Effectiveness of Non-Invasive Methods in Studying Jaguar (Panthera onca) Hair
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Considerations
2.2. Study Area
2.3. Hair Collection
2.4. Electron Microscopy
2.5. Molecular Analysis
2.6. Analysis Isotopic
2.7. Analysis of Essential and Toxic Metallic Elements
3. Results
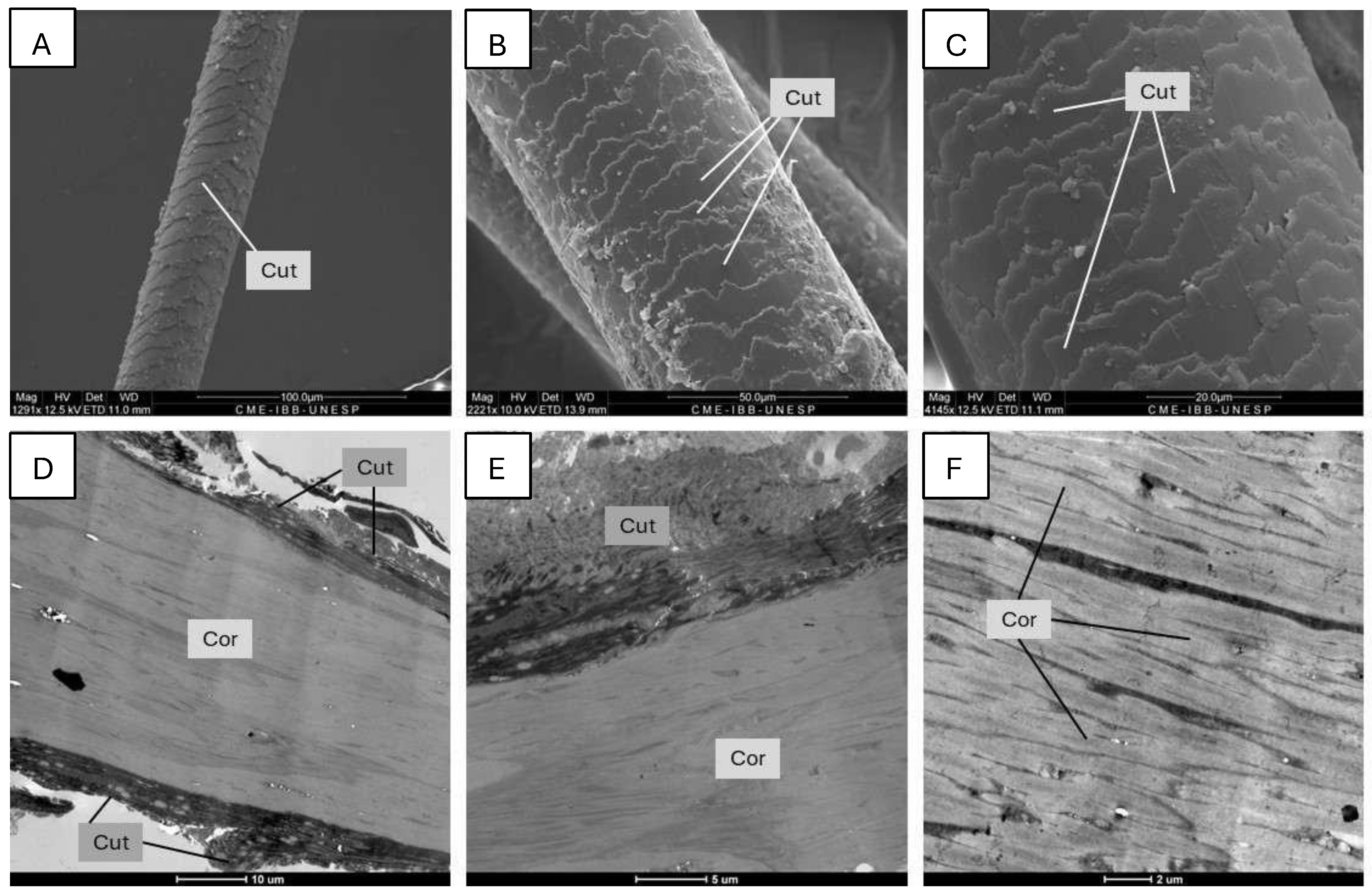
3.1. Electron Microscopy
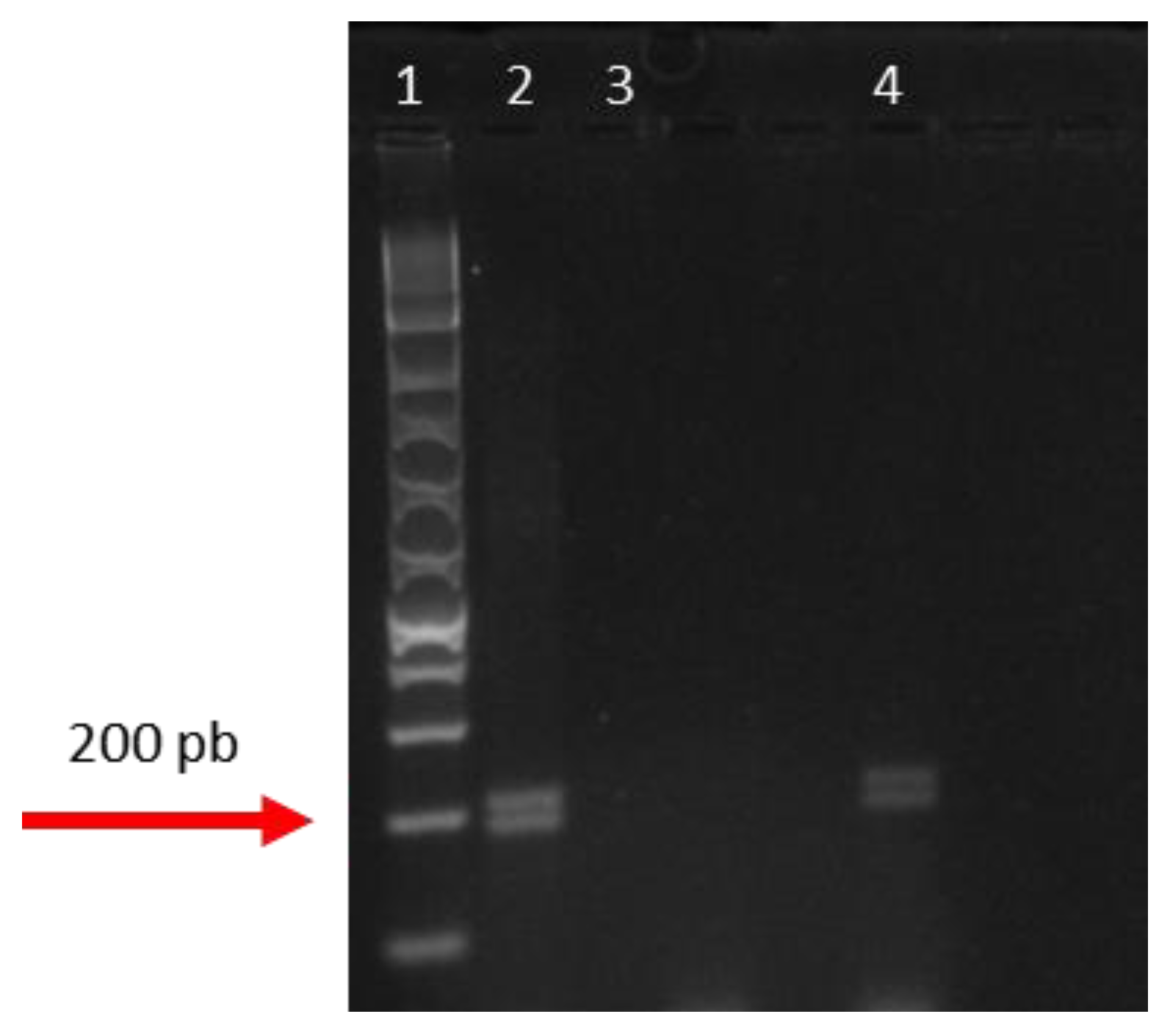
3.2. Molecular Analysis
3.3. Essential and Heavy Metals
3.4. Stable Isotopes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SEM | Scanning Electron Microscopy |
| TEM | Transmission Electron Microscopy |
| DNA | Deoxyribonucleic Acid |
| PCR | Polymerase Chain Reaction |
| AML | Amelogenin Gene |
| AMELY | Amelogenin Y-linked |
| AMELX | Amelogenin X-linked |
| bp | Base Pairs |
| TAE | Tris–Acetic Acid–EDTA Buffer |
| CF-IRMS | Continuous Flow Isotope Ratio Mass Spectrometry |
| VPDB | Vienna Pee Dee Belemnite |
| δ13C | Delta Carbon-13 |
| δ15N | Delta Nitrogen-15 |
| TOC | Total Organic Carbon |
| TN | Total Nitrogen |
| Cu | Copper |
| Cr | Chromium |
| Fe | Iron |
| Zn | Zinc |
| Pb | Lead |
| As | Arsenic |
| Hg | Mercury |
| Cd | Cadmium |
| Mn | Manganese |
| LCO | Hollow Cathode Lamp |
| CEUA | Ethics Committee on the Use of Animals |
| SISBIO | Biodiversity Authorization and Information System |
| APC | Article Processing Charge |
| Unesp | São Paulo State University “Júlio de Mesquita Filho” |
| IBB | Institute of Biosciences |
References
- Pilgrim, K.L.; McKelvey, K.S.; Riddle, A.E.; Schwartz, M.K. Sex identification of felids based on noninvasive genetic samples. Mol. Ecol. Notes 2005, 5, 60–61. [Google Scholar] [CrossRef]
- Tremori, T.; Monteiro Garcia, F.; Montoya Flórez, L.M.; Picado Gonçalves, B.; Ferraz de Camargo, B.; Gwinnett, C.; Teixeira, C.; Sousa Rocha, N. Hair analysis of mammals of Brazilian wildlife for forensics purposes. Open J. Anim. Sci. 2018, 8, 335–345. [Google Scholar] [CrossRef]
- Tremori, T.M.; Antonio, L.U.; Cardena, M.M.S.G.; Gwinnett, C.; Davidson, A.; do Amaral, J.B.; Fridman, C.; Rocha, N.S. Forensic genetics associated with hair analysis as a tool for jaguar (Panthera onca) identification. Glob. Ecol. Conserv. 2024, 52, e02956. [Google Scholar] [CrossRef]
- Dobrzański, Z.; Filistowicz, A.; Przysiecki, P.; Nowicki, S.; Walkowiak, K.; Czyż, K. Mercury bioaccumulation in hair and skin of arctic foxes (Vulpes lagopus) and silver foxes (Vulpes vulpes) in rural and urbanized regions. Czech J. Anim. Sci. 2014, 59, 480–487. [Google Scholar] [CrossRef]
- Lopes, M.C.B.; de Carvalho, G.O.; Bernardo, R.R.; Macedo, J.; Lino, A.S.; Ramalho, E.E.; Kasper, D.; Meire, R.O.; Torres, J.P.M.; Malm, O. Total mercury in wild animals born in protected areas in central Brazil Amazon. Acta Amaz. 2020, 50, 142–148. [Google Scholar] [CrossRef]
- Montillo, M.; Caslini, C.; Peric, T.; Prandi, A.; Netto, P.; Tubaro, F.; Pedrotti, L.; Bianchi, A.; Mattiello, S. Analysis of 19 minerals and cortisol in red deer hair from two different areas of the Stelvio National Park: A preliminary study. Animals 2019, 9, 492. [Google Scholar] [CrossRef]
- Picone, M.; Distefano, G.G.; Zangrando, R.; Gambaro, A.; Ghirardini, A.V. Neonicotinoids and pharmaceuticals in hair of the red fox (Vulpes vulpes) from the Cavallino-Treporti peninsula, Italy. Environ. Res. 2023, 228, 115837. [Google Scholar] [CrossRef]
- Richards, N.L.; Cook, G.; Simpson, V.; Hall, S.; Harrison, N.; Scott, K.S. Qualitative detection of NSAIDs diclofenac and ibuprofen in the hair of Eurasian otters (Lutra lutra) inhabiting UK waterways using GC-MS. Eur. J. Wildl. Res. 2011, 57, 1107–1114. [Google Scholar] [CrossRef]
- Calamari, C.V.; Viau, P.; Nichi, M.; Martins, G.S.; Sobral, G.; Dias, J.M.; de Oliveira, C.A. Hair as a non-invasive alternative matrix: Sources of variation in testosterone levels. Domest. Anim. Endocrinol. 2020, 72, 106477. [Google Scholar] [CrossRef]
- Cattet, M.; Stenhouse, G.B.; Janz, D.M.; Kapronczai, L.; Erlenbach, J.A.; Jansen, H.T.; Nelson, O.L.; Robbins, C.T.; Boulanger, J. Quantification of reproductive hormones in the hair of captive adult grizzly bears and its application as indicators of sex and reproductive state. Conserv. Physiol. 2017, 5, cox032. [Google Scholar] [CrossRef]
- Heimbürge, S.; Kanitz, E.; Otten, W. The use of hair cortisol for stress assessment in animals. Gen. Comp. Endocrinol. 2019, 270, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Ayuso-Sáinz, J.J.; Belinchón-Lorenzo, S.; Fernández-Cotrina, J.; Jimenez, M.; Orduña-Domingo, A.; Quinto, E.J. Detection of Leishmania infantum kinetoplast DNA by real-time PCR in hair of wild rabbits. Med. Weter. 2022, 78, 91–94. [Google Scholar]
- Muñoz-Madrid, R.; Belinchón-Lorenzo, S.; Iniesta, V.; Fernández-Cotrina, J.; Parejo, J.C.; Serrano, F.J.; Monroy, I.; Baz, V.; Gómez-Luque, A.; Gómez-Nieto, L.C. First detection of Leishmania infantum kinetoplast DNA in the wild mammal hair: Application of qPCR method to determine potential parasite reservoirs. Acta Trop. 2013, 128, 706–709. [Google Scholar] [CrossRef]
- Koehler, G.; Schmidt-Küntzel, A.; Marker, L.; Hobson, K.A. Tracing the origins of cheetah cubs in the illegal wildlife trade: Enhancements based on the use of δ18O hair measurements. Front. Ecol. Evol. 2023, 11, 1058985. [Google Scholar] [CrossRef]
- Hopkins, J.B.; Whittington, J.; Clevenger, A.P.; Sawaya, M.A.; Clair, C.C.S. Stable isotopes reveal rail-associated behavior in a threatened carnivore. Isot. Environ. Health Stud. 2014, 50, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Mowat, G.; Heard, D.C.; Curtis, P.J. Combining stable isotope ratios with elemental concentrations to improve the estimation of terrestrial carnivore diets. Glob. Ecol. Conserv. 2023, 45, e02507. [Google Scholar] [CrossRef]
- García-Alaníz, N.; Naranjo, E.J.; Mallory, F.F. Hair-snares: A non-invasive method for monitoring feline populations in the Lacandon Jungle, Mexico. Trop. Conserv. Sci. 2010, 3, 403–411. [Google Scholar] [CrossRef]
- McDaniel, G.W.; McKelvey, K.S.; Squires, J.R.; Ruggiero, L.F. Effectiveness of lures and hair snares for detecting lynx. In Wildlife Society Bulletin; Wiley: Hoboken, NJ, USA, 2000; pp. 119–123. [Google Scholar]
- Petisco, J.S.E.; Sánchez-Carrasco, P.; Fernández-García, J.L. The wildcat (Felis s. silvestris) in the Mediterranean forest: Sightings through camera traps and non-invasive hair collection for genetic purposes. Vet. Res. Commun. 2024, 48, 2309–2320. [Google Scholar] [CrossRef]
- Portella, T.P.; Bilski, D.R.; Passos, F.C.; Pie, M.R. Assessing the efficacy of hair snares as a method for noninvasive sampling of neotropical felids. Zoology 2013, 30, 49–54. [Google Scholar] [CrossRef]
- Sawaya, M.A.; Stetz, J.B.; Clevenger, A.P.; Gibeau, M.L.; Kalinowski, S.T. Estimating grizzly and black bear population abundance and trends in Banff National Park using noninvasive genetic sampling. PLoS ONE 2012, 7, e34777. [Google Scholar] [CrossRef]
- Schilling, A.K.; Mazzamuto, M.V.; Romeo, C. A review of non-invasive sampling in wildlife health and disease research: What is new? Animals 2022, 12, 1719. [Google Scholar] [CrossRef] [PubMed]
- Hunter, L. Carnivores of the World; Princeton University Press: Princeton, NJ, USA, 2018; Volume 117. [Google Scholar]
- Jędrzejewski, W.; Boron, V.; Garrido, E.P.; Hoogesteijn, R.; Abarca, M.; Romero, A.P.; Viloria, Á.; Lampo, M.; Marquina, F.; Velásquez, G. Jaguars (Panthera onca) in the Plains of Colombia and Venezuela: Estimating distribution and population size by combining different modeling approaches. In Neotropical Mammals: Hierarchical Analysis of Occupancy and Abundance; Springer: Berlin/Heidelberg, Germany, 2023; pp. 197–235. [Google Scholar] [CrossRef]
- Rodríguez-Soto, C.; Monroy-Vilchis, O.; Zarco-González, M.M. Corridors for jaguar (Panthera onca) in Mexico: Conservation strategies. J. Nat. Conserv. 2013, 21, 438–443. [Google Scholar] [CrossRef]
- Quigley, H.; Foster, R.; Petracca, L.; Payan, E.; Salom, R.; Harmsen, B. The IUCN Red List of Threatened Species 2017: e.T15953A123791436. Panthera onca. 2017. (errata version published in 2018). Available online: https://dx.doi.org/10.2305/IUCN.UK.2017-3.RLTS.T15953A50658693.en (accessed on 20 March 2025).
- Scudeler, E.L.; de Carvalho, S.F.; Garcia, A.S.G.; Santorum, M.; Padovani, C.R.; dos Santos, D.C. Midgut and fat body: Multisystemic action of pyriproxyfen on non-target organism Ceraeochrysa claveri (Neuroptera: Chrysopidae). Environ. Pollut. 2022, 293, 118580. [Google Scholar] [CrossRef] [PubMed]
- Kantek, D.L.Z.; Trinca, C.S.; Tortato, F.; Devlin, A.L.; de Azevedo, F.C.C.; Cavalcanti, S.; Silveira, L.; Miyazaki, S.S.; Junior, P.G.C.; May-Junior, J.A.; et al. Jaguars from the Brazilian Pantanal: Low genetic structure, male-biased dispersal, and implications for long-term conservation. Biol. Conserv. 2021, 259, 109153. [Google Scholar] [CrossRef]
- Bradham, J.; Jorge, M.L.S.P.; Pedrosa, F.; Keuroghlian, A.; Costa, V.E.; Bercê, W.; Galetti, M. Spatial isotopic dietary plasticity of a Neotropical Forest ungulate: The white-lipped peccary (Tayassu pecari). J. Mammal. 2019, 100, 464–474. [Google Scholar] [CrossRef]
- Provecto Analítica. Manual de Operação e Métodos Orientativos Para Utilização Com o Digestor de Amostras (Versão 1.1) [Manual Técnico]. Scribd. 2013. Available online: https://pt.scribd.com/document/107336640/Manual-Dgt-v-1-1 (accessed on 20 March 2025).
- Elmer, P. Analytical Methods for Atomic Absorption Spectroscopy; The Perkin-Elmer Corporation: Eden Praire, MN, USA, 1996; p. 132. [Google Scholar]
- Miranda, G.H.; Rodrigues, F.H. Guia de Identificação de Pelos de Mamíferos Brasileiros; Brasília–Ciências Forenses, Publisher Editora da Academia Brasileira de Ciências Forenses: Brasília, Brasil, 2014. [Google Scholar]
- Souza, F.C.; Azevedo, F.C.C. Hair as a tool for identification of predators and prey: A study based on scats of jaguars (Panthera onca) and pumas (Puma concolor). Biota Neotrop. 2020, 21, e20201044. [Google Scholar] [CrossRef]
- Pozebon, D.; Dressler, V.L.; Curtius, A.J. Hair analysis: A review of procedures for trace elements determination and applications. Química Nova 1999, 22, 838–846. [Google Scholar] [CrossRef]
- López-Alonso, M.; Miranda, M.; Castillo, C.; Hernández, J.; Benedito, J.L. Toxic and essential metals in liver, kidney, and muscle of pigs at slaughter in Galicia, north-west Spain. Food Addit. Contam. 2007, 24, 943–954. [Google Scholar] [CrossRef]
- Ogilvy, C.; Constantine, R.; Bury, S.J.; Carroll, E.L. Diet variation in a critically endangered marine predator revealed with stable isotope analysis. R. Soc. Open Sci. 2022, 9, 220470. [Google Scholar] [CrossRef]

| Metals | As | Cd | Cr | Hg | Hg | Mn | Pb | Zn |
|---|---|---|---|---|---|---|---|---|
| Detection limits | <0.05 | <0.002 | <0.004 | <0.01 | <0.009 | <0.005 | <0.05 | <0.02 |
| Quantification limits | 0.5 | 0.02 | 0.04 | 0.1 | 0.09 | 0.05 | 0.5 | 0.2 |
| Metals | As | Cd | Cr | Cu | Hg | Mn | Pb | Zn |
|---|---|---|---|---|---|---|---|---|
| Concentrations | <0.05 | 9.3 | 0.48 | 15.9 | 0.08 | 66.2 | 0.2 | 155.0 |
| * References (Human hair) | 0.03–25.0 | 0.04–5.3 | 0.08–2.5 | 6.0–293.0 | 0.3–12.2 | 0.04–24.0 | 0.004–95.0 | 53.7–327.0 |
| ID | Est. | Amount (mg) | NT (%) | δ 15 N (‰) | COT (%) | δ 13 C (‰) |
|---|---|---|---|---|---|---|
| OP 1 | 1 | 0.394 | 12.38 | 11.12 | 45.81 | −22.02 |
| OP 2 | 1 | 0.349 | 14.69 | 11.15 | 41.06 | −20.20 |
| OP 3 | 1 | 0.601 | 14.92 | 11.30 | 40.95 | −20.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, L.P.; Raad, P.; Santos, D.C.d.; Almeida, A.A.; Costa, V.E.; Mota, L.S.L.S.d. Effectiveness of Non-Invasive Methods in Studying Jaguar (Panthera onca) Hair. Animals 2025, 15, 1415. https://doi.org/10.3390/ani15101415
Rodrigues LP, Raad P, Santos DCd, Almeida AA, Costa VE, Mota LSLSd. Effectiveness of Non-Invasive Methods in Studying Jaguar (Panthera onca) Hair. Animals. 2025; 15(10):1415. https://doi.org/10.3390/ani15101415
Chicago/Turabian StyleRodrigues, Larissa Pereira, Paul Raad, Daniela Carvalho dos Santos, Alaor Aparecido Almeida, Vladimir Eliodoro Costa, and Ligia Souza Lima Silveira da Mota. 2025. "Effectiveness of Non-Invasive Methods in Studying Jaguar (Panthera onca) Hair" Animals 15, no. 10: 1415. https://doi.org/10.3390/ani15101415
APA StyleRodrigues, L. P., Raad, P., Santos, D. C. d., Almeida, A. A., Costa, V. E., & Mota, L. S. L. S. d. (2025). Effectiveness of Non-Invasive Methods in Studying Jaguar (Panthera onca) Hair. Animals, 15(10), 1415. https://doi.org/10.3390/ani15101415






