Structure and Kinship Using the Linear Mixed Model: Theory and Extensions. PLoS ONE 2013, 8, e75707. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Tier, B. “SNP Snappy”: A Strategy for Fast Genome-Wide Association Studies Fitting a Full Mixed Model. Genetics 2012, 190, 275–277. [Google Scholar] [CrossRef]
- Zhou, X.; Carbonetto, P.; Stephens, M. Polygenic Modeling with Bayesian Sparse Linear Mixed Models. PLoS Genet. 2013, 9, e1003264. [Google Scholar] [CrossRef]
- Ricard, A.; Danvy, S.; Legarra, A. Computation of Deregressed Proofs for Genomic Selection When Own Phenotypes Exist with an Application in French Show-Jumping Horses. J. Anim. Sci. 2013, 91, 1076–1085. [Google Scholar] [CrossRef]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Fernando, R.L.; Vitezica, Z.; Okimoto, R.; Wing, T.; Hawken, R.; Muir, W.M. Genome-Wide Association Mapping Including Phenotypes from Relatives without Genotypes in a Single-Step (SsGWAS) for 6-Week Body Weight in Broiler Chickens. Front. Genet. 2014, 5, 134. [Google Scholar] [CrossRef]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-Wide Association Mapping Including Phenotypes from Relatives without Genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef]
- Silva, E.F.P.; Gaia, R.C.; Mulim, H.A.; Pinto, L.F.B.; Iung, L.H.S.; Brito, L.F.; Pedrosa, V.B. Genome-Wide Association Study of Conformation Traits in Brazilian Holstein Cattle. Animals 2024, 14, 2472. [Google Scholar] [CrossRef]
- Howie, B.; Fuchsberger, C.; Stephens, M.; Marchini, J.; Abecasis, G.R. Fast and Accurate Genotype Imputation in Genome-Wide Association Studies through Pre-Phasing. Nat. Genet. 2012, 44, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Edea, Z.; Dessie, T.; Dadi, H.; Do, K.-T.; Kim, K.-S. Genetic Diversity and Population Structure of Ethiopian Sheep Populations Revealed by High-Density SNP Markers. Front. Genet. 2017, 8, 218. [Google Scholar] [CrossRef]
- Shafer, A.B.A.; Peart, C.R.; Tusso, S.; Maayan, I.; Brelsford, A.; Wheat, C.W.; Wolf, J.B.W. Bioinformatic Processing of RAD-seq Data Dramatically Impacts Downstream Population Genetic Inference. Methods Ecol. Evol. 2017, 8, 907–917. [Google Scholar] [CrossRef]
- Hong, Y.; Ye, J.; Dong, L.; Li, Y.; Yan, L.; Cai, G.; Liu, D.; Tan, C.; Wu, Z. Genome-Wide Association Study for Body Length, Body Height, and Total Teat Number in Large White Pigs. Front. Genet. 2021, 12, 650370. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Luan, T.; Woolliams, J.A.; Lien, S.; Kent, M.; Svendsen, M.; Meuwissen, T.H.E. The Accuracy of Genomic Selection in Norwegian Red Cattle Assessed by Cross-Validation. Genetics 2009, 183, 1119–1126. [Google Scholar] [CrossRef]
- Henderson, C.R. Best Linear Unbiased Prediction of Breeding Values Not in the Model for Records. J. Dairy Sci. 1977, 60, 783–787. [Google Scholar] [CrossRef]
- Henderson, C.R. Estimation of Variances in Animal Model and Reduced Animal Model for Single Traits and Single Records. J. Dairy Sci. 1986, 69, 1394–1402. [Google Scholar] [CrossRef]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot Topic: A Unified Approach to Utilize Phenotypic, Full Pedigree, and Genomic Information for Genetic Evaluation of Holstein Final Score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- VanRaden, P.M.; Van Tassell, C.P.; Wiggans, G.R.; Sonstegard, T.S.; Schnabel, R.D.; Taylor, J.F.; Schenkel, F.S. Invited review: Reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 2009, 92, 16–24. [Google Scholar] [CrossRef]
- Aguilar, I.; Misztal, I.; Legarra, A.; Tsuruta, S. Efficient Computation of the Genomic Relationship Matrix and Other Matrices Used in Single-Step Evaluation: Matrix Computation Genomic Selection. J. Anim. Breed. Genet. 2011, 128, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Tiezzi, F.; Parker-Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. A Genome-Wide Association Study for Clinical Mastitis in First Parity US Holstein Cows Using Single-Step Approach and Genomic Matrix Re-Weighting Procedure. PLoS ONE 2015, 10, e0114919. [Google Scholar] [CrossRef] [PubMed]
- Parker Gaddis, K.L.; Megonigal, J.H.; Clay, J.S.; Wolfe, C.W. Genome-Wide Association Study for Ketosis in US Jerseys Using Producer-Recorded Data. J. Dairy Sci. 2018, 101, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I.; Lourenco, D.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs; UGA Animal Breeding and Genetics Group: Athens, GA, USA, 2014; p. 143. [Google Scholar]
- Dyer, S.C.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Barrera-Enriquez, V.P.; Becker, A.; Bennett, R.; Beracochea, M.; Berry, A.; et al. Ensembl 2025. Nucleic Acids Res. 2025, 53, D948–D957. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Imboonta, N.; Rydhmer, L.; Tumwasorn, S. Genetic Parameters and Trends for Production and Reproduction Traits in Thai Landrace Sows. Livest. Sci. 2007, 111, 70–79. [Google Scholar] [CrossRef]
- Lundgren, H.; Canario, L.; Grandinson, K.; Lundeheim, N.; Zumbach, B.; Vangen, O.; Rydhmer, L. Genetic Analysis of Reproductive Performance in Landrace Sows and Its Correlation to Piglet Growth. Livest. Sci. 2010, 128, 173–178. [Google Scholar] [CrossRef]
- Camargo, E.G.; Marques, D.B.D.; de Figueiredo, E.A.P.; e Silva, F.F.; Lopes, P.S. Genetic Study of Litter Size and Litter Uniformity in Landrace Pigs. Rev. Bras. Zootec. 2020, 49, e20180295. [Google Scholar] [CrossRef]
- Damgaard, L.H.; Rydhmer, L.; Løvendahl, P.; Grandinson, K. Genetic Parameters for Within-Litter Variation in Piglet Birth Weight and Change in within-Litter Variation during Suckling1. J. Anim. Sci. 2003, 81, 604–610. [Google Scholar] [CrossRef]
- Lopez, B.I.; Kim, T.H.; Makumbe, M.T.; Song, C.W.; Seo, K.S. Variance Components Estimation for Farrowing Traits of Three Purebred Pigs in Korea. Asian-Australas. J. Anim. Sci. 2017, 30, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Roehe, R.; Kennedy, B.W. Estimation of Genetic Parameters for Litter Size in Canadian Yorkshire and Landrace Swine with Each Parity of Farrowing Treated as a Different Trait. J. Anim. Sci. 1995, 73, 2959. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Song, K.-D.; Lee, H.-K.; Cho, K.-H.; Park, H.-C.; Park, K.-D. Genetic Parameters of Reproductive and Meat Quality Traits in Korean Berkshire Pigs. Asian-Australas. J. Anim. Sci. 2015, 28, 1388–1393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, S.; Bidanel, J.-P.; Burlot, T.; Legault, C.; Naveau, J. Genetic Parameters and Genetic Trends in the Chinese × European Tiameslan Composite Pig Line. II. Genetic Trends. Genet. Sel. Evol. 2000, 32, 57. [Google Scholar] [CrossRef]
- Satoh, M. Comparison of Genetic Improvement for Litter Size at Birth by Direct and Indirect Selection in Swine Herd. Anim. Sci. J. 2006, 77, 566–573. [Google Scholar] [CrossRef]
- VanRaden, P.M. Symposium Review: How to Implement Genomic Selection. J. Dairy Sci. 2020, 103, 5291–5301. [Google Scholar] [CrossRef]
- Misztal, I.; Lourenco, D.; Legarra, A. Current Status of Genomic Evaluation. J. Anim. Sci. 2020, 98, skaa101. [Google Scholar] [CrossRef]
- Legarra, A.; Christensen, O.F.; Aguilar, I.; Misztal, I. Single Step, a General Approach for Genomic Selection. Livest. Sci. 2014, 166, 54–65. [Google Scholar] [CrossRef]
- Teissier, M.; Larroque, H.; Robert-Granié, C. Weighted Single-Step Genomic BLUP Improves Accuracy of Genomic Breeding Values for Protein Content in French Dairy Goats: A Quantitative Trait Influenced by a Major Gene. Genet. Sel. Evol. 2018, 50, 31. [Google Scholar] [CrossRef]
- Pszczola, M.; Strabel, T.; Mulder, H.A.; Calus, M.P.L. Reliability of Direct Genomic Values for Animals with Different Relationships within and to the Reference Population. J. Dairy Sci. 2012, 95, 389–400. [Google Scholar] [CrossRef]
- Naj, A.C. Genotype Imputation in Genome-Wide Association Studies. Curr. Protoc. Hum. Genet. 2019, 102, e84. [Google Scholar] [CrossRef] [PubMed]
- Asoro, F.G.; Newell, M.A.; Beavis, W.D.; Scott, M.P.; Jannink, J. Accuracy and Training Population Design for Genomic Selection on Quantitative Traits in Elite North American Oats. Plant Genome 2011, 4, 132–144. [Google Scholar] [CrossRef]
- Hinckley, M.; Vaccari, S.; Horner, K.; Chen, R.; Conti, M. The G-Protein-Coupled Receptors GPR3 and GPR12 Are Involved in CAMP Signaling and Maintenance of Meiotic Arrest in Rodent Oocytes. Dev. Biol. 2005, 287, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.J.; Laun, A.S.; Song, Z.-H. Cannabidiol, a Novel Inverse Agonist for GPR12. Biochem. Biophys. Res. Commun. 2017, 493, 451–454. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, X.; Chen, S.; Jia, W.; Ma, X.; Wang, J.; Qian, Y.; Lei, D.; Liu, H.; Pan, X. GPR12 Inhibits Migration and Promotes Apoptosis in Esophageal Cancer and Hypopharyngeal Cancer Cells. Thorac. Cancer 2021, 12, 1525–1535. [Google Scholar] [CrossRef]
- Fujii, A.; Masuda, T.; Iwata, M.; Tobo, T.; Wakiyama, H.; Koike, K.; Kosai, K.; Nakano, T.; Kuramitsu, S.; Kitagawa, A.; et al. The Novel Driver Gene ASAP2 Is a Potential Druggable Target in Pancreatic Cancer. Cancer Sci. 2021, 112, 1655–1668. [Google Scholar] [CrossRef]
- Tekola-Ayele, F.; Zeng, X.; Ouidir, M.; Workalemahu, T.; Zhang, C.; Delahaye, F.; Wapner, R. DNA Methylation Loci in Placenta Associated with Birthweight and Expression of Genes Relevant for Early Development and Adult Diseases. Clin. Epigenet. 2020, 12, 78. [Google Scholar] [CrossRef]
- Tebani, A.; Jotanovic, J.; Hekmati, N.; Sivertsson, Å.; Gudjonsson, O.; Edén Engström, B.; Wikström, J.; Uhlèn, M.; Casar-Borota, O.; Pontén, F. Annotation of Pituitary Neuroendocrine Tumors with Genome-Wide Expression Analysis. Acta Neuropathol. Commun. 2021, 9, 181. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, K.; Cui, J.; Xiong, J.; Wu, H.; Peng, T.; Guo, Y. Circ-MBOAT2 Knockdown Represses Tumor Progression and Glutamine Catabolism by MiR-433-3p/GOT1 Axis in Pancreatic Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 124. [Google Scholar] [CrossRef]
- Firestein, R.; Bass, A.J.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Guney, I.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 Is a Colorectal Cancer Oncogene That Regulates β-Catenin Activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef]
- Teng, Y.; Mei, Y.; Hawthorn, L.; Cowell, J.K. WASF3 Regulates MiR-200 Inactivation by ZEB1 through Suppression of KISS1 Leading to Increased Invasiveness in Breast Cancer Cells. Oncogene 2014, 33, 203–211. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aron, R.; Pellegrini, P.; Green, E.W.; Maddison, D.C.; Opoku-Nsiah, K.; Oliveira, A.O.; Wong, J.S.; Daub, A.C.; Giorgini, F.; Muchowski, P.; et al. Deubiquitinase Usp12 Functions Noncatalytically to Induce Autophagy and Confer Neuroprotection in Models of Huntington’s Disease. Nat. Commun. 2018, 9, 3191. [Google Scholar] [CrossRef] [PubMed]
- Wangsa, D.; Braun, R.; Stuelten, C.H.; Brown, M.; Bauer, K.M.; Emons, G.; Weston, L.A.; Hu, Y.; Yang, H.H.; Vila-Casadesús, M.; et al. Induced Chromosomal Aneuploidy Results in Global and Consistent Deregulation of the Transcriptome of Cancer Cells. Neoplasia 2019, 21, 721–729. [Google Scholar] [CrossRef]
- Haag, N.; Tan, E.-C.; Begemann, M.; Buschmann, L.; Kraft, F.; Holschbach, P.; Lai, A.H.M.; Brett, M.; Mochida, G.H.; DiTroia, S.; et al. Biallelic Loss-of-Function Variants in WDR11 Are Associated with Microcephaly and Intellectual Disability. Eur. J. Hum. Genet. 2021, 29, 1663–1668. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Peiris, M.N.; Donoghue, D.J. Functions of FGFR2 Corrupted by Translocations in Intrahepatic Cholangiocarcinoma. Cytokine Growth Factor Rev. 2020, 52, 56–67. [Google Scholar] [CrossRef]
- Sinderewicz, E.; Grycmacher, K.; Boruszewska, D.; Kowalczyk-Zięba, I.; Staszkiewicz, J.; Ślężak, T.; Woclawek-Potocka, I. Bovine Ovarian Follicular Growth and Development Correlate with Lysophosphatidic Acid Expression. Theriogenology 2018, 106, 1–14. [Google Scholar] [CrossRef]
- Lian, Y.-F.; Huang, Y.-L.; Zhang, Y.-J.; Chen, D.-M.; Wang, J.-L.; Wei, H.; Bi, Y.-H.; Jiang, Z.-W.; Li, P.; Chen, M.-S.; et al. CACYBP Enhances Cytoplasmic Retention of P27 Kip1 to Promote Hepatocellular Carcinoma Progression in the Absence of RNF41 Mediated Degradation. Theranostics 2019, 9, 8392–8408. [Google Scholar] [CrossRef]
- Jackson, C.B.; Huemer, M.; Bolognini, R.; Martin, F.; Szinnai, G.; Donner, B.C.; Richter, U.; Battersby, B.J.; Nuoffer, J.-M.; Suomalainen, A.; et al. A Variant in MRPS14 (US14m) Causes Perinatal Hypertrophic Cardiomyopathy with Neonatal Lactic Acidosis, Growth Retardation, Dysmorphic Features and Neurological Involvement. Hum. Mol. Genet. 2019, 28, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Lévy, J.; Jung-Klawitter, S.; Bakhtiari, S.; Monteiro, F.; Maroofian, R.; Bierhals, T.; Hempel, M.; Elmaleh-Bergès, M.; Kitajima, J.P.; et al. Loss of TNR Causes a Nonprogressive Neurodevelopmental Disorder with Spasticity and Transient Opisthotonus. Genet. Med. 2020, 22, 1061–1068. [Google Scholar] [CrossRef]
- Guillen-Ahlers, H.; Erbe, C.B.; Chevalier, F.D.; Montoya, M.J.; Zimmerman, K.D.; Langefeld, C.D.; Olivier, M.; Runge, C.L. TMTC2 Variant Associated with Sensorineural Hearing Loss and Auditory Neuropathy Spectrum Disorder in a Family Dyad. Mol. Genet. Genomic Med. 2018, 6, 653–659. [Google Scholar] [CrossRef]
- Jacquemin, V.; Antoine, M.; Duerinckx, S.; Massart, A.; Desir, J.; Perazzolo, C.; Cassart, M.; Thomas, D.; Segers, V.; Lecomte, S.; et al. TrkA Mediates Effect of Novel KIDINS220 Mutation in Human Brain Ventriculomegaly. Hum. Mol. Genet. 2020, 29, 3757–3764. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).