Simple Summary
Olive mill waste-water (OMWW) is a liquid waste produced by the olive oil industry that has been recently regarded as a good source of polyphenols. Phenolic molecules are among the most active secondary molecules in the gut for their antioxidant, anti-inflammatory and antimicrobial effects. They may also contribute to positively changing the distribution of gut microbial species, but their effects have not been widely explored in pigs. The intestinal porcine epithelial cell line IPEC-J2 represents a good model for the study of innate immunity and inflammatory response in animal intestinal diseases and has already been used to investigate the effect of phytogenic feed additives on swine intestinal epithelium. This study aimed to evaluate the in vitro effects of an OMWW extract enriched in polyphenols on Salmonella typhimurium (S. typhimurium) infection in IPEC-J2 cells. Polyphenols extracted from OMWW showed the ability to regulate the host–pathogen interaction by decreasing S. typhimurium invasiveness and modulating the expression of many innate immune genes.
Abstract
The dietary supplementation of olive oil by-products, including olive mill waste-water (OMWW) in animal diets, is a novel application that allows for their re-utilization and recycling and could potentially decrease the use of antibiotics, antimicrobial resistance risk in livestock species, and the occurrence of intestinal diseases. Salmonella serovar typhimurium is one of the most widespread intestinal pathogens in the world, causing enterocolitis in pigs. The aim of this study was to investigate the effect of an OMWW extract enriched in polyphenols (hydroxytyrosol and tyrosol) in the immune response of an intestinal porcine epithelial cell line (IPEC-J2) following S. typhimurium infection. Cells were pre-treated with OMWW-extract polyphenols (OMWW-EP, 0.35 and 1.4 µg) for 24 h and then infected with S. typhimurium for 1 h. We evaluated bacterial invasiveness and assayed IPEC-J2 gene expression with RT-qPCR and cytokine release with an ELISA test. The obtained results showed that OMWW-EP (1.4 µg) significantly reduced S. typhimurium invasiveness; 0.35 µg decreased the IPEC-J2 gene expression of IL1B, MYD88, DEFB1 and DEFB4A, while 1.4 µg down-regulated IL1B and DEFB4A and increased TGFB1. The cytokine content was unchanged in infected cells. This is the first study demonstrating the in vitro immunomodulatory and antimicrobial activity of OMWW extracts enriched in polyphenols, suggesting a protective role of OMWW polyphenols on the pig intestine and their potential application as feed supplements in farm animals such as pigs.
1. Introduction
The extraction of olive oil produces a series of by-products, including olive mill waste-water (OMWW)—olive vegetation water diluted in the water used during the oil-extraction process. This by-product is characterized by a high organic material load, ranging from 36.07 g/L to 230 g/L, and a content of phenolic compounds that varies from 0.9 to 30.5 g/L [1,2]. The large amount of this by-product (30 million m3), produced every year in the Mediterranean basin, contributes to environmental pollution due to the high presence of organic compounds, including phenolic ones [3,4,5,6]. This by-product comprises about 50% of the total phenolic compounds of the olive fruit [7], with different phenolic types, mainly tyrosol, hydroxytyrosol, verbascoside and oleuropein [8,9,10], which are highly known for their antioxidant, antimicrobial and anti-inflammatory activities [11,12,13,14,15]. The supplementation of olive by-products, including OMWW, as a source of polyphenols in animal diets potentially represents an innovative strategy for olive oil waste recycling, in line with the current concept of the circular economy [16,17,18,19,20,21,22].
In the swine industry, the use of antibiotics can favor the occurrence of antimicrobial resistance in bacteria of the pig intestinal microbiome, therefore increasing the risk of severe intestinal diseases and impairing the pig’s growth performance, especially at the weaning stage [23,24,25]. For this reason, this habit has been limited in various countries. Among pig intestinal diseases, salmonellosis is one of the most common and it represents a severe problem for the swine industry worldwide [26]. Salmonella enterica serovar typhimurium (S. typhimurium) is the agent of a very widespread enterocolitis form, which can be subclinical, but it can also be associated with a reduction in both productive performance and average daily gain in pigs [27,28].
In order to restrict the use of antibiotics, novel feeding strategies are required to modulate intestinal and immunological functions, as well as to improve the development and health of the swine gastrointestinal tract [23]. Given the correlation between bioactive molecules, such as polyphenols, and the pig intestinal microbiota and immune response to enteric diseases, their use can have a good impact on pig gut health [23]. The health benefits of polyphenols derive from their antioxidant, anti-inflammatory, and/or gene-regulating effects in tissues. Several studies showed that they help decrease the risk of many diseases, including intestinal ones, but the mechanisms correlated are not clear and need further investigation [29,30,31]. At present, they can be considered among the most active secondary bioactive molecules in the gut, contributing to beneficial changes in the distribution of gut microbial species, reducing pathogenic bacteria, and/or promoting the growth of probiotics [29,32,33]. A number of in vivo studies demonstrated that the administration of dietary polyphenols resulted in a reduction of pathogenic species and an increase in probiotic species in the intestinal microbiota of rats, pigs, and calves [32,34,35,36,37]. Olive oil by-products rich in polyphenols (e.g., olive leaf extract) were able to interfere with the growth of intestinal bacteria, including Salmonella [38,39]. Compared to human and laboratory animals (e.g., rats and mice), responses to polyphenols have been less explored in farm animals, such as pigs [29]. However, it has been recently demonstrated that the supplementation of natural polyphenols in piglets could contribute to alleviating weaning stress and improve intestinal barrier function, thus providing a nutritional strategy to protect intestinal health [40,41]. Other studies examined changes in the pig gut microbiome after the consumption of plant polyphenols, thanks to their ability to reduce oxidative stress and inflammation [42,43] and modulate immune cells and gut microbiota composition [29,44,45,46]. This action contributes to an improvement in intestinal bacterial function, decreases the release of microbial components into the circulation, and stimulates host immune response [47].
A suitable in vitro model to assess the immunomodulatory properties of polyphenols is represented by the porcine jejunal epithelial cell line IPEC-J2. This continuous cell line provides a valuable model to study both innate immunity and inflammatory responses in human and animal intestinal diseases [26,48,49,50,51,52]. Indeed, IPEC-J2 cells are intestinal porcine enterocytes isolated from the jejunum of an unsuckled neonatal pig, which showed the ability to express and produce cytokines, toll-like receptors (TLRs), defensins, and mucins [53]. In particular, these cells spontaneously secrete the pro-inflammatory chemokine IL-8 and possess ideal characteristics for in vitro studies on host–intestinal pathogen interactions [49,50,54]. Indeed, the primary host-cell barrier against pathogens is represented by the mucosal innate immune system, which is characterized by toll-like receptor (TLR) pathways, NF-kB signaling activation (with inflammatory cytokine release), and Type-I Interferon (IFN) responses [49,55]. Moreover, gastrointestinal tract homeostasis can be maintained when the immune response against commensal bacteria is controlled. When this equilibrium is compromised, excessive immune response causes an inflammatory condition [56]. Besides epithelial cells’ mechanical function, their role in gut microbiota homeostasis was recently recognized, as they are involved in maintaining the balance between host microbial components and gut immune cells [57]. In addition, IPEC-J2 cells mime the physiological characteristics of intestinal cells and have therefore been employed in several studies on Salmonella infections [53,58], providing valuable information on host responses to this bacteria. In fact, invasion with S. typhimurium in IPEC-J2 cells was comparable to that occurring in porcine ileal mucosal explants [59]. This cell line has been employed in studies focused on pigs’ innate immune response to dietary treatments [60], which can be regarded as a reference for in vitro studies of innate immunity in neonatal intra-epithelial cells after dietary stimuli [48,60]. These cells showed high morphological and functional similarities to porcine enterocytes in vivo; therefore, they were employed to evaluate the effects of phytogenic feed additives on swine intestinal epithelium [61]. In recent years, various plant-feed additives have demonstrated antioxidant, antimicrobial, and anti-inflammatory actions and other supporting barrier functions in IPEC-J2 cells [62,63,64,65].
With this study, we aimed to investigate in vitro the IPEC-J2 response to S. typhimurium infection after a pre-treatment with OMWW-extract polyphenols (OMWW-EP), to evaluate the influence on bacterial invasion and immune cells’ gene expression.
2. Materials and Methods
2.1. Olive Mill Waste-Water Extract and Composition
The OMWW extract enriched in polyphenols (hydroxytyrosol and tyrosol) was provided by Stymon Natural Products P.C., Patras, Greece (www.stymon.com, accessed on 21 December 2023). This product derives from OMWW of the olive (Olea Europaea L.) variety Koroneiki and is produced based on a unique patent (Patent number 1,010,150 IOBE (INT.CL.2021.01) A23L 19/00 A23L 33/105, Stymonphen Liquid) using only green technologies. Its total polyphenol content was equal to 15,000 ± 592 mg/kg, according to the Folin–Ciocalteu method [66]; hydroxytyrosol and tyrosol were the main phenolic compounds (8784 mg/kg and 1638 mg/kg, respectively), detected by HPLC-DAD [67]. The stock solution was filtered, vortexed, and diluted in phosphate-buffered saline (PBS, Euroclone, Milan, Italy) to reach 1400 µg/mL; from this, different scalar concentrations of polyphenols (0.35; 0.7; 7; 14; 70; 140 µg) were obtained for the successive analyses by diluting them in complete culture medium.
2.2. Cell Cultures
Porcine jejunal epithelial cells (IPEC-J2, IZSLER Cell Bank code BS CL 205) were grown in a mixture (1:1) of Dulbecco’s Modified Eagle (DMEM) (Euroclone, Milan, Italy) and Nutrient Mixture F-12 (F12) (Euroclone, Milan, Italy) enriched with 10% Fetal Bovine Serum (FBS, GIBCOTM, Thermofisher Scientific, Milan, Italy), 1% L-glutamine solution (Euroclone, Milan, Italy) and 1% penicillin/streptomycin solution (Euroclone, Milan, Italy) and kept in culture at 37 °C under 5% CO2.
2.2.1. Cell Viability
First, to determine the most suitable amount of OMWW extract to be used on IPEC-J2 cells, we tested different scalar phenolic dosages using a 2,3-bis-(2-methoxy-4-nitro-5-sulphophenyl)-2H-tetrazolium-5-carboxanilide (XTT) assay, according to the manufacturer’s instructions (XTT Cell Viability Assay, Cell Signaling Technology, Milan, Italy). In brief, IPEC-J2 cells were plated on 96-well plates (100 µL per well, 0.1 × 105) in complete culture medium and incubated for 24 h at 37 °C under 5% CO2 until confluence. The day after, the seeding cells on the 96-well plates were exposed to different doses of OMWW-EP (0.35, 0.7, 1.4, 14, 70, 140 µg), and untreated cells were employed as a negative control. Two independent experiments were performed, each including four technical replicates (four wells) for each of the seven experimental conditions: untreated cells (control) and cells treated with six different doses of OMWW-EP (0.35, 0.7, 1.4, 14, 70, and 140 µg). An XTT assay was performed at 24 h and at the end of the treatments, and the cell culture medium was removed and replaced with 100 µL of the fresh DMEM/F12 medium supplemented with XTT detected solution (1:50). The plates were then incubated again at 37 °C under 5% CO2 for 2 h, and the absorbance was measured at 450 nm using a multimode microplate reader (Glomax, Promega, Milan, Italy). This assay was performed two times for each phenolic concentration.
2.2.2. Bacterial Invasion
An isolate of S. typhimurium strain (ATCC 14028) was used to evaluate bacterial invasion in IPEC-J2 cells. In three independent experiments, IPEC-J2 cells were seeded into a 12-well plate (1 mL per well, 1.5 × 105 cells/mL) and incubated until confluence. Cells were treated with OMWW-EP (0.35 μg and 1.4 µg) for 24 h. S. typhimurium was stored at −80 °C until use, then thawed and grown overnight (18–24 h at 37 °C) in Brain Heart Infusion (BHI) (Sigma, Saint Louis, MO, USA). Then, it was sub-cultured in BHI and incubated for 2 h at 37 ± 1 °C to obtain a mid-log phase culture. The strain was pelleted and re-suspended in DMEM/F12 and L-glutamine medium to obtain a concentration of 108 CFU/mL and used to infect pig intestinal IPEC-J2 cells pre-treated with 0.35 μg and 1.4 µg of OMWW-EP for 24 h; infected cells without polyphenolic pre-treatment were used as comparison, while uninfected cells were employed as a negative control. For each of the three independent experiments, one plate was used, employing one well for each replicate, resulting in four replicates for each experimental condition: cells without phenolic pre-treatment and infected (ST); cells pre-treated with two OMWW-EP dosages and infected (ST + 0.35 µg POL; ST + 1.4 µg POL). Cells were stimulated with 1 mL/well of bacterial suspension at 108 CFU/mL and incubated at 37 °C under 5% CO2 for 1 h. Then, monolayers were washed five times with DMEM/F12 and L-glutamine medium (1 mL/well) and treated with 1 mL of colistin 300 μg/mL at 37 °C under 5% CO2 for 2 h to remove all extracellular bacteria. Cells were washed four times with medium and lysed by adding 200 μL/well 1% of Triton X-100 (Merck KgaA, Darmstadt, Germany) in PBS (Euroclone, Milan, Italy) at room temperature for 5 min (min); then, they were blocked by adding 800 μL of PBS to each well. The resulting cell suspension was vortexed, serially diluted in PBS, and seeded on XLD (Sigma, Saint Louis, MO, USA); then, it was incubated for 24–48 h at 37 °C for intracellular bacterial counts.
2.2.3. Modulation of the Immune Response
Cells from the IPEC-J2 line were seeded into 12-well plates (1 mL per well, 3 × 105 cells/mL) and then incubated at 37 °C under 5% CO2 until confluence. Two experimental designs were applied: the first one to evaluate the effect of OMWW-EP on IPEC-J2 gene expression and cytokine release, and the second one to investigate cellular pathways modulated by OMWW pre-treatment behind host–pathogen interactions in S. typhimurium infection. For the first experiment, cells were treated with OMWW-EP at 0.35 µg, 0.7 µg, 1.4 µg or 7 µg for 24 h, alongside untreated controls. A total of three independent experiments comprising two replicates each (one well for each replicate) were used for each experimental condition: cells with medium only (control); cells treated with OMWW-EP (0.35 µg POL, 0.7 µg POL, 1.4 µg POL and 7 µg POL). For the second, cells were treated with OMWW-EP at 0.35 µg or 1.4 µg for 24 h, then infected with 1 mL of 108 CFU/mL S. typhimurium suspension and incubated at 37 °C under 5% CO2 for 1 h. Moreover, cells infected with S. typhimurium without OMWW-EP pre-treatment were used as a control of the infection, and cells with the medium only were used as an untreated and uninfected control. To summarize, four independent experiments, including three replicates (one well for each replicate), were used for each experimental condition: cells with medium only (control); infected cells only (ST); cells pre-treated with polyphenols (0.35 and 1.4 µg); and infected (0.35 µg POL + ST; 1.4 µg POL + ST). After the first incubation, cells were washed five times and again incubated in their medium at 37 °C under 5% CO2 for 3 h. The resulting IPEC-J2 cell supernatants were stored at −80 °C until our evaluation of the cytokine contents. In parallel, cells were lysed with 400 µL of RLT Buffer (Qiagen, Hilden, Germany) and, after incubation for 10 min (min) at room temperature, collected and stored at −80 °C until RNA extraction and RT-q PCR analysis.
2.3. RNA Extraction and Reverse Transcription Quantitative PCR (RT-qPCR)
Total RNA was extracted from the cells described in Section 2.2.3 for both the experimental designs using Rneasy Mini Kit (Qiagen s.r.l., Milan, Italy) in the Qiacube System (Qiagen s.r.l., Milan, Italy), in accordance with the manufacturer’s instructions. The quality of extraction was assessed using a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). The same amount of RNA for each sample (250 ng) was reverse-transcribed into cDNA using a iScript cDNA Synthesis Kit (Bio-Rad, Milan, Italy). Amplification was performed on a CFX96TM Real-Time System (Bio-Rad, Milan, Italy) using SoFastTM Eva Green Supermix (Bio-Rad, Milan, Italy) following a protocol previously described [48]. Primers of target genes, coding for C-X-C motif chemokine ligand 8 (CXCL8), interleukin 1 beta (IL1B), IL18, nitric oxide synthase 2 (NOS2), nuclear factor kappa B subunit 1 (NFKB1), RELA proto-oncogene (NFKB/p65), toll-like receptor 4 (TLR4), toll-like receptor 5 (TLR5), myeloid differentiation primary response gene 88 (MYD88), transforming growth factor beta 1 (TGFB1), beta defensin 1 (DEFB1), beta defensin 2 (DEFB4A), and reference genes glyceraldehyde 3-phosphate dehydrogenase (GADPH) and hypoxanthine phosphoribosyltransferase 1 (HPRT1) were described in previous studies (Table 1). The relative normalized expression of the selected genes was assessed using the 2−ΔΔCt method [68], comparing different conditions. Samples scored negatively when the Ct was ≥39.

Table 1.
Primer set sequences of target and reference genes.
2.4. Cytokine Quantification
The cytokine content was investigated in culture supernatants of IPEC-J2 described in Section 2.2.3, using both experimental designs. Cells were treated for 24 h with OMWW-EP (0.35 and 1.4 µg) without infection. Moreover, after OMWW pre-treatment, cells were infected (1 h) with S. typhimurium after a polyphenolic pre-treatment, alongside the corresponding controls. The culture medium was changed, and the cells were incubated for 3 h at 37 °C under 5% CO2. Then, culture supernatants were collected, centrifuged (at 2500× g for 3 min), and kept at -80 °C until use. Levels of GM-CSF, IL-1α, IL-1β, IL-1Ra, IL-6, IL-8, IL-10, IL-18 were determined using the Porcine Cytokine/Chemokine Magnetic Bead Panel Multiplex assay (Merck Millipore, Darmstadt, Germany) and a Bioplex MAGPIX Multiplex Reader (Bio-Rad, Hercules, CA, USA), following the manufacturer’s instructions [48].
2.5. Statistical Analyses
A Kolmogorov–Smirnov test was conducted to check for Gaussian distribution in the data sets, concerning the viability assay, gene expression, cells invasion, and protein release. Data showing Gaussian distributions were checked for significant differences by one-way ANOVA or unpaired T-test. Results failing the Kolmogorov–Smirnov test were checked for significant differences by non-parametric Kruskal–Wallis test, followed by a Dunn’s Multiple Comparison post-hoc test. The significance threshold was set at p < 0.05 (Prism 5, GraphPad Software, GraphPad Software Inc., San Diego, CA, USA).
3. Results
3.1. Cell Viability
Cells from the IPEC-J2 line cells were exposed to scalar doses of OMWW-EP (0.35, 0.7, 1.4, 7, 14, 70, 140 µg), and 24 h later, viability was measured through an XTT assay. The XTT viability test showed that treatment with OMWW-EP at 140 μg and 70 μg induced a statistically significant (p < 0.0001) decrease in IPEC-J2 viability after 24 h (Figure 1) OMWW-EP exposition. The other concentrations tested did not show a significant effect. The two doses (0.35, 1.4 µg) not affecting IPEC-J2 viability were therefore selected for the following experiments.
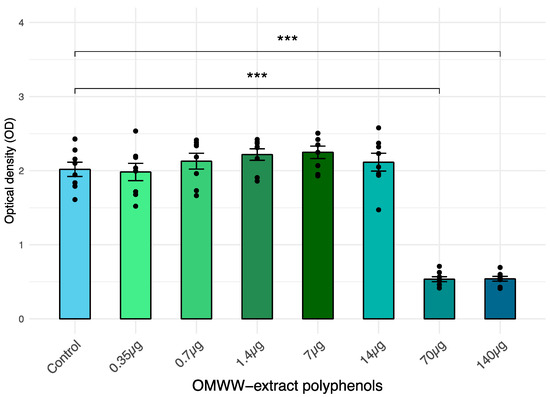
Figure 1.
IPEC-J2 viability after a 24 h exposition to the extract of liquid olive mill waste-water (OMWW) polyphenols (OMWW-EP: 0.35 μg; 0.7 μg; 1.4 μg; 7 µg; 14 µg; 70 µg; 140 µg). Cell viability was determined with XTT test. The number of living cells is expressed as optical density (OD) ± standard error (SE) and dots indicate samples included in each group. Statistical difference was calculated for all groups vs. Control (untreated cells): *** p < 0.001.
3.2. Salmonella Typhimurium Invasiveness
A significant (p < 0.05) decrease in S. typhimurium invasiveness into IPEC-J2 cells (p < 0.05; log10 CFU/3 × 105 cells) after an exposure to OMWW-EP of 1.4 μg for 24 h was demonstrated when compared with controls (untreated infected cells). CFU data were converted into log10 values (Figure 2).
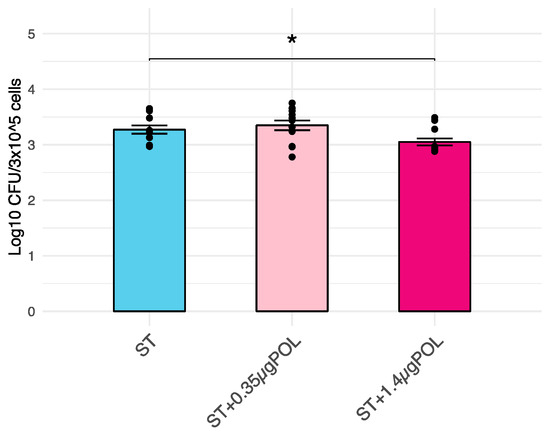
Figure 2.
Effects of OMWW-extract polyphenols (OMWW-EP) on S. typhimurium penetration into IPEC-J2 cells. Data are expressed as log10 CFU of penetrated, intracellular ST/3 × 105 cells. The mean value of five replicates + standard error is presented, and dots indicate samples included in each group. The significant difference between S. typhimurium infected cells and pretreated with different concentrations of OMWW-EP (ST + 0.35 µg POL—pink; ST + 1.4 µg POL—fuchsia) and S. typhimurium infected cells (ST—blue) is indicated by * (* p < 0.05).
3.3. Modulation of Immune Response
The immunomodulant effect of OMWW-EP at two dosages (0.35 µg and 1.4 µg) was monitored through RT-qPCR and ELISA tests.
3.3.1. OMWW-Extract Polyphenols’ Effect on IPEC-J2 Gene Expression and Cytokine Release
The effect of OMWW-EP (0.35 μg and 1.4 μg) treatment for 24 h on IPEC-J2 cells was monitored through RT-qPCR. A panel of seven genes was analyzed (Table 1), and the levels of treated cells were compared to untreated control cells. Moreover, complete results for the other polyphenol dosages are reported in the Supplementary Figure S1. A significant decrease in CXCL8 (p < 0.001), IL18 (p < 0.05), and MYD88 (p < 0.001) and a significant increase in NOS2 (p < 0.05) were observed in cells exposed to 0.35 µg of OMWW-EP (Figure 3). The treatment with 1.4 µg of OMWW-EP triggered a significant decrease in CXCL8 (p < 0.001) and MYD88 (p < 0.001). The other genes under study were not significantly modulated (Figure 3).
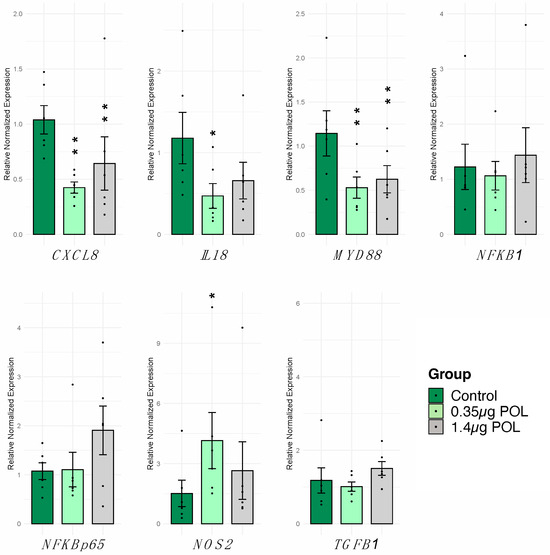
Figure 3.
Effects of 24 h OMWW-extract polyphenols on IPEC-J2 gene expression. The RT-qPCR analysis was performed to evaluate CXCL8, IL18, MYD88, NFKB1, NFKB/p65, TGFB1 and NOS2 gene expression. Data are presented as bar plots displaying the mean value of normalized expression, standard error as error bars and dots indicate samples included in each group. For each gene and cytokine, differences between treated with polyphenols (0.35 μg POL—light green; 1.4 μg POL—gray) vs. untreated (Control—dark green) cells were evaluated through one-way ANOVA followed by a Dunnett’s test or a Kruskal–Wallis test followed by Dunn’s multiple comparison test; * p < 0.05, ** p < 0.01.
3.3.2. OMWW-Extract Polyphenols’ Effect on IPEC-J2 Cytokine Release
In parallel, the impact of scalar doses of OMWW-EP (0.35, 1.4 μg) on cytokine levels in IPEC-J2 culture supernatants was investigated using multiplex ELISA. Eight cytokines were tested: IL-1α, IL-1β, IL-1Ra, IL-6, IL-8, IL-10, IL-18 and GM-CSF (Figure 4). The levels of GM-CSF and IL-β were below the assay detection limit. Exposure to polyphenols did not alter the levels of IL-1α, IL-1Ra, and IL-10 in IPEC-J2 culture supernatants (Figure 4). In agreement with the RT-qPCR data, these compounds decreased the levels of the pro-inflammatory cytokines IL-6, IL-8, and IL-18, although a statistically significant difference was observed only for the latter (Figure 4).
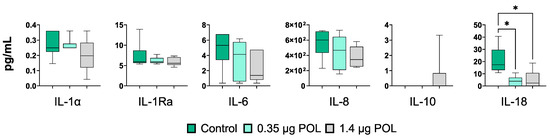
Figure 4.
Effects of OMWW-extract polyphenols on IPEC-J2 cytokine release through multiplex ELISA measuring cytokine contents in culture supernatants (IL-1α, IL-1Ra, IL-6, IL-8, IL-10, IL-18). Data are presented as box and whisker plots displaying median and interquartile range (boxes) and minimum and maximum values (whiskers). For each gene and cytokine, differences between treated with polyphenols (0.35 μg POL—light green; 1.4 μg POL—gray) and untreated (Control—dark green) cells were evaluated through one-way ANOVA followed by a Dunnett’s test or a Kruskal–Wallis test followed by Dunn’s multiple comparison test; * p < 0.05.
3.3.3. OMWW-Extract Polyphenols’ and S. typhimurium Infection Effects on IPEC-J2 Gene Expression
The effect of OMWW-EP pre-treatment for 24 h in IPEC-J2 responses to S. typhimurium infection (for 1 h) was evaluated. First, a panel of 10 genes was analyzed through RT-qPCR (Table 1). We observed a significant increase in CXCL8 (p < 0.0001), MYD88 (p < 0.0016), DEFB1 (p < 0.0001), and DEFB4A (p < 0.0044) in cells not exposed to OMWW (control) in response to S. typhimurium infection (Figure 5).
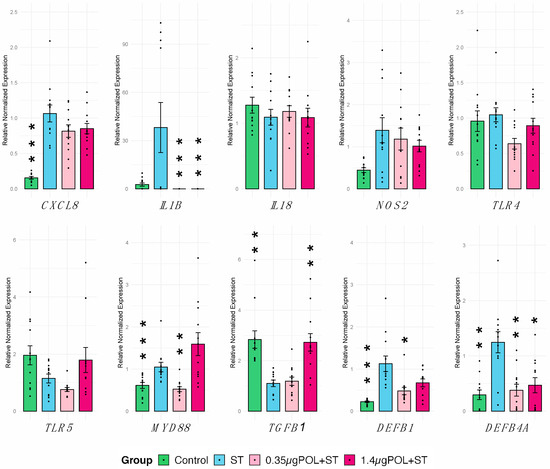
Figure 5.
Gene expression of IPEC-J2 cells in response to S. typhimurium infection with OMWW-extract polyphenol (OMWW-EP) pre-treatment. The tested conditions for IPEC-J2 cells were: uninfected and untreated cells (Control—dark green), infected with S. typhimurium (ST—blue), pre-treated with 0.35 µg OMWW-EP and infected (0.35 µg POL + ST—pink), and pre-treated with 1.4 µg OMWW-EP and infected (1.5 µg POL + ST—fuchsia). Data are reported as mean value and standard error, and dots indicate samples included in each group. Statistical tests were carried out comparing all conditions vs. ST. Significant differences: * p < 0.05, ** p < 0.01, *** p <0.001.
Then we investigated the impact of OMWW-EP in IPEC-J2 ability to respond to S. typhimurium infection. Different results were obtained depending on polyphenol dosages. The pre-treatment of infected cells with 0.35 µg induced a significant decrease in IL1B (p = 0.019), MYD88 (p = 0.062), DEFB1 (p < 0.0001) and DEFB4A (p = 0.0012) compared to untreated infected cells, whereas with a pre-treatment of infected cells using 1.4 µg showed a significant decrease in IL1B (p = 0.019) and DEFB4A (p = 0.023) and a significant increase in TGFB1 (p = 0.008) (Figure 5). Other genes under study were not significantly modulated (Figure 5).
3.3.4. OMWW-Extract Polyphenols’ and S. typhimurium Infection Effects on IPEC-J2 Cytokine Production
To further investigate the immunomodulatory properties of OMWW-EP, we assayed the cytokine contents in the supernatants of un-infected and untreated IPEC-J2 and S. typhimurium-infected cells (ST) pretreated or not with OMWW-EP (0.35 μg or 1.4 μg) (Figure 6). S. typhimurium infection triggered an enhanced release of pro-inflammatory cytokines, such as IL-1α (p = 0.02927), IL-6 (p < 0.0001), and IL-8 (p = 0.0006) (Figure 6). OMWW-EP did not affect the cytokine content in S. typhimurium-infected IPEC supernatants (Figure 6). Values of GM-CSF and IL-1β were below the reference range values.
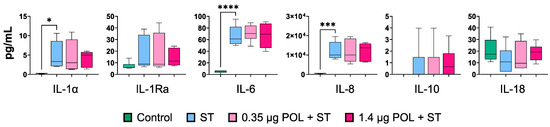
Figure 6.
Cytokine release by IPEC-J2 cells in response to S. typhimurium infection after OMWW-extract polyphenol (OMWW-EP) pre-treatment. The tested conditions for IPEC-J2 cells were: uninfected and untreated cells (Control—dark green), infected with S. typhimurium (ST—blue), pre-treated with 0.35 µg OMWW-EP and infected (0.35 µg POL + ST—pink), and pre-treated with 1.4 µg OMWW-EP and infected (1.5 µg POL + ST—fuchsia). For each cytokine, differences between ST-infected cells and the other conditions were evaluated through one-way ANOVA followed by a Dunnett’s test or a Kruskal–Wallis test followed by Dunn’s multiple comparison test; * p < 0.05, *** p < 0.001; **** p < 0.0001.
4. Discussion
Gut epithelial cells have a predominant role as the first defense from pathogenic insults [53,57]. Thus, the obtained results regarding the modulation of immune genes in the intestinal epithelium after treatment with polyphenols are a prodromal step to feed supplementation with polyphenols in livestock species. The IPEC-J2 cell line was chosen on the basis of our previous studies [48,49,50,52]. Indeed, it represents a good model for investigating epithelial immune response in pigs, in order to evaluate the ability of OMWW polyphenols to modulate the in vitro gut immunological response to S. typhimurium infection.
Our screening of different amounts of OMWW-extract polyphenols (OMWW-EP) carried out in the first part of the study through the IPEC-J2 viability test allowed us to choose the appropriate dosages for the successive analyses (Figure 1). We then investigated the effect of pre-treatment with OMWW-EP (0.35 and 1.4 μg) on IPEC-J2 cells for 24 h with or without an infective insult and found that exposure to these compounds triggered a decreased expression of CXCL8, IL18, and MYD88 genes; IL-18 release; and an up-regulation of NOS2 gene expression (Figure 3 and Figure 4). The anti-inflammatory effect of polyphenols is due to complex cellular mechanisms that are still not clear, but most of them have been correlated with the NF-kB pathway [45]. CXCL8 is a pro-inflammatory chemokine (IL-8)-encoding gene whose expression could be regulated by the TLR4/MyD88/NF-kB pathway. In our study, OMWW-EP seemed to exert anti-inflammatory action by decreasing MYD88 (a gene with a pivotal role in NF-kB activation) and therefore CXCL8 gene expression. Indeed, in other studies, dietary supplementation with grape seed cake (another by-product rich in polyphenols) was shown to significantly reduce MYD88 gene expression in the colon of Dextran Sulfate Sodium (DSS)-treated piglets [70], and Li et al. [71] demonstrated the ability of other natural polyphenols (the flavonoids quercetin and catechin) to restore the increased expression of MYD88 in LPS-stimulated murine macrophage RAW 264.7 cells.
In addition, our data demonstrated the ability of OMWW-EP to decrease both the expression and secretion of IL-18. IL-18 is a member of the IL-1 family, with an important role in the inflammatory response [72,73,74,75,76,77]. Its release must be tightly controlled, in order to avoid the development of auto-inflammatory diseases [73].
These data suggest a possible effect of OMWW polyphenols on host–pathogens interaction, which was successively tested in vitro using S. typhimurium assay. In this way, the ability of a pre-treatment with different dosages of OMWW-EP (0.35 and 1.4 µg) to decrease S. typhimurium invasiveness and modulate immune response related-genes in S. typhimurium-infected cells was assessed. First of all, our data confirmed S. typhimurium’s ability to penetrate IPEC-J2 cells (Figure 2), which is known to be related to the up-regulation of the pro-inflammatory molecule CXCL8 [49], as we found (Figure 5 and Figure 6). In our analysis, we additionally found that S. typhimurium’s invasion of IPEC-J2 significantly increases the expression of MYD88 gene (Figure 5) encoding for the MyD88 adaptor protein, which is the mediator of NF-kB activation, essential for the stimulation of pro-inflammatory gene expressions. Not surprisingly, we therefore observed the increased release of other pro-inflammatory cytokines (IL-1α, IL-6, IL-18) after S. typhimurium infection, in accordance with a previous study [49] (Figure 6). It is known that mucosal bacteria are able to stimulate the transcription of pro-inflammatory genes through epithelial cell invasion, interacting with many receptors such as TLR or acting on NF-kB [44]. Surface-expressed TLRs are activated by the pathogen-associated molecular patterns (PAMPs), which are microbe structures, exploiting the adaptor molecule MyD88 and stimulating NF-kB translocation into the nucleus [49]. The activation of the nuclear factor NF-kB leads to the increased transcription of pro-inflammatory mediators (such as the cytokines IL-8, IL-1B, IL-6 and TNF) [78], as shown in our experiment (Figure 6).
Interestingly, the pre-treatment with OMWW extract enriched in polyphenols reduced S. typhimurium invasiveness. Thus, we tried to highlight molecules influencing this host–pathogen interaction, modulated by the polyphenol treatment. Firstly, the expression of two TLRs (TLR4 and TLR5) was investigated, but no effects on S. typhimurium invasion after OMWW-EP pre-treatment were observed.
As for the effects on pro-inflammatory cytokines and related pathways, we observed a down-regulation of IL1B in S. typhimurium-infected IPEC-J2 cells after OMWW-EP pre-treatment (both dosages). A down-regulation of MYD88 for the 0.35 µg group was detected as well.
The expression of MYD88 was also investigated and showed a decrease in S. typhimurium-infected cells in the 0.35 µg pre-treatment group, while MYD88 gene expression was raised in cells infected and not pre-treated with OMWW-EP. The pre-treatment with 0.35 µg OMWW-EP probably prevented the activation of NF-kB and pro-inflammatory mediators through the down-regulation of this adaptor molecule-encoding gene (MYD88).
Moreover, inflammasome-induced cell death contributes to host control of S. typhimurium infection. Species differences in inflammasomes may contribute to zoonotic immune tolerance. Inflammasomes are molecular platforms that promote the maturation of the proinflammatory cytokines IL-1β and IL-18. During enteric Salmonella infection, the activation of caspase-1 and the production of IL-1β and IL-18 provide a protective host response [79]. The inflammasome activation could be mediated by MyD88, but there are other pathways in the activation signaling: various PAMPs, DAMPs, or intracellular changes induce the formation of the NLRP3 inflammasome composed of NLRP3 as a PRR, pro-caspase-1, and adapter proteins such as the apoptosis-associated speck-like protein containing a caspase recruitment domain [80].
At both dosages (0.35 and 1.4 µg), the polyphenolic pre-treatment induced a down-regulation of pro-inflammatory cytokine IL1B, which is also involved in the inflammasome reaction, together with IL-18 [73,74,75,76,77]. The combined effect of OMWW-EP on IL-18, which induced gene expression and reduced cytokine release (without Salmonella infection), leads the authors to suppose that this is the pathway through which OMWW-EP potentially protects IPEC-J2 cells against S. typhimurium infection.
In line with our results, the ability of the polyphenol resveratrol to potentially protect the intestinal barrier against deoxynivalenol (DON)-induced dysfunction and Escherichia coli (E. coli) translocation in IPEC-J2 cells [64] and S. typhimurium infection was demonstrated. Several in vitro studies concerning intestinal cells demonstrated that plant extracts rich in polyphenols or isolated molecules can limit induced-inflammation processes [30,81,82,83,84]. It was also shown that natural polyphenols can modulate inflammasome activation [77], interfering with the production (both at mRNA and protein levels) of pro-inflammatory mediators [30]. Moreover, it has been demonstrated that IL1-β is reduced by polyphenols such as curcumin and resveratrol [84,85,86]. Other good sources of polyphenols, i.e., dietary grape seed cake, decreased IL1B gene expression and protein concentration in fattening pigs’ spleens [87]. Feeding weaned pigs with polyphenol-rich plant products (grape seed, grape marc meal extract, and spent hops) down-regulated various pro-inflammatory cytokines, including IL-1β [44], in the intestine, and the oleuropein glycoside polyphenol significantly decreased the release of IL1-β in LPS-stimulated human whole-blood cultures [88].
Meanwhile, we did not observe differences between OMWW-EP-treated and untreated IPEC-J2 cells concerning the expression and release of other pro-inflammatory cytokines in response to S. typhimurium infection. This probably relates to the fact that these inflammatory molecules are primarily stimulated by TLR4 receptors, whose expression seemed to not be significantly modulated by polyphenols. We also might speculate that OMWW-EP could reduce the levels of pro-inflammatory cytokine release in response to Salmonella if a lower infective dose is used.
Not only pro-inflammatory but also anti-inflammatory cytokines such as IL-10 and TGF-β were tested. In particular, TGF-β can dampen the inflammatory effects of cytokines such as IL-1β, IL-12, TNF [89].
In our experiments, we observed that S. typhimurium infection determined a decrease in TGFB1 expression in IPEC-J2 cells. This is not surprising, considering a recent study by Qin et al. [90], who mimed the bacterial infection process with an LPS stimulus in human Caco-2 colon cells. The authors observed a down-regulation of several genes involved in the inflammation response linked to TGF-beta signaling pathways. Interestingly, the pre-treatment with OMWW-EP (1.4 µg) induced the up-regulation of TGFB1 in infected cells compared to cells that were infected without OMWW-EP pre-treatment. The TGFB1 gene encodes the TGF-beta superfamily ligands and binds different TGF-beta receptors, regulating gene expression as well as cell growth, proliferation, and differentiation [91]. It is produced by different cell types, including the intestinal cells [92], and is a cytokine involved in the homeostasis of the epithelial barrier, which is normalized by up-regulating the expression of tight junction proteins [93]. The up-regulation of this gene in our experiments may be correlated with a possible inhibitory action of bacterial replication inside the cells, in line with Huang et al. [91]., who demonstrated that, in pigs, the inhibition of S. typhimurium intracellular replication can be associated with TGFB1 increase. Moreover, in a study by Nallathambi et al. [93], a polyphenol-rich grape seed extract was able to enhance TGFB1 expression in Caco-2 cells, in line with the observed increase in tight junction protein expression.
Finally, the expression of genes coding for antimicrobial peptides (AMPs), released during early response to invading pathogens, was investigated. These molecules show efficacy in disrupting both the Gram-positive and Gram-negative bacterial membranes and are also expressed in epithelial cells of the gastrointestinal tract [26]. Beta-defensins are known AMPs also involved in the maintenance of the homeostasis in the gut microbiota, regulating its composition and thus protecting from microbial pathogens [48,75,94], and it is well known that IPEC-J2 cells express beta-defensin genes [26]. Our results showed that S. typhimurium invasion up-regulated DEFB1 and DEFB4A gene expressions in IPEC-J2 cells, as reported by previous studies [26,49]. DEFB4A expression was also increased in another porcine ileum epithelial cell line (IPI-2I) after infection with S. typhimurium DT104 [26]. Furthermore, it was demonstrated that E. coli adhesion increases the expression of DEFB1 and DEFB4A in IPEC-J2 cells [95]. In this study, pre-treatment with OMWW-EP seemed to induce a return to the basal expression of DEFB1 (at the 0.35 µg dosage) and DEFB4A (at both dosages) after S. typhimurium infection, counteracting the effect of bacterial invasion and potentially restoring gut homeostasis.
5. Conclusions
Our results confirmed the potential ability of OMWW-EP to modulate host–pathogen interactions in pigs by inducing an alteration of S. typhimurium invasiveness. In particular, our data showed a significant reduction of S. typhimurium’s ability to invade cells pre-treated with 1.4 µg of OMWW-EP compared to untreated IPEC-J2 cells. Furthermore, pre-treatment (independently from the dosage) with OMWW-EP modulated several innate immune-response genes influencing the S. typhimurium invasiveness in IPEC-J2, exhibiting potential antimicrobial activity by decreasing intracellular bacterial replication. This is the first study performed in an in vitro swine intestinal model that suggests a potential protective role of OMWW polyphenols in the pig intestine, paving the way for in vivo studies to confirm these promising results; increasing our knowledge of related molecular mechanisms; and making the possible use of this by-product feasible for livestock animal welfare and health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani14040564/s1, Figure S1: Effect of 24 h OMWW-extract polyphenols on IPEC-J2 gene expression.
Author Contributions
Conceptualization, M.T.-M., E.R. and K.C.; methodology, F.F. (Flavia Ferlisi), C.G.D.C., F.F. (Floriana Fruscione); validation, K.C.; formal analysis, E.R., C.G.D.C., F.F. (Flavia Ferlisi) and S.M.; investigation, F.F. (Flavia Ferlisi), C.G.D.C., G.F., S.Z., F.F. (Floriana Fruscione) and R.G.; resources, E.R.; data curation, E.R. and S.M.; writing—original draft preparation, F.F. (Flavia Ferlisi) and K.C.; writing—review and editing, G.F., S.Z., C.G.D.C., S.M., R.G., M.T.-M. and E.R.; visualization, S.M.; supervision, K.C., E.R. and M.T.-M.; project administration, M.T.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in article and Supplementary Materials.
Acknowledgments
The authors would like to thank Paraskevas-Domenico Pettas (Chemist, CEO of Stymon Natural Product P.C.) for providing the OMWW polyphenolic extract and Gianluca Alunni for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Capasso, R.; Cristinzio, G.; Evidente, A.; Scognamiglio, F. Isolation, Spectroscopy and Selective Phytotoxic Effects of Polyphenols from Vegetable Waste Waters. Phytochemistry 1992, 31, 4125–4128. [Google Scholar] [CrossRef]
- Mekki, A.; Dhouib, A.; Sayadi, S. Changes in Microbial and Soil Properties Following Amendment with Treated and Untreated Olive Mill Wastewater. Microbiol. Res. 2006, 161, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Ben Sassi, A.; Boularbah, A.; Jaouad, A.; Walker, G.; Boussaid, A. A Comparison of Olive Oil Mill Wastewaters (OMW) from Three Different Processes in Morocco. Process Biochem. 2006, 41, 74–78. [Google Scholar] [CrossRef]
- Casa, R.; D’Annibale, A.; Pieruccetti, F.; Stazi, S.R.; Giovannozzi Sermanni, G.; Lo Cascio, B. Reduction of the Phenolic Components in Olive-Mill Wastewater by an Enzymatic Treatment and Its Impact on Durum Wheat (Triticum durum Desf.) Germinability. Chemosphere 2003, 50, 959–966. [Google Scholar] [CrossRef] [PubMed]
- El Hadrami, A.; Belaqziz, M.; El Hassni, M.; Hanifi, S.; Abbad, A.; Capasso, R.; Gianfreda, L.; El Hadrami, I. Physico-chemical characterization and effects of olive oil mill wastewaters fertirrigation on the growth of some Mediterranean crops. J. Agron. 2004, 3, 247–254. [Google Scholar] [CrossRef]
- Quaratino, D.; D’Annibale, A.; Federici, F.; Cereti, C.F.; Rossini, F.; Fenice, M. Enzyme and Fungal Treatments and a Combination Thereof Reduce Olive Mill Wastewater Phytotoxicity on Zea mays L. Seeds. Chemosphere 2007, 66, 1627–1633. [Google Scholar] [CrossRef] [PubMed]
- Rodis, P.S.; Karathanos, V.T.; Mantzavinou, A. Partitioning of Olive Oil Antioxidants between Oil and Water Phases. J. Agric. Food Chem. 2002, 50, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Bertin, L.; Ferri, F.; Scoma, A.; Marchetti, L.; Fava, F. Recovery of High Added Value Natural Polyphenols from Actual Olive Mill Wastewater through Solid Phase Extraction. Chem. Eng. J. 2011, 171, 1287. [Google Scholar] [CrossRef]
- De Marco, E.; Savarese, M.; Paduano, A.; Sacchi, R. Characterization and Fractionation of Phenolic Compounds Extracted from Olive Oil Mill Wastewaters. Food Chem. 2007, 104, 858–867. [Google Scholar] [CrossRef]
- Varricchio, E.; Coccia, E.; Orso, G.; Lombardi, V.; Imperatore, R.; Vito, P.; Paolucci, M. Influence of Polyphenols from Olive Mill Wastewater on the Gastrointestinal Tract, Alveolar Macrophages and Blood Leukocytes of Pigs. Ital. J. Anim. Sci. 2019, 18, 574–586. [Google Scholar] [CrossRef]
- Branciari, R.; Ranucci, D.; Ortenzi, R.; Roila, R.; Trabalza-Marinucci, M.; Servili, M.; Papa, P.; Galarini, R.; Valiani, A. Dietary Administration of Olive Mill Wastewater Extract Reduces Campylobacter Spp. Prevalence in Broiler Chickens. Sustainability 2016, 8, 837. [Google Scholar] [CrossRef]
- Hume, D.A.; Whitelaw, C.B.A.; Archibald, A.L. The Future of Animal Production: Improving Productivity and Sustainability. J. Agric. Sci. 2011, 149, 9–16. [Google Scholar] [CrossRef]
- Obied, H.K.; Bedgood, D.R.; Prenzler, P.D.; Robards, K. Bioscreening of Australian Olive Mill Waste Extracts: Biophenol Content, Antioxidant, Antimicrobial and Molluscicidal Activities. Food Chem. Toxicol. 2007, 45, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Servili, M.; Sordini, B.; Esposto, S.; Urbani, S.; Veneziani, G.; Di Maio, I.; Selvaggini, R.; Taticchi, A. Biological Activities of Phenolic Compounds of Extra Virgin Olive Oil. Antioxidants 2013, 3, 1–23. [Google Scholar] [CrossRef]
- Veneziani, G.; Novelli, E.; Esposto, S.; Taticchi, A.; Servili, M. Applications of Recovered Bioactive Compounds in Food Products. In Olive Mill Waste: Recent Advances for Sustainable Management; Academic Press: Cambridge, MA, USA, 2017; pp. 231–253. ISBN 978-0-12-805314-0. [Google Scholar]
- Salomone, R.; Ioppolo, G. Environmental Impacts of Olive Oil Production: A Life Cycle Assessment Case Study in the Province of Messina (Sicily). J. Clean. Prod. 2012, 28, 88–100. [Google Scholar] [CrossRef]
- Vastolo, A.; Calabrò, S.; Cutrignelli, M.I. A Review on the Use of Agro-Industrial CO-Products in Animals’ Diets. Ital. J. Anim. Sci. 2022, 21, 577–594. [Google Scholar] [CrossRef]
- Branciari, R.; Galarini, R.; Giusepponi, D.; Trabalza-Marinucci, M.; Forte, C.; Roila, R.; Miraglia, D.; Servili, M.; Acuti, G.; Valiani, A. Oxidative Status and Presence of Bioactive Compounds in Meat from Chickens Fed Polyphenols Extracted from Olive Oil Industry Waste. Sustainability 2017, 9, 1566. [Google Scholar] [CrossRef]
- Sabino, M.; Cappelli, K.; Capomaccio, S.; Pascucci, L.; Biasato, I.; Verini-Supplizi, A.; Valiani, A.; Trabalza-Marinucci, M. Dietary Supplementation with Olive Mill Wastewaters Induces Modifications on Chicken Jejunum Epithelial Cell Transcriptome and Modulates Jejunum Morphology. BMC Genom. 2018, 19, 576. [Google Scholar] [CrossRef]
- Ferlisi, F.; Tang, J.; Cappelli, K.; Trabalza-Marinucci, M. Dietary Supplementation with Olive Oil Co-Products Rich in Polyphenols: A Novel Nutraceutical Approach in Monogastric Animal Nutrition. Front. Vet. Sci. 2023, 10, 1272274. [Google Scholar] [CrossRef]
- Maranesi, M.; Dall’Aglio, C.; Acuti, G.; Cappelli, K.; Trabalza Marinucci, M.; Galarini, R.; Suvieri, C.; Zerani, M. Effects of Dietary Polyphenols from Olive Mill Waste Waters on Inflammatory and Apoptotic Effectors in Rabbit Ovary. Animals 2021, 11, 1727. [Google Scholar] [CrossRef]
- Cappelli, K.; Ferlisi, F.; Mecocci, S.; Maranesi, M.; Trabalza-Marinucci, M.; Zerani, M.; Dal Bosco, A.; Acuti, G. Dietary Supplementation of Olive Mill Waste Water Polyphenols in Rabbits: Evaluation of the Potential Effects on Hepatic Apoptosis, Inflammation and Metabolism through RT-qPCR Approach. Animals 2021, 11, 2932. [Google Scholar] [CrossRef] [PubMed]
- Lessard, M.; Talbot, G.; Bergeron, N.; Verso, L.L.; Morissette, B.; Yergeau, É.; Matte, J.J.; Bissonnette, N.; Blais, M.; Gong, J.; et al. Weaning Diet Supplemented with Health-Promoting Feed Additives Influences Microbiota and Immune Response in Piglets Challenged with Salmonella. Vet. Immunol. Immunopathol. 2023, 255, 110533. [Google Scholar] [CrossRef] [PubMed]
- Lian, S.; Lin, X.; Zhan, F.; Shen, X.; Liang, Y.; Li, C. Transcriptome Analysis Reveals the Multiple Functions of pBD2 in IPEC-J2 Cells against E. coli. Int. J. Mol. Sci. 2022, 23, 9754. [Google Scholar] [CrossRef]
- Van den Bogaard, A.E.; Stobberingh, E.E. Antibiotic Usage in Animals. Drugs 1999, 58, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Veldhuizen, E.J.A.; Koomen, I.; Ultee, T.; van Dijk, A.; Haagsman, H.P. Salmonella Serovar Specific Upregulation of Porcine Defensins 1 and 2 in a Jejunal Epithelial Cell Line. Vet. Microbiol. 2009, 136, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Farzan, A.; Friendship, R.M. A Clinical Field Trial to Evaluate the Efficacy of Vaccination in Controlling Salmonella Infection and the Association of Salmonella-Shedding and Weight Gain in Pigs. Can. J. Vet. Res. 2010, 74, 258–263. [Google Scholar] [PubMed]
- Naberhaus, S.A.; Krull, A.C.; Arruda, B.L.; Arruda, P.; Sahin, O.; Schwartz, K.J.; Burrough, E.R.; Magstadt, D.R.; Matias Ferreyra, F.; Gatto, I.R.H.; et al. Pathogenicity and Competitive Fitness of Salmonella enterica Serovar 4,[5],12:i:- Compared to Salmonella Typhimurium and Salmonella Derby in Swine. Front. Vet. Sci. 2019, 6, 502. [Google Scholar] [CrossRef]
- Scott, M.B.; Styring, A.K.; McCullagh, J.S.O. Polyphenols: Bioavailability, Microbiome Interactions and Cellular Effects on Health in Humans and Animals. Pathogens 2022, 11, 770. [Google Scholar] [CrossRef]
- Romier, B.; Schneider, Y.-J.; Larondelle, Y.; During, A. Dietary Polyphenols Can Modulate the Intestinal Inflammatory Response. Nutr. Rev. 2009, 67, 363–378. [Google Scholar] [CrossRef]
- Zhang, Y.; Mu, T.; Deng, X.; Guo, R.; Xia, B.; Jiang, L.; Wu, Z.; Liu, M. New Insights of Biological Functions of Natural Polyphenols in Inflammatory Intestinal Diseases. Int. J. Mol. Sci. 2023, 24, 9581. [Google Scholar] [CrossRef]
- Axling, U.; Olsson, C.; Xu, J.; Fernandez, C.; Larsson, S.; Ström, K.; Ahrné, S.; Holm, C.; Molin, G.; Berger, K. Green Tea Powder and Lactobacillus plantarum Affect Gut Microbiota, Lipid Metabolism and Inflammation in High-Fat Fed C57BL/6J Mice. Nutr. Metab. 2012, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Tassou, C.C.; Nychas, G.J.; Board, R.G. Effect of Phenolic Compounds and Oleuropein on the Germination of Bacillus cereus T Spores. Biotechnol. Appl. Biochem. 1991, 13, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Anhê, F.F.; Pilon, G.; Roy, D.; Desjardins, Y.; Levy, E.; Marette, A. Triggering Akkermansia with Dietary Polyphenols: A New Weapon to Combat the Metabolic Syndrome? Gut Microbes 2016, 7, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, M.; Muñoz-González, I.; Cueva, C.; Jiménez-Girón, A.; Sánchez-Patán, F.; Santos-Buelga, C.; Moreno-Arribas, M.V.; Bartolomé, B. A Survey of Modulation of Gut Microbiota by Dietary Polyphenols. BioMed Res. Int. 2015, 2015, e850902. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Orita, N.; Hatano, S.; Ichikawa, H.; Hara, Y.; Matsumoto, N.; Kimura, Y.; Terada, A.; Mitsuoka, T. Effect of Tea Polyphenols on Fecal Flora and Fecal Metabolic Products of Pigs. J. Vet. Med. Sci. 1995, 57, 45–49. [Google Scholar] [CrossRef]
- Ishihara, N.; Chu, D.-C.; Akachi, S.; Juneja, L.R. Improvement of Intestinal Microflora Balance and Prevention of Digestive and Respiratory Organ Diseases in Calves by Green Tea Extracts. Livest. Prod. Sci. 2001, 68, 217–229. [Google Scholar] [CrossRef]
- Liu, Y. Assessment of the Antimicrobial Activity of Olive Leaf Extract Against Foodborne Bacterial Pathogens. Front. Microbiol. 2017, 8, 113. [Google Scholar] [CrossRef]
- Sudjana, A.N.; D’Orazio, C.; Ryan, V.; Rasool, N.; Ng, J.; Islam, N.; Riley, T.V.; Hammer, K.A. Antimicrobial Activity of Commercial Olea europaea (Olive) Leaf Extract. Int. J. Antimicrob. Agents 2009, 33, 461–463. [Google Scholar] [CrossRef]
- Chen, J.; Yu, B.; Chen, D.; Huang, Z.; Mao, X.; Zheng, P.; Yu, J.; Luo, J.; He, J. Chlorogenic Acid Improves Intestinal Barrier Functions by Suppressing Mucosa Inflammation and Improving Antioxidant Capacity in Weaned Pigs. J. Nutr. Biochem. 2018, 59, 84–92. [Google Scholar] [CrossRef]
- Chen, X.; Zeng, Z.; Huang, Z.; Chen, D.; He, J.; Chen, H.; Yu, B.; Yu, J.; Luo, J.; Luo, Y.; et al. Effects of Dietary Resveratrol Supplementation on Immunity, Antioxidative Capacity and Intestinal Barrier Function in Weaning Piglets. Anim. Biotechnol. 2021, 32, 240–245. [Google Scholar] [CrossRef]
- Ahasan, A.S.M.L.; Invernizzi, G.; Farina, G.; Pilotto, A.; Barbé, F.; Bontempo, V.; Rossi, R.; Bellagamba, F.; Lecchi, C.; Savoini, G.; et al. The Effects of Superoxide Dismutase-Rich Melon Pulp Concentrate on Inflammation, Antioxidant Status and Growth Performance of Challenged Post-Weaning Piglets. Animal 2019, 13, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.J.; Jiang, X.R.; Mantovani, G.; Lumbreras, A.E.V.; Comi, M.; Alborali, G.; Savoini, G.; Dell’Orto, V.; Bontempo, V. Modulation of Plasma Antioxidant Activity in Weaned Piglets by Plant Polyphenols; Taylor & Francis: Abingdon, UK, 2014. [Google Scholar]
- Fiesel, A.; Gessner, D.K.; Most, E.; Eder, K. Effects of Dietary Polyphenol-Rich Plant Products from Grape or Hop on Pro-Inflammatory Gene Expression in the Intestine, Nutrient Digestibility and Faecal Microbiota of Weaned Pigs. BMC Vet. Res. 2014, 10, 196. [Google Scholar] [CrossRef] [PubMed]
- Gessner, D.K.; Ringseis, R.; Eder, K. Potential of Plant Polyphenols to Combat Oxidative Stress and Inflammatory Processes in Farm Animals. J. Anim. Physiol. Anim. Nutr. 2017, 101, 605–628. [Google Scholar] [CrossRef] [PubMed]
- Mahfuz, S.; Shang, Q.; Piao, X. Phenolic Compounds as Natural Feed Additives in Poultry and Swine Diets: A Review. J. Anim. Sci. Biotechnol. 2021, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Mosele, J.I.; Macià, A.; Motilva, M.-J. Metabolic and Microbial Modulation of the Large Intestine Ecosystem by Non-Absorbed Diet Phenolic Compounds: A Review. Molecules 2015, 20, 17429–17468. [Google Scholar] [CrossRef] [PubMed]
- Mecocci, S.; De Paolis, L.; Fruscione, F.; Pietrucci, D.; De Ciucis, C.G.; Giudici, S.D.; Franzoni, G.; Chillemi, G.; Cappelli, K.; Razzuoli, E. In Vitro Evaluation of Immunomodulatory Activities of Goat Milk Extracellular Vesicles (mEVs) in a Model of Gut Inflammation. Res. Vet. Sci. 2022, 152, 546–556. [Google Scholar] [CrossRef]
- Razzuoli, E.; Amadori, M.; Lazzara, F.; Bilato, D.; Ferraris, M.; Vito, G.; Ferrari, A. Salmonella Serovar-Specific Interaction with Jejunal Epithelial Cells. Vet. Microbiol. 2017, 207, 219–225. [Google Scholar] [CrossRef]
- Razzuoli, E.; Villa, R.; Amadori, M. IPEC-J2 Cells as Reporter System of the Anti-Inflammatory Control Actions of Interferon-Alpha. J. Interferon Cytokine Res. 2013, 33, 597–605. [Google Scholar] [CrossRef]
- Sonntag, A.-K.; Bielaszewska, M.; Mellmann, A.; Dierksen, N.; Schierack, P.; Wieler, L.H.; Schmidt, M.A.; Karch, H. Shiga Toxin 2e-Producing Escherichia coli Isolates from Humans and Pigs Differ in Their Virulence Profiles and Interactions with Intestinal Epithelial Cells. Appl. Environ. Microbiol. 2005, 71, 8855–8863. [Google Scholar] [CrossRef]
- De Ciucis, C.G.; Fruscione, F.; De Paolis, L.; Mecocci, S.; Zinellu, S.; Guardone, L.; Franzoni, G.; Cappelli, K.; Razzuoli, E. Toll-like Receptors and Cytokine Modulation by Goat Milk Extracellular Vesicles in a Model of Intestinal Inflammation. Int. J. Mol. Sci. 2023, 24, 11096. [Google Scholar] [CrossRef]
- Mariani, V.; Palermo, S.; Fiorentini, S.; Lanubile, A.; Giuffra, E. Gene Expression Study of Two Widely Used Pig Intestinal Epithelial Cell Lines: IPEC-J2 and IPI-2I. Vet. Immunol. Immunopathol. 2009, 131, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Schierack, P.; Nordhoff, M.; Pollmann, M.; Weyrauch, K.D.; Amasheh, S.; Lodemann, U.; Jores, J.; Tachu, B.; Kleta, S.; Blikslager, A.; et al. Characterization of a Porcine Intestinal Epithelial Cell Line for in Vitro Studies of Microbial Pathogenesis in Swine. Histochem. Cell Biol. 2006, 125, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.A.; Artis, D. Intestinal Bacteria and the Regulation of Immune Cell Homeostasis. Annu. Rev. Immunol. 2010, 28, 623–667. [Google Scholar] [CrossRef] [PubMed]
- Dejban, P.; Nikravangolsefid, N.; Chamanara, M.; Dehpour, A.; Rashidian, A. The Role of Medicinal Products in the Treatment of Inflammatory Bowel Diseases (IBD) through Inhibition of TLR4/NF-kappaB Pathway. Phytother. Res. PTR 2021, 35, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Okumura, R.; Takeda, K. Roles of Intestinal Epithelial Cells in the Maintenance of Gut Homeostasis. Exp. Mol. Med. 2017, 49, e338. [Google Scholar] [CrossRef] [PubMed]
- Scharek, L.; Tedin, K. The Porcine Immune System—Differences Compared to Man and Mouse and Possible Consequences for Infections by Salmonella Serovars. Berl. Munch. Tierarztl. Wochenschr. 2007, 120, 347–354. [Google Scholar] [CrossRef]
- Skjolaas, K.A.; Burkey, T.E.; Dritz, S.S.; Minton, J.E. Effects of Salmonella enterica Serovars Typhimurium (ST) and Choleraesuis (SC) on Chemokine and Cytokine Expression in Swine Ileum and Jejunal Epithelial Cells. Vet. Immunol. Immunopathol. 2006, 111, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Støy, A.C.F.; Heegaard, P.M.H.; Sangild, P.T.; Østergaard, M.V.; Skovgaard, K. Gene Expression Analysis of the IPEC-J2 Cell Line: A Simple Model for the Inflammation-Sensitive Preterm Intestine. Int. Sch. Res. Not. 2013, 2013, 980651. [Google Scholar] [CrossRef][Green Version]
- Marks, H.; Grześkowiak, Ł.; Martinez-Vallespin, B.; Dietz, H.; Zentek, J. Porcine and Chicken Intestinal Epithelial Cell Models for Screening Phytogenic Feed Additives-Chances and Limitations in Use as Alternatives to Feeding Trials. Microorganisms 2022, 10, 629. [Google Scholar] [CrossRef]
- Hejna, M.; Kovanda, L.; Rossi, L.; Liu, Y. Mint Oils: In Vitro Ability to Perform Anti-Inflammatory, Antioxidant, and Antimicrobial Activities and to Enhance Intestinal Barrier Integrity. Antioxidants 2021, 10, 1004. [Google Scholar] [CrossRef]
- Li, E.; Horn, N.; Ajuwon, K.M. Mechanisms of Deoxynivalenol-Induced Endocytosis and Degradation of Tight Junction Proteins in Jejunal IPEC-J2 Cells Involve Selective Activation of the MAPK Pathways. Arch. Toxicol. 2021, 95, 2065–2079. [Google Scholar] [CrossRef]
- Ling, K.-H.; Wan, M.L.Y.; El-Nezami, H.; Wang, M. Protective Capacity of Resveratrol, a Natural Polyphenolic Compound, against Deoxynivalenol-Induced Intestinal Barrier Dysfunction and Bacterial Translocation. Chem. Res. Toxicol. 2016, 29, 823–833. [Google Scholar] [CrossRef]
- Pomothy, J.M.; Barna, R.F.; Pászti, E.A.; Babiczky, Á.; Szóládi, Á.; Jerzsele, Á.; Gere, E.P. Beneficial Effects of Rosmarinic Acid on IPEC-J2 Cells Exposed to the Combination of Deoxynivalenol and T-2 Toxin. Mediat. Inflamm. 2020, 2020, 8880651. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- International Olive Council. Determination of Biophenols in Olive Oils by HPLC. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2022/10/COI-T20-Doc.-29-REV-1-2017-Eng.pdf (accessed on 21 December 2023).
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Razzuoli, E.; Vencia, W.; Modesto, P.; Franzoni, G.; Giudici, S.D.; Parisi, E.; Ferrari, A.; Amadori, M. Yersinia enterocolitica-Specific Modulation of Innate Immune Responses in Jejunal Epithelial Cells. Vet. Microbiol. 2020, 242, 108596. [Google Scholar] [CrossRef] [PubMed]
- Pistol, G.C.; Bulgaru, C.V.; Marin, D.E.; Oancea, A.G.; Taranu, I. Dietary Grape Seed Meal Bioactive Compounds Alleviate Epithelial Dysfunctions and Attenuates Inflammation in Colon of DSS-Treated Piglets. Foods 2021, 10, 530. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, F.; Liu, X.; Liu, J.; Li, D. Synergistic Anti-Inflammatory Effects of Quercetin and Catechin via Inhibiting Activation of TLR4–MyD88-Mediated NF-κB and MAPK Signaling Pathways. Phytother. Res. 2019, 33, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, M.; Ceballos-Olvera, I.; del Barrio, L.; Re, F. Role of the Inflammasome, IL-1β, and IL-18 in Bacterial Infections. Sci. World J. 2011, 11, 2037–2050. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Overview of the IL-1 Family in Innate Inflammation and Acquired Immunity. Immunol. Rev. 2018, 281, 8–27. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, N.; Kurata, M.; Yamamoto, T.; Morikawa, S.; Masumoto, J. The Role of Interleukin-1 in General Pathology. Inflamm. Regen. 2019, 39, 12. [Google Scholar] [CrossRef]
- Olcum, M.; Tastan, B.; Ercan, I.; Eltutan, I.B.; Genc, S. Inhibitory Effects of Phytochemicals on NLRP3 Inflammasome Activation: A Review. Phytomedicine 2020, 75, 153238. [Google Scholar] [CrossRef]
- Serrano-López, J.; Martín-Antonio, B. Inflammaging, an Imbalanced Immune Response that Needs to Be Restored for Cancer Prevention and Treatment in the Elderly. Cells 2021, 10, 2562. [Google Scholar] [CrossRef]
- Wang, T.; Xu, H.; Dong, R.; Wu, S.; Guo, Y.; Wang, D. Effectiveness of Targeting the NLRP3 Inflammasome by Using Natural Polyphenols: A Systematic Review of Implications on Health Effects. Food Res. Int. 2023, 165, 112567. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.-T.; Yan, W.-H.; Cao, Y.; Yan, J.-K.; Cai, W. Neutralization of IL-6 and TNF-α Ameliorates Intestinal Permeability in DSS-Induced Colitis. Cytokine 2016, 83, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L. Role of Inflammasomes in Salmonella Infection. Front. Microbiol. 2011, 2, 8. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed]
- Gessner, D.K.; Ringseis, R.; Siebers, M.; Keller, J.; Kloster, J.; Wen, G.; Eder, K. Inhibition of the Pro-Inflammatory NF-κB Pathway by a Grape Seed and Grape Marc Meal Extract in Intestinal Epithelial Cells. J. Anim. Physiol. Anim. Nutr. 2012, 96, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- González, R.; Ballester, I.; López-Posadas, R.; Suárez, M.D.; Zarzuelo, A.; Martínez-Augustin, O.; Medina, F.S.D. Effects of Flavonoids and Other Polyphenols on Inflammation. Crit. Rev. Food Sci. Nutr. 2011, 51, 331–362. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Tyagi, A.K.; Deshmukh-Taskar, P.; Hinojosa, M.; Prasad, S.; Aggarwal, B.B. Downregulation of Tumor Necrosis Factor and Other Proinflammatory Biomarkers by Polyphenols. Arch. Biochem. Biophys. 2014, 559, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.R.; Villegas, I.; La Casa, C.; de la Lastra, C.A. Resveratrol, a Polyphenol Found in Grapes, Suppresses Oxidative Damage and Stimulates Apoptosis during Early Colonic Inflammation in Rats. Biochem. Pharmacol. 2004, 67, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Camuesco, D.; Comalada, M.; Concha, A.; Nieto, A.; Sierra, S.; Xaus, J.; Zarzuelo, A.; Gálvez, J. Intestinal Anti-Inflammatory Activity of Combined Quercitrin and Dietary Olive Oil Supplemented with Fish Oil, Rich in EPA and DHA (n-3) Polyunsaturated Fatty Acids, in Rats with DSS-Induced Colitis. Clin. Nutr. 2006, 25, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Salh, B.; Assi, K.; Templeman, V.; Parhar, K.; Owen, D.; Gómez-Muñoz, A.; Jacobson, K. Curcumin Attenuates DNB-Induced Murine Colitis. Am. J. Physiol.-Gastrointest. Liver Physiol. 2003, 285, G235–G243. [Google Scholar] [CrossRef] [PubMed]
- Taranu, I.; Habeanu, M.; Gras, M.A.; Pistol, G.C.; Lefter, N.; Palade, M.; Ropota, M.; Chedea, V.S.; Marin, D.E. Assessment of the Effect of Grape Seed Cake Inclusion in the Diet of Healthy Fattening-Finishing Pigs. J. Anim. Physiol. Anim. Nutr. 2018, 102, e30–e42. [Google Scholar] [CrossRef]
- Britti, D.; Impellizzeri, D.; Procopio, A.; Cuzzocrea, S.; Britti, D.; Impellizzeri, D.; Procopio, A.; Cuzzocrea, S. Oleuropein an Olive Oil Compound in Acute and Chronic Inflammation Models: Facts and Perspectives. In Olive Germplasm—The Olive Cultivation, Table Olive and Olive Oil Industry in Italy; IntechOpen: London, UK, 2012; ISBN 978-953-51-0883-2. [Google Scholar]
- Letterio, J.J.; Roberts, A.B. Regulation of Immune Responses by TGF-Beta. Annu. Rev. Immunol. 1998, 16, 137–161. [Google Scholar] [CrossRef]
- Qin, G.; Zhao, Y.; Gan, Y.; Yu, X.; Zhao, Y.; Peng, H.; Fang, S. Alterations in Gene Expressions of Caco-2 Cell Responses to LPS and Ploy(I:C) Stimulation. PeerJ 2023, 11, e15459. [Google Scholar] [CrossRef]
- Huang, T.; Huang, X.; Shi, B.; Wang, F.; Feng, W.; Yao, M. Regulators of Salmonella-Host Interaction Identified by Peripheral Blood Transcriptome Profiling: Roles of TGFB1 and TRP53 in Intracellular Salmonella Replication in Pigs. Vet. Res. 2018, 49, 121. [Google Scholar] [CrossRef]
- Howe, K.L.; Reardon, C.; Wang, A.; Nazli, A.; McKay, D.M. Transforming Growth Factor-β Regulation of Epithelial Tight Junction Proteins Enhances Barrier Function and Blocks Enterohemorrhagic Escherichia coli O157:H7-Induced Increased Permeability. Am. J. Pathol. 2005, 167, 1587–1597. [Google Scholar] [CrossRef]
- Nallathambi, R.; Poulev, A.; Zuk, J.B.; Raskin, I. Proanthocyanidin-Rich Grape Seed Extract Reduces Inflammation and Oxidative Stress and Restores Tight Junction Barrier Function in Caco-2 Colon Cells. Nutrients 2020, 12, 1623. [Google Scholar] [CrossRef]
- Laurindo, L.F.; Santos, A.R.d.O.d.; Carvalho, A.C.A.d.; Bechara, M.D.; Guiguer, E.L.; Goulart, R.d.A.; Vargas Sinatora, R.; Araújo, A.C.; Barbalho, S.M. Phytochemicals and Regulation of NF-kB in Inflammatory Bowel Diseases: An Overview of In Vitro and In Vivo Effects. Metabolites 2023, 13, 96. [Google Scholar] [CrossRef] [PubMed]
- Mecocci, S.; De Paolis, L.; Zoccola, R.; Fruscione, F.; De Ciucis, C.G.; Chiaradia, E.; Moccia, V.; Tognoloni, A.; Pascucci, L.; Zoppi, S.; et al. Antimicrobial and Immunomodulatory Potential of Cow Colostrum Extracellular Vesicles (ColosEVs) in an Intestinal In Vitro Model. Biomedicines 2022, 10, 3264. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).