1. Introduction
Utilizing molecular genetics techniques is a powerful approach for selecting and breeding animals with superior reproductive traits [
1,
2]. Numerous single nucleotide polymorphisms (SNPs) affecting goat litter size have been identified [
3,
4], yet most studies focus on single molecular markers, with limited research on multi-gene polymerization. Some traits or functions of biological organisms are controlled by multiple genes and belong to a regulatory network consisting of multiple genes and their products. Therefore, the effect of single genetic markers is relatively small and often cannot be practically applied in production, and polygenic polymerization is the first step to open the regulatory network of complex traits. In this study, the gonadotropin-releasing hormone receptor (
GnRHR), bone morphogenetic protein 6 (
BMP6) and follicle-stimulating hormone (
FSH) receptor (
FSHR) genes, which were the most prevalent, were used as candidate genes.
GnRHR gene is a high-affinity G-protein-coupled receptor of the pituitary gonadotropin membrane, which is located on chromosome 6 of sheep and is composed of 3 exons and 2 introns [
5,
6].
GnRHR gene as an essential mediator in the Gonadotropin-releasing hormone (
GnRH) signaling pathway,
GnRHR coupled to
GnRH, plays an important role in activating the downstream pathway after stimulating a series of cascades to regulate reproduction [
7]. In addition,
GnRHR can transduce the signal of
GnRH produced in the hypothalamus, regulate the synthesis and release of
FSH and LH in the anterior pituitary, thereby promoting the growth, maturation and regulation of the reproductive performance of the gonad tissue.
BMP6 gene is a member of the bone morphogenetic proteins (BMPs), which are members of the
TGF-β superfamily.
BMP6 cDNA was first isolated from mouse embryos by Lyons et al. [
8]. Recent studies have found that the
BMP6 gene directly participates in the regulation of the synthesis and secretion of reproduction-related hormones by inhibiting or promoting the activity of related enzymes or hormones in the body, thus affecting the formation and development of reproductive organs. For example, the
BMP6 gene can stimulate the synthesis of
FSH via the autocrine or paracrine pathway [
9,
10,
11]. El-Halawany et al. investigated the effect of
BMP6 gene polymorphisms on Egyptian sheep fecundity and its transcript expression in ovarian cells, but the result showed that the mutations of
BMP6 had no significance for litter size [
12].
The
FSHR gene, which contains 10 exons and 9 introns, is a glycoprotein family member of the G-protein-coupled receptor super-family, which mainly encodes the transmembrane domain and the intracellular domain in the 10th exon [
13].
FSHR cDNA was first cloned from rat Sertoli cells in 1990 and its genetic mutations may affect the ability of
FSH signal transduction [
14]. More importantly,
FSHR plays an important role in regulating the growth, development, differentiation and maturation of animal follicles. Borgbo et al. found that
FSHR deficiency in Homo sapiens can lead to primary infertility [
15]. Several studies have demonstrated that mutations of the
FSHR gene were significantly associated with litter size in Hu sheep, Small Tailed Han sheep and Jining Grey goats [
16,
17,
18]. Additionally, one study showed that the ovine
FSHR gene was expressed widely and was significantly higher in sexual glands (testicle and ovary) than in other tissues [
19].
This study analyzed four goat breeds: Chongqing black goat (CQ), Chuanzhong black goat (CZ), Leizhou goat (LZ), and Nubian goat (NS). Among them, NS were imported from abroad and then cultivated by local people, while the other three breeds are local goats in China. GnRHR, BMP6 and FSHR genes have been extensively studied in pigs, sheep and other livestock, but most of them are based on the effects of a single-marker on fecundity. So far, no reports have been published on the analysis of the pyramiding effect of these three genes in goats. Although the polygene breeding of plants has achieved initial success, there were almost no reports of success on goat breeding. The aim of this study is to detect the SNPs of GnRHR, BMP6, and FSHR genes in four goat breeds, investigate their relationship with litter size, and analyze the polygene pyramiding effect on goat prolificacy, providing a basis for molecular marker selection for goat litter-size traits.
2. Materials and Methods
2.1. Experimental Materials
A total of 959 does were collected, including 290 LZ from Wuzhou company Shengzhou breeding goat farm (Wuzhou, China), 391 NS from Lezhu breeding goat farm of Xinxing County WENS Xinwang Goat Industry Co., Ltd. (Yunfu, China), 91 CQ and 187 CZ from Liangdong breeding goat farm of Xinxing County WENS Xinwang Goat Industry Co., Ltd. (Yunfu, China). All goats were raised in the same managed conditions. Venous jugular blood samples (5 mL per doe) were collected from these does in 2017, using acid citrate dextrose as an anticoagulant, and stored at −20 °C until DNA extraction; at the same time, the data on litter size in the first, second, third, or fourth parity of each individual were recorded in detail.
In addition, 8 CZ does were selected from Liangdong breeding goat farm of Xinxing County WENS Xinwang Goat Industry Co., Ltd. (Yunfu, China), which were equally divided into a non-prolific group (single lambs in three to four consecutive parities) and a prolific group (multiple lambs in three to four consecutive parities). All goats, aged 3–4 years, healthy, were in the artificial insemination system. Twelve tissue samples (heart, liver, spleen, lung, kidney, dorsal longest muscle, hypothalamus, pituitary, ovary, oviduct, uterus and breast) were taken from different parts of each goat, and immediately placed in liquid nitrogen and then stored at −80 °C until RNA extraction.
2.2. DNA Extraction
Genomic DNA was extracted from whole blood with Tissue DNA Kit D3396 kit, and detected by electrophoresis on 1% agarose gels (TaKaRa, Osaka, Japan); then, the concentration and purity of DNA fragment was detected by nanodrop spectrophotometer, and kept at −20 °C.
2.3. RNA Extraction and cDNA Synthesis
Total RNA was extracted from caprine tissues using a kit according to the instructions of the manufacturer (Total RNA Kit R6731, OMEGA, Bienne, Switzerland). RNA integrity was ascertained by agarose gel electrophoresis, and the purity and concentration of RNA were determined by spectrophotometer. A measure of 1 μg of total RNA was used for reverse transcription in a final volume of 20 μL according to the manufacturer’s recommendations (PrimeScriptTM RT reagent Kit with gDNA Eraser, TaKaRa, Japan). Synthesized cDNA was stored at −20 °C until further use.
2.4. Primers Design
Using Primer Primier 5.0 software, design universal primers and real-time PCR primers for
GnRHR,
BMP6,
FSHR, and
β-actin genes based on GenBank sequences (the accession number of goat genomic
GnRHR,
BMP6 and
FSHR are NC_019463.2, NC_019477.2, and NC_019460.2, respectively) were obtained and synthesized by Beijing Huada gene Biotechnology Co., Ltd. (Beijing, China). All primer information is listed in
Table 1.
2.5. Genotyping of SNPs
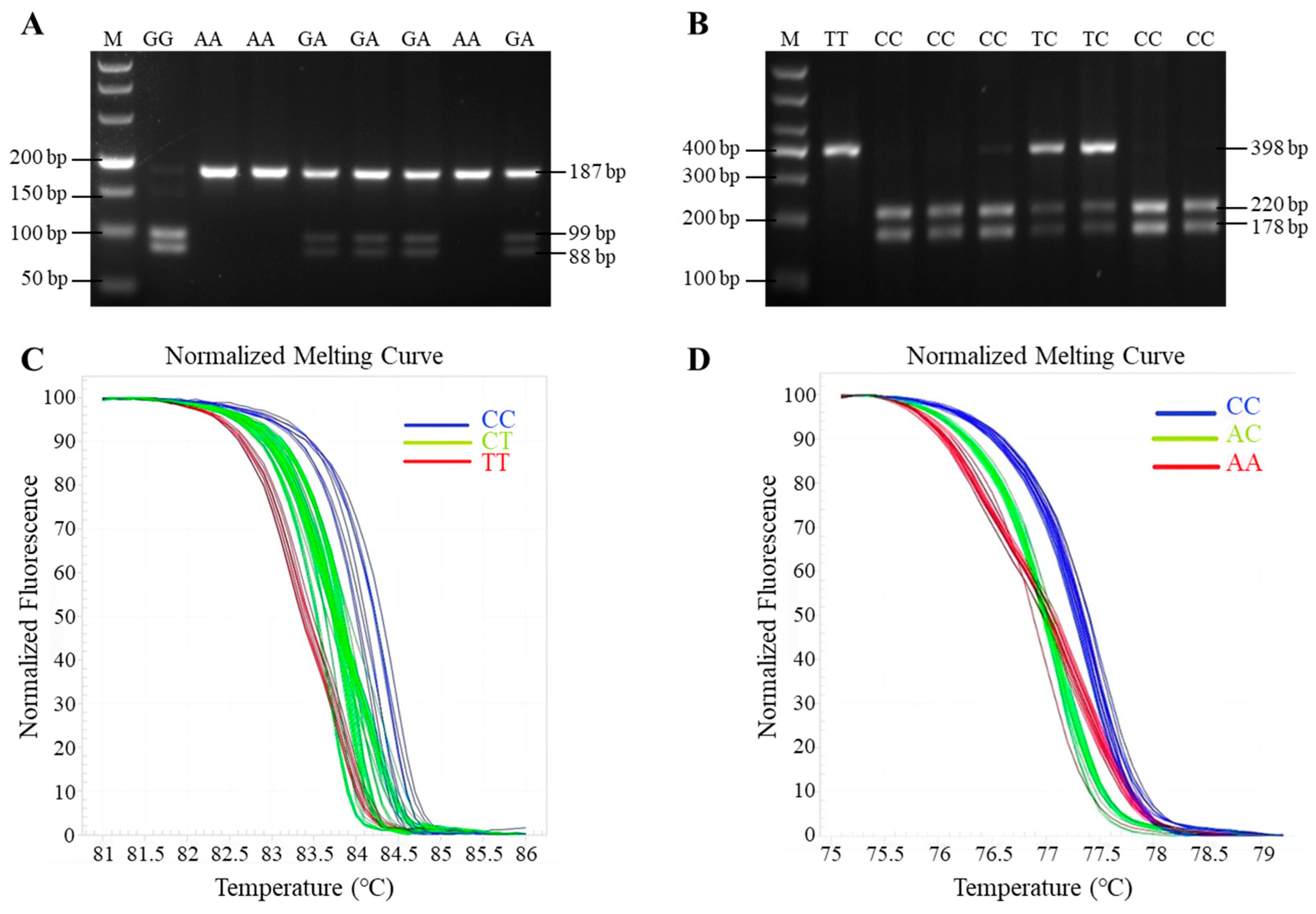
PCR was carried out in a 10 µL reaction mixture containing 5 µL Taq DNA polymerase (Takara), 3.4 µL of ddH2O, 1 µL of goat genomic DNA, and 0.3 µL each of forward and reverse primer, respectively. The PCR amplification procedure was as follows: pre-denaturation at 94 °C for 2 min; followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s; extension at 72 °C for 5 min; and with a final preservation at 4 °C for 10 min. The PCR products of GnRHR and BMP6 genes were digested separately with MspI (Thermo, Waltham, MA, USA) and HhaI (Thermo, Waltham, MA, USA) at 37 °C for 30 min in a 15 µL reaction mixture. The digestion system was as follows: 1 µL 10 × buffer, 8.5 µL of ddH2O, 0.5 µL restriction endonuclease (10 U/µL), and 5 µL PCR product. Then, the resultant fragments were separated by electrophoresis on 3% agarose gels.
HRM of FSHR gene was carried out in 10 µL reaction mixture containing 5 µL EvaGreen® dye (Takara), 3.4 µL RNase-free water, 1 µL of goat genomic DNA, and 0.3 µL upstream and downstream primers, respectively. The HRM procedure was as follows: pre-denaturation at 94 °C for 2 min; followed by 40 cycles of denaturation at 94 °C for 10 s, annealing at 60 °C for 10 s, and extension at 72 °C for 5 s; finally, the fluorescence changes were monitored continuously during the procedure at 95 °C for 15 s, 55 °C 15 s, 55–95 °C (0.1 °C/s temperature rise rate), and the melting curve was obtained; the whole process takes about 10 min.
2.6. Reaction System and Conditions for qPCR
The cDNA of all tissue samples was used for qPCR analysis. qPCR amplification was performed in 20 µL of reaction mixture containing 10 µL of SYBR
® Green qPCR Master Mix (2×), 0.4 µL of ROX Reference Dye II (50×), 0.4 µL each of forward and reverse primer, 2 µL of cDNA, and 6.8 µL of ddH
2O. PCR amplification was performed in triplicate wells, using the following procedure: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 10 s. Then, the melting curve was obtained, and the fluorescence changes were monitored continuously at 58 °C. The results were analyzed with the 2
−ΔΔCt method, using
β-actin as the reference gene. The primers are listed in
Table 1.
2.7. Statistical Analysis
Statistical analysis was conducted using GraphPad Prism version 8.0 (GraphPad Software, San Diego, CA, USA), with data presented as means ± SEM. Significant differences between two groups were determined using an unpaired two-tailed Student’s t-test or the Mann–Whitney U test for non-normally distributed samples. For data sets involving more than two groups, one-way ANOVA followed by the least significant difference (LSD) test or the non-parametric Kruskal–Wallis test was used, with SPSS 24.0 (SPSS Inc., Chicago, IL, USA). A p-value of less than 0.05 was considered statistically significant.
4. Discussion
In the present study, we selected the caprine
GnRHR,
BMP6 and
FSHR genes as candidate genes to analyze the effect of single-marker and multi-marker aggregation on litter size. The
GnRHR gene plays a vital role in the regulation of reproductive performance in goats. Several studies indicated that does carrying the mutation of the
GnRHR gene had significantly higher litter sizes compared with wild-type individuals [
20,
21,
22]. The caprine
BMP6 gene is crucial in promoting normal fertility and skeletal growth in mice [
23,
24,
25]. In the control study of sheep and rats, it was found that
BMP6 inhibited the progesterone production of both ovarian granulosa cells and stimulated the proliferation or survival of granulosa cells in rats. In contrast, it had no effect on the proliferation of ovarian granulosa cells but could inhibit its differentiation [
10]. In addition,
BMP6 can maintain a lower apoptotic rate in cumulus cells by forming a local concentration gradient [
26]. Numerous reports have shown that the
FSHR has a central role in animal follicular development and litter traits [
27,
28,
29]. Guo et al. research showed that allele C of 5′ regulatory region of the
FSHR gene was a potential marker for improving litter size in goats [
17]. Additionally, the g.47C > T mutation was found in the 5′ flanking region of the
FSHR gene in the Small Tailed Han sheep and Hu sheep, and was significantly associated with litter size [
19]. Overall, all these three genes could be selected as candidate genes for improving litter-size traits in goat husbandry.
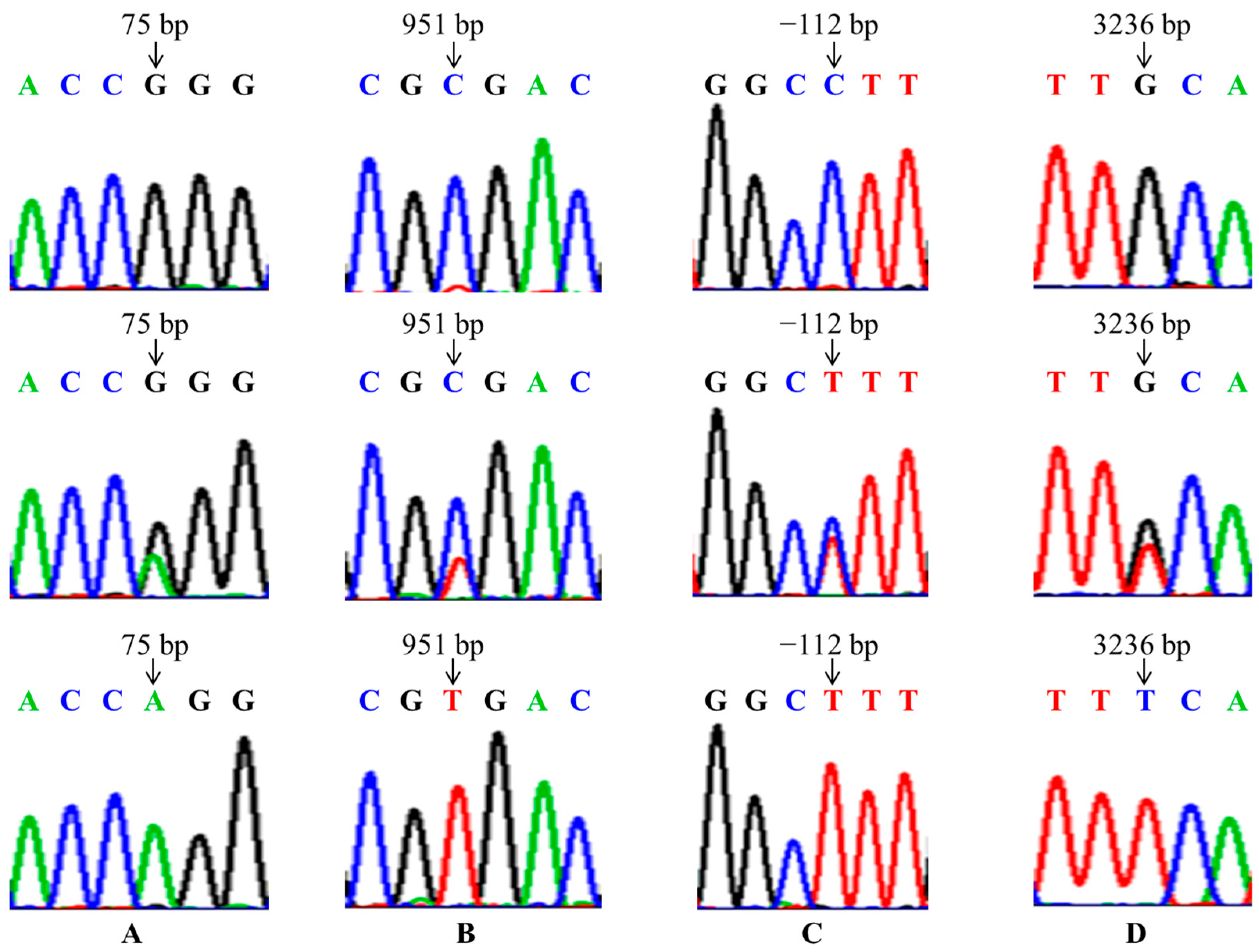
In the present study, the mutations of GnRHR (g.75G > A), FSHR (g.-112C > T, g.3236C > A) and BMP6 (g.951T > C) were found in four goat breeds, and were significantly associated with litter size in the CZ, CQ and DS breeds, so we can select the dominant genotypes to improve the fecundity of these three goat breeds in breeding work. But these four SNPs had no significant association with litter size in the NS. This may be because the NS had just been introduced into the field from Yunnan, and the growth environment was quite different, so its reproductive performance was still very unstable. Nonetheless, it may also be due to the small sample size or the distribution of the samples not being wide enough. Therefore, in order to make the research results more universal, we can further expand the sample size and sample distribution range in the following study.
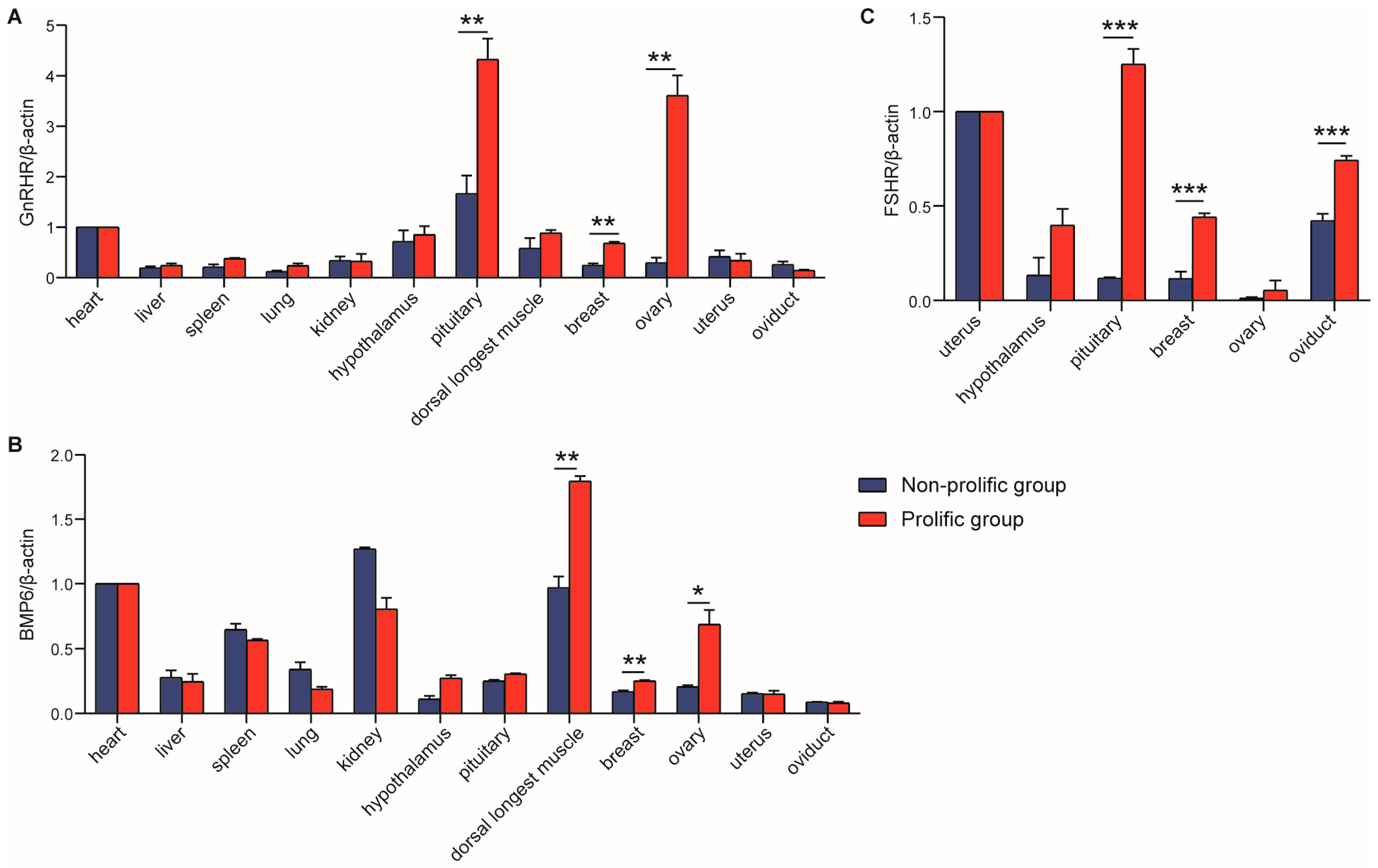
Interestingly, the expression of
BMP6 in ovarian cells suggests that
BMP6 would be the most likely ligand regulating follicular maturation and ovulation rate through BMPR-IB [
12]. In addition, studies have shown that the expression level of
GnRHR mRNA is highest in the pituitary tissues of rats, pigs, cattle and sheep [
30], which was consistent with our study. In this study,
GnRHR and
BMP6 were detected in all 12 tissues, which implied that they played a role in promoting differentiation in many body tissues. The highest expression of
GnRHR and
BMP6 was in the pituitary and longest dorsal muscle, respectively. Moreover, the
GnRHR mRNA expression level in breast and ovary in the prolific group was significantly higher than that in the non-prolific group;
BMP6 mRNA expression level in the longest dorsal muscle, breast and ovary in the prolific group was prominently higher than that in the non-prolific group. The results of Pan et al. showed that ovine
FSHR was expressed widely in detected tissues and was significantly higher in sexual glands (testicle and ovary) than in other tissues (heart, liver, spleen, lung, kidney, rumen, duodenum, muscle, fat, hypothalamus and pituitary) [
19]. However, in our study,
FSHR was checked only in reproduction-related tissues, and the expression level in the prolific group was significantly higher than the non-prolific group in the pituitary. This evidence suggested that
FSHR might play a different role in goats and sheep. To some extent, the expression of candidate genes in the ovary is positively correlated with the reproductive performance of mammals [
31,
32], and the function of the ovary is generally regulated by the pituitary gland. In this study, the high expression of
GnRHR and
FSHR in the pituitary of the prolific group may promote the development and maturation of the follicle; it was also explained by the fact that they could regulate the ovulation rate of the ovary by their expression level, and then participate in the mechanism of regulating litter size in animals [
33]. All these results further confirmed that these three genes are associated with the litter-size traits of goats.
So far, some studies have reported the effect of polygene pyramiding on litter size in sheep. Hanrahan et al. found that the ewes with mutations in both GDF9and BMP15 had a greater ovulation rate than those with either mutation alone in Cambridge and Belclare Sheep [
34]. In the research of Chu et al., the Small Tailed Han sheep carried genotype BB/G+ (BMPR-IB/BMP-15) showed a greater litter size than those with only one mutation [
35]. The result of Wang et al. indicated that mutations among
BMPR-IB,
BMP15 and
FSHR genes were found and the association analysis revealed that sheep with multiple markers had a higher litter size than those with single marker or two markers, and the combination genotype BB/G+/CC of
BMPR-IB,
BMP15 and
FSHR genes was considered the superior genotype [
18], which is consistent with the present study. There were four SNPs detected in the current experimental populations. Polygene polymerizing effect analysis revealed that the CQ and CZ with combination genotypes AATTCC and AACCAACC, and LZ with combination genotypes AAAACC and AACCCC had more kids than any other combination genotypes or those with only one predominant genotype among the does. Moreover, the combination genotypes AATTCC, AACCAACC and AAAACC were the combination of the predominant genotypes of
GnRHR,
BMP6 and
FSHR genes, indicating that polygene polymerizing may have a greater effect on contributing to the litter size of goats, and the combination genotypes AATTCC, AACCAACC, AAAACC and AACCCC could be considered as the superior genotypes and used as molecular markers in breeding work.
Obviously, it is unrealistic to predict that the does that are selected will have a high litter size in the future if they rely solely on molecular markers to detect the genotype of the base. There are many factors that affect the litter size of does, including temperature, nutrition, disease, and stress response. If the does carry a dominant genotype, it can only show that the does have the potential for high fecundity, and their potential is also influenced by external factors during the process of growth and development. If the environment is conducive to the growth and development of the does, the potential of their prolificacy is likely to be exerted, otherwise their potential will be buried. It can be seen that in order to give full play to the role of molecular markers related to litter size in production and breeding, we must pay attention to the feeding and management level of the goats at the same time, so as to get better breeding effects.