Gut Microbiota Variation in Aging Dogs with Osteoarthritis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Ethics Statement
2.2. Animals and Diets
2.3. Data Recording and Samples Collections
2.4. Microbiota Analysis
2.5. Bioinformatic
2.6. Statistical Analysis
3. Results
3.1. Rarefaction Curve and Relative Abundance of Whole Population
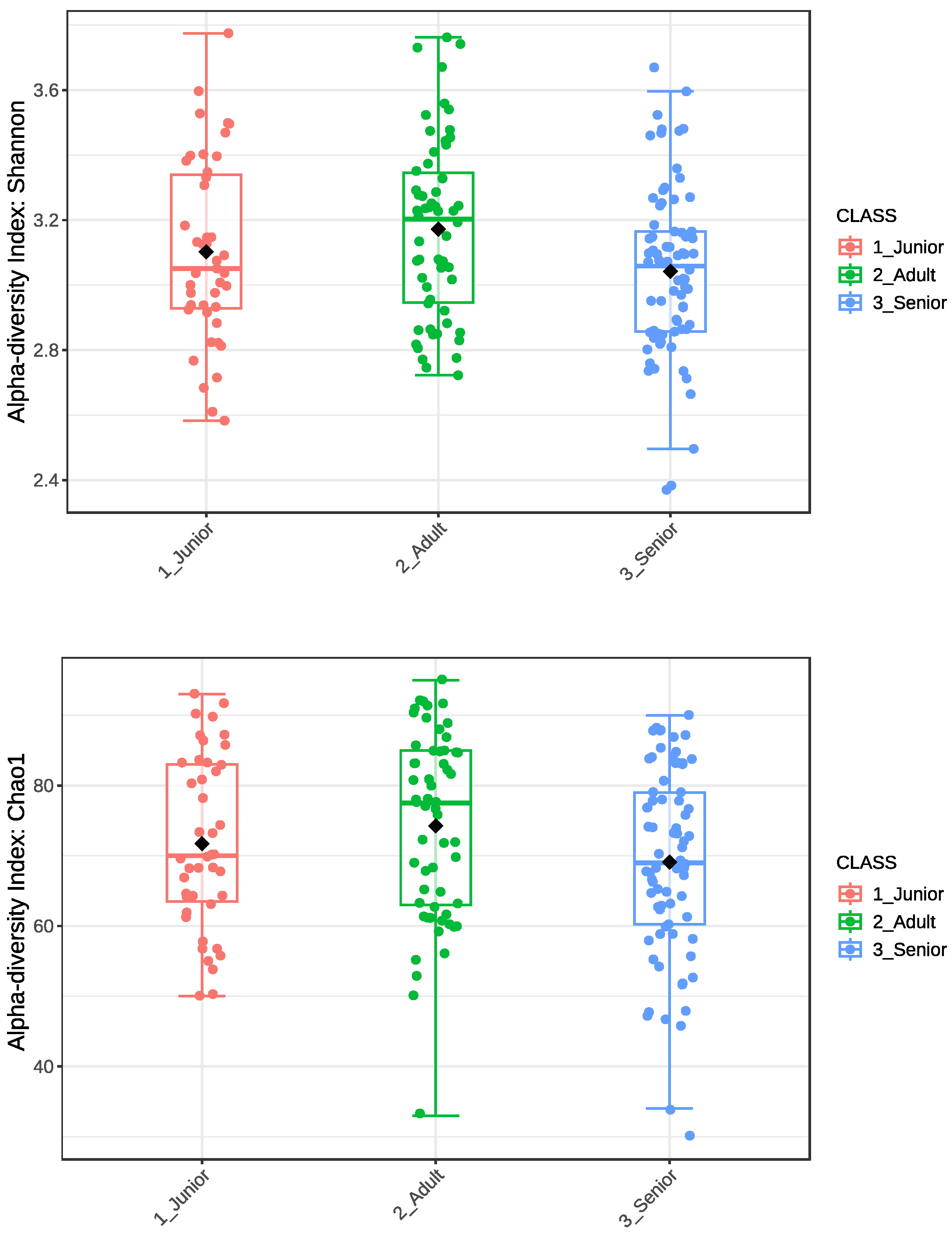
3.2. Alpha Diversity
3.3. Beta Diversity
3.4. LEfSe Analysis
4. Discussion
4.1. Microbial Diversity
4.2. Microbial Signatures Associated with Age and OA
4.3. Microbiota and Osteoarthritis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bajinka, O.; Darboe, A.; Tan, Y.; Abdelhalim, K.A.; Cham, L.B. Gut microbiota and the human gut physiological changes. Ann. Microbiol. 2020, 70, 65. [Google Scholar] [CrossRef]
- Wen, L.; Duffy, A. Factors Influencing the gut microbiota, inflammation, and type 2 diabetes. J. Nutr. 2017, 147, 1468S–1475S. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef]
- Pilla, R.; Suchodolski, J.S. The gut microbiome of dogs and cats, and the influence of diet. Vet. Clin. N. Am. Small Anim. Pr. 2021, 51, 605–621. [Google Scholar] [CrossRef]
- Saraswati, S.; Sitaraman, R. Aging and the human gut microbiota from correlation to causality. Front. Microbiol. 2014, 5, 764. [Google Scholar]
- Biragyn, A.; Ferrucci, L. Gut dysbiosis: A potential link between increased cancer risk in ageing and inflammaging. Lancet Oncol. 2018, 19, e295–e304. [Google Scholar] [CrossRef] [PubMed]
- Badal, V.D.; Vaccariello, E.D.; Murray, E.R.; Yu, K.E.; Knight, R.; Jeste, D.V.; Nguyen, T.T. The gut microbiome, aging, and longevity: A systematic review. Nutrients 2020, 12, 3759. [Google Scholar] [CrossRef]
- Kumar, M.; Babaei, P.; Ji, B.; Nielsen, J. Human gut microbiota and healthy aging: Recent developments and future prospective. Nutr. Healthy Aging 2016, 4, 3–16. [Google Scholar] [CrossRef]
- Masuoka, H.; Shimada, K.; Kiyosue-Yasuda, T.; Kiyosue, M.; Oishi, Y.; Kimura, S.; Yamada, A.; Hirayama, K. Transition of the intestinal microbiota of dogs with age. Biosci. Microbiota Food Health 2016, 36, 27–31. [Google Scholar] [CrossRef]
- Garrigues, Q.; Apper, E.; Chastant, S.; Mila, H. Gut microbiota development in the growing dog: A dynamic process influenced by maternal, environmental and host factors. Front. Vet. Sci. 2022, 9, 964649. [Google Scholar] [CrossRef]
- Guard, B.C.; Mila, H.; Steiner, J.M.; Mariani, C.; Suchodolski, J.S.; Chastant-Maillard, S. Characterization of the fecal microbiome during neonatal and early pediatric development in puppies. PLoS ONE 2017, 12, e0175718. [Google Scholar] [CrossRef] [PubMed]
- Blake, A.B.; Cigarroa, A.; Klein, H.L.; Khattab, M.R.; Keating, T.; Coevering, P.V.D.; Lidbury, J.A.; Steiner, J.M.; Suchodolski, J.S. Developmental stages in microbiota, bile acids, and clostridial species in healthy puppies. J. Vet. Intern. Med. 2020, 34, 2345–2356. [Google Scholar] [CrossRef] [PubMed]
- Scanzello, C.R.; Plaas, A.; Crow, M.K. Innate immune system activation in osteoarthritis: Is osteoarthritis a chronic wound? Curr. Opin. Rheumatol. 2008, 20, 565–572. [Google Scholar] [CrossRef]
- Sandell, L.J. Etiology of osteoarthritis: Genetics and synovial joint development. Nat. Rev. Rheumatol. 2012, 8, 77–89. [Google Scholar] [CrossRef]
- Fox, S.M. Pathophysiology of osteoarthritic pain. In Chronic Pain in Small Animal Medicine; Northcott, J., Ed.; Manson Publishing Ltd.: London, UK, 2011; pp. 74–96. [Google Scholar]
- Pet Population PFMA. Available online: https://www.pfma.org.uk/pet-population-2022 (accessed on 7 July 2023).
- APPA. 2019–2020 APPA National Pet Owners Survey. Available online: https://petsplusmag.com/appa-releases-findings-from-new-2019-2020-national-pet-owners-survey (accessed on 28 March 2019).
- Johnston, S.A. Osteoarthritis. Joint anatomy, physiology, and pathobiology. Vet. Clin. N. Am. Small Anim. Pract. 1997, 27, 699–723. [Google Scholar] [CrossRef]
- O’Neill, D.G.; Church, D.B.; McGreevy, P.D.; Thomson, P.C.; Brodbelt, D.C. Prevalence of disorders recorded in dogs attending primary-care veterinary practices in England. PLoS ONE 2014, 9, e90501. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.L.; O’Neill, D.G.; Brodbelt, D.C.; Church, D.B.; Meeson, R.L.; Sargan, D.; Summers, J.F.; Zulch, H.; Collins, L.M. Prevalence, duration and risk factors for appendicular osteoarthritis in a UK dog population under primary veterinary care. Sci. Rep. 2018, 8, 5641. [Google Scholar] [CrossRef]
- Clements, D.N.; Carter, S.D.; Innes, J.F.; Ollier, W.E. Genetic basis of secondary osteoarthritis in dogs with joint dysplasia. Am. J. Vet. Res. 2006, 67, 909–918. [Google Scholar] [CrossRef]
- Belshaw, Z.; Dean, R.; Asher, L. “You can be blind because of loving them so much”: The impact on owners in the United Kingdom of living with a dog with osteoarthritis. BMC Vet. Res. 2020, 16, 190. [Google Scholar] [CrossRef]
- Belshaw, Z.; Dean, R.; Asher, L. Slower, shorter, sadder: A qualitative study exploring how dog walks change when the canine participant develops osteoarthritis. BMC Vet. Res. 2020, 16, 85. [Google Scholar] [CrossRef]
- Gore, M.; Lana, S.E.; Bishop, G.A. Colorado State University, Pet Hospice Program. Vet. Clin. N. Am. Small Anim. Pract. 2019, 49, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Wilke, V.; Robinson, D.; Evans, R.; Rothschild, M.; Conzemius, M. Estimate of the annual economic impact of treatment of cranial cruciate ligament injury in dogs in the United States. J. Am. Vet. Med. Assoc. 2005, 227, 1604–1607. [Google Scholar] [CrossRef]
- Cachon, T.; Frykman, O.; Innes, J.F.; Lascelles, B.D.X.; Okumura, M.; Sousa, P.; Staffieri, F.; Steagall, P.V.; Van Ryssen, B. COAST Development Group’s international consensus guidelines for the treatment of canine osteoarthritis. Front. Vet. Sci. 2023, 10, 1137888. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- National Research Council. Nutrient Requirements of Dogs and Cats; The National Academies Press: Washington, DC, USA, 2006. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Available online: https://www.ukpetfood.org/resource/new-pfma-pet-population-data-highlights-pet-peak-but-the-number-of-owners-giving-up-their-pet-is-huge-concern.html (accessed on 31 January 2025).
- Available online: https://ftp.microbio.me/greengenes_release/current/ (accessed on 31 January 2025).
- ICNP. Available online: https://www.the-icsp.org/index.php/code-of-nomenclatur (accessed on 31 January 2025).
- Available online: https://www.microbiomeanalyst.ca/ (accessed on 31 January 2025).
- Lu, Y.; Zhou, G.; Ewald, J.; Pang, Z.; Shiri, T.; Xia, J. Microbiome Analyst 2.0: Comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res. 2023, 51, W310–W318. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Mizukami, K.; Uchiyama, J.; Igarashi, H.; Murakami, H.; Osumi, T.; Shima, A.; Ishiahra, G.; Nasukawa, T.; Une, Y.; Sakaguchi, M. Age-related analysis of the gut microbiome in a purebred dog colony. FEMS Microbiol. Lett. 2019, 366, fnz095. [Google Scholar] [CrossRef] [PubMed]
- Omatsu, T.; Omura, M.; Katayama, Y.; Kimura, T.; Okumura, M.; Okumura, A.; Murata, Y.; Mizutani, T. Molecular diversity of the faecal microbiota of Toy Poodles in Japan. J. Vet. Med. Sci. 2018, 80, 749–754. [Google Scholar] [CrossRef]
- Fernández-Pinteño, A.; Pilla, R.; Manteca, X.; Suchodolski, J.; Torre, C.; Salas-Mani, A. Age-associated changes in intestinal health biomarkers in dogs. Front. Vet. Sci. 2023, 10, 1213287. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stevens, C.; Norris, S.; Arbeeva, L.; Carter, S.; Enomoto, M.; Nelson, A.E.; Lascelles, B.D.X. Gut Microbiome and Osteoarthritis: Insights from the Naturally Occurring Canine Model of Osteoarthritis. Arthritis Rheumatol. 2024, 76, 1758–1763. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cintio, M.; Scarsella, E.; Sgorlon, S.; Sandri, M.; Stefanon, B. Gut Microbiome of Healthy and Arthritic Dogs. Vet. Sci. 2020, 7, 92. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fernández-Pinteño, A.; Pilla, R.; Suchodolski, J.; Apper, E.; Torre, C.; Salas-Mani, A.; Manteca, X. Age-Related Changes in Gut Health and Behavioral Biomarkers in a Beagle Dog Population. Animals 2025, 15, 234. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Honneffer, J.B.; Minamoto, Y.; Suchodolski, J.S. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J. Gastroenterol. 2014, 20, 16489–16497. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Suchodolski, J.S.; Dowd, S.E.; Wilke, V.; Steiner, J.M.; Jergens, A.E. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS ONE 2012, 7, e39333. [Google Scholar] [CrossRef]
- Lapébie, P.; Lombard, V.; Drula, E.; Terrapon, N.; Henrissat, B. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 2019, 10, 2043. [Google Scholar] [CrossRef]
- Bermingham, E.N.; Maclean, P.; Thomas, D.G.; Cave, N.J.; Young, W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ 2017, 5, e3019. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vázquez-Baeza, Y.; Hyde, E.; Suchodolski, J.; Knight, R. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat. Microbiol. 2016, 1, 16177. [Google Scholar] [CrossRef]
- Jackson, M.A.; Jeffery, I.B.; Beaumont, M.; Bell, J.T.; Clark, A.G.; Ley, R.E.; O’toole, P.W.; Spector, T.D.; Steves, C.J. Signatures of early frailty in the gut microbiota. Genome Med. 2016, 8, 8. [Google Scholar] [CrossRef]
- Lin, D.; Peters, B.A.; Friedlander, C.; Freiman, H.J.; Goedert, J.J.; Sinha, R.; Miller, G.; Bernstein, M.A.; Hayes, R.B.; Ahn, J. Association of dietary fibre intake and gut microbiota in adults. Br. J. Nutr. 2018, 120, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- You, I.; Kim, M.J. Comparison of Gut Microbiota of 96 Healthy Dogs by Individual Traits: Breed, Age, and Body Condition Score. Animals 2021, 11, 2432. [Google Scholar] [CrossRef]
- Boer, C.G.; Radjabzadeh, D.; Medina-Gomez, C.; Garmaeva, S.; Schiphof, D.; Arp, P.; Koet, T.; Kurilshikov, A.; Fu, J.; Ikram, M.A.; et al. Intestinal microbiome composition and its relation to joint pain and inflammation. Nat. Commun. 2019, 10, 4881. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.H.; Yang, Y.Q.; Cao, R.R.; Bo, L.; Lei, S.-F. The causal role of gut microbiota in development of osteoarthritis. Osteoarthr. Cartil. 2021, 29, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, J.; Li, B.; Zeng, B.; Chou, C.-H.; Zheng, X.; Xie, J.; Li, H.; Hao, Y.; Chen, G.; et al. Faecal microbiota transplantation from metabolically compromised human donors accelerates osteoarthritis in mice. Ann. Rheum. Dis. 2020, 79, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Osumi, T.; Mizukami, K.; Fukuyama, T.; Shima, A.; Unno, A.; Takemura-Uchiyama, I.; Une, Y.; Murakami, H.; Sakaguchi, M. Characterization of the oral and faecal microbiota associated with atopic dermatitis in dogs selected from a purebred Shiba Inu colony. Lett. Appl. Microbiol. 2022, 75, 1607–1616. [Google Scholar] [CrossRef]





| Age | Sex | Healthy | Osteoarthritis | Other | Total |
|---|---|---|---|---|---|
| Junior | Fs | 11 | 0 | 0 | 11 |
| Adult | 17 | 16 | 2 | 35 | |
| Senior | 4 | 41 | 7 | 52 | |
| Junior | Mc | 19 | 0 | 13 | 32 |
| Adult | 13 | 8 | 2 | 24 | |
| Senior | 5 | 16 | 1 | 21 | |
| Junior | Total | 30 | 0 | 13 | 43 |
| Adult | 30 | 24 | 4 | 58 | |
| Senior | 9 | 57 | 8 | 74 | |
| Total | 69 | 81 | 25 | 175 |
| Healthy | Osteo-Arthritic | Total | |||
|---|---|---|---|---|---|
| Fs | Mc | Fs | Mc | ||
| Junior | 10 | 16 | 0 | 0 | 26 |
| Adult | 14 | 12 | 16 | 7 | 49 |
| Senior | 4 | 5 | 37 | 15 | 61 |
| Total | 28 | 33 | 53 | 22 | 136 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balouei, F.; Rivera, C.d.; Paradis, A.; Stefanon, B.; Kelly, S.; McCarthy, N.; Mongillo, P. Gut Microbiota Variation in Aging Dogs with Osteoarthritis. Animals 2025, 15, 1619. https://doi.org/10.3390/ani15111619
Balouei F, Rivera Cd, Paradis A, Stefanon B, Kelly S, McCarthy N, Mongillo P. Gut Microbiota Variation in Aging Dogs with Osteoarthritis. Animals. 2025; 15(11):1619. https://doi.org/10.3390/ani15111619
Chicago/Turabian StyleBalouei, Fatemeh, Christina de Rivera, Andrea Paradis, Bruno Stefanon, Stephanie Kelly, Noelle McCarthy, and Paolo Mongillo. 2025. "Gut Microbiota Variation in Aging Dogs with Osteoarthritis" Animals 15, no. 11: 1619. https://doi.org/10.3390/ani15111619
APA StyleBalouei, F., Rivera, C. d., Paradis, A., Stefanon, B., Kelly, S., McCarthy, N., & Mongillo, P. (2025). Gut Microbiota Variation in Aging Dogs with Osteoarthritis. Animals, 15(11), 1619. https://doi.org/10.3390/ani15111619







