Expression and Variations in EPO Associated with Oxygen Metabolism in Tibetan Sheep
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Objects and Sample Collection
2.2. Blood Gas Indicator Measurement
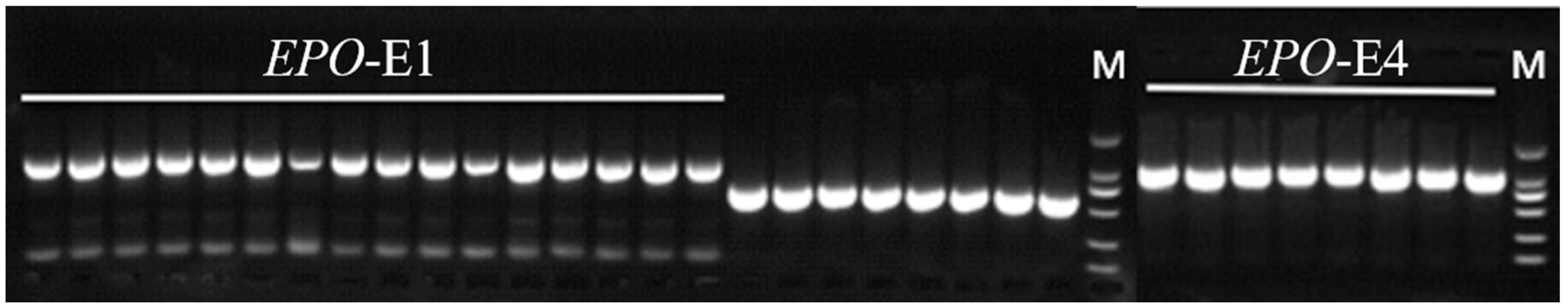
2.3. Primers for PCR and RT-qPCR
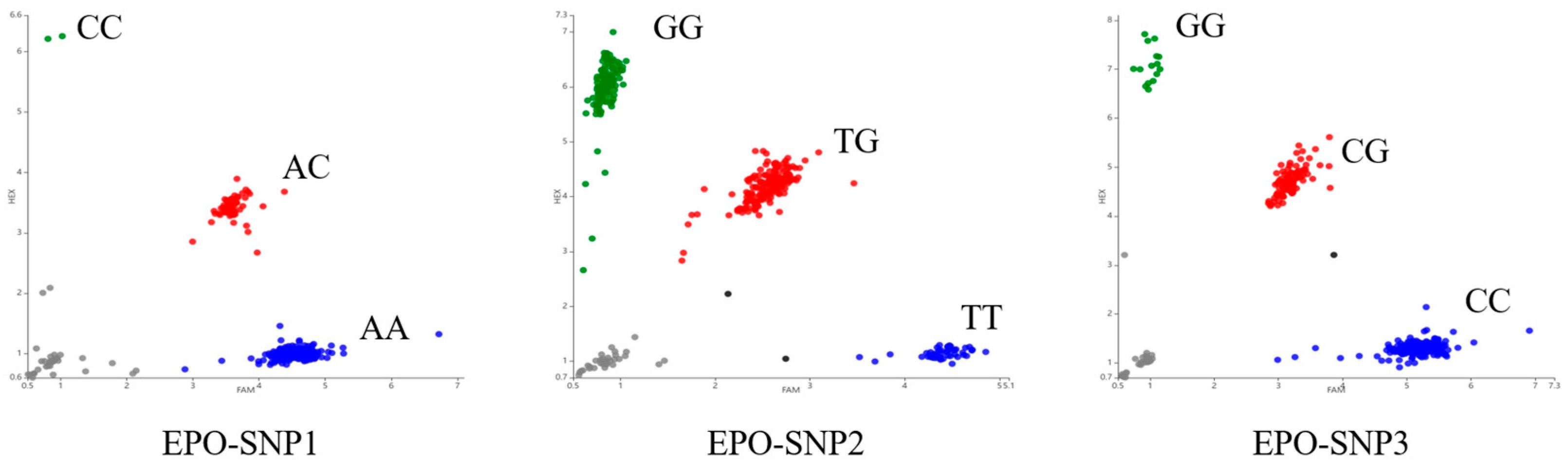
2.4. Genotyping
2.5. Statistical Analyses
3. Results
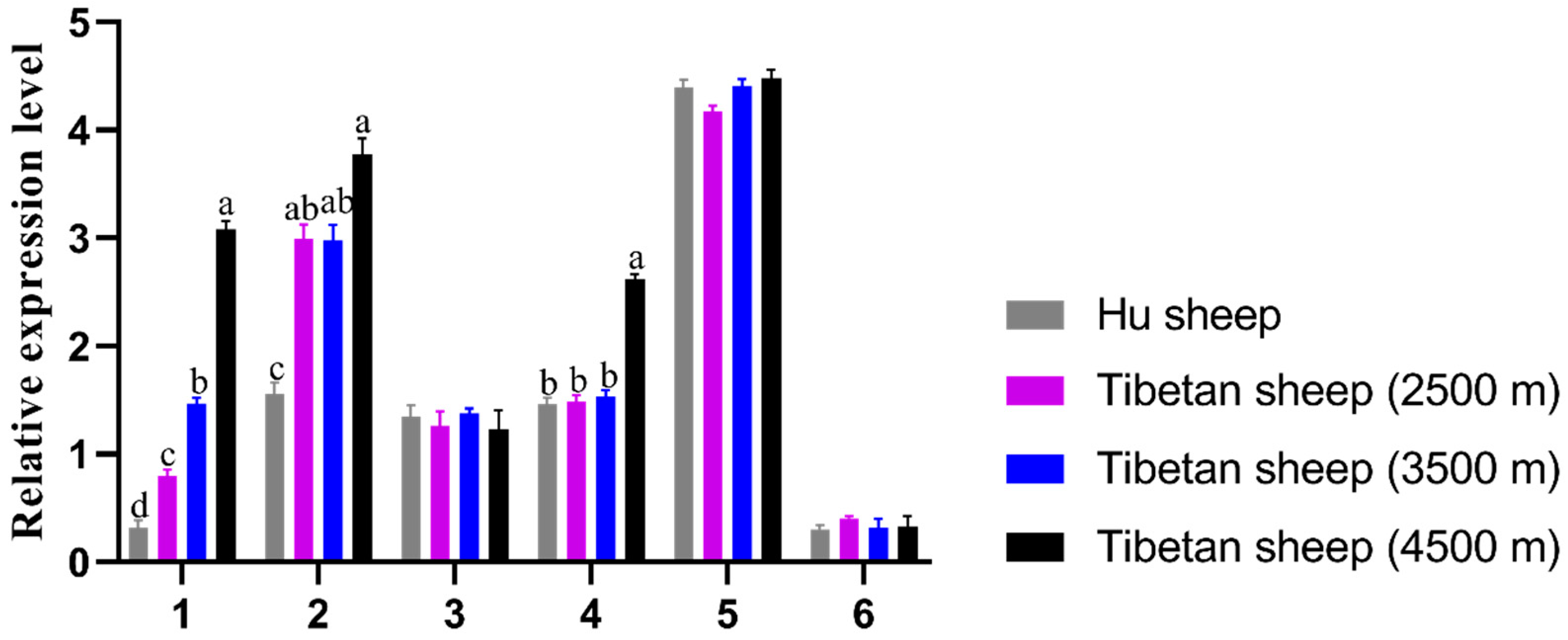
3.1. Tissue Expression Characteristics of EPO Gene
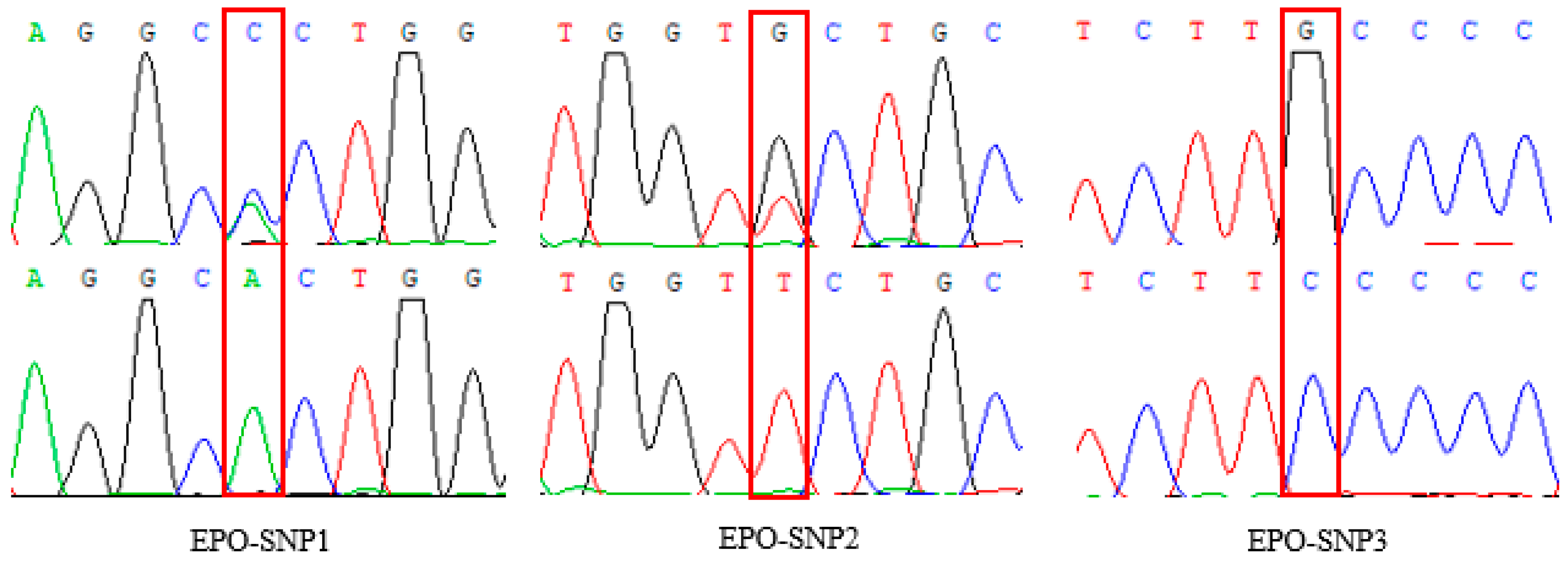
3.2. Variation of EPO in Tibetan Sheep
3.3. Population Genetic Diversity Analysis of EPO Gene
3.4. Effect of Variation in EPO on Blood Gas
3.5. Analysis Linkage Disequilibrium
3.6. Correlation Analysis between Haplotype Combination and Blood Gas
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsiftsoglou, A.S. Erythropoietin (EPO) as a Key Regulator of Erythropoiesis, Bone Remodeling and Endothelial Transdifferentiation of Multipotent Mesenchymal Stem Cells (MSCs): Implications in Regenerative Medicine. Cells 2021, 10, 2140. [Google Scholar] [CrossRef]
- Chatterjee, P.K. Pleiotropic renal actions of erythropoietin. Lancet 2005, 365, 1890–1892. [Google Scholar] [CrossRef] [PubMed]
- Maiese, K.; Li, F.; Chong, Z.Z. New avenues of exploration for erythropoietin. JAMA 2005, 293, 90–95. [Google Scholar] [CrossRef]
- Jelkmann, W. Molecular biology of erythropoietin. Intern. Med. 2004, 43, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Lappin, T.R.; Lee, F.S. Update on mutations in the HIF: EPO pathway and their role in erythrocytosis. Blood Rev. 2019, 37, 100590. [Google Scholar] [CrossRef]
- Storti, F.; Santambrogio, S.; Crowther, L.M.; Otto, T.; Abreu-Rodríguez, I.; Kaufmann, M.; Hu, C.J.; Dame, C.; Fandrey, J.; Wenger, R.H.; et al. A novel distal upstream hypoxia response element regulating oxygen-dependent erythropoietin gene expression. Haematologica 2014, 99, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Hirano, I.; Suzuki, N.; Yamazaki, S.; Sekine, H.; Minegishi, N.; Shimizu, R.; Yamamoto, M. Renal Anemia Model Mouse Established by Transgenic Rescue with an Erythropoietin Gene Lacking Kidney-Specific Regulatory Elements. Mol. Cell Biol. 2017, 37, e00451-16. [Google Scholar] [CrossRef]
- Deji, B.Z.; Shang, P.; Danzeng, W.J.; Zhang, H.; Qiangba, Y.Z. Expression and hypoxia adaptation analysis of the EPO gene in different tissues of plateau Tibetan pigs. Genet. Mol. Res. 2015, 14, 1700–1706. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, T.; Zhang, Y.Y.; Xu, J.; Li, M.; Zhang, Q.; Qiao, Z.; Yang, K. Expression and distribution of erythropoietin, vascular endothelial growth factor (VEGF) and VEGF receptor 2 in small intestine of yaks at different ages. Folia Morphol. 2023, 82, 683–695. [Google Scholar] [CrossRef]
- Scholz, H.; Schurek, H.J.; Eckardt, K.U.; Bauer, C. Role of erythropoietin in adaptation to hypoxia. Experientia 1990, 46, 1197–1201. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Q.; Liu, Y.; Xu, Q. Characterization of EPO H131S as a key mutation site in the hypoxia-adaptive evolution of Gymnocypris dobula. Fish. Physiol. Biochem. 2022, 48, 723–733. [Google Scholar] [CrossRef]
- Pang, K.; Lennikov, A.; Yang, M. Hypoxia adaptation in the cornea: Current animal models and underlying mechanisms. Anim. Model. Exp. Med. 2021, 4, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Vočanec, D.; Prijatelj, T.; Debeljak, N.; Kunej, T. Genetic variants of erythropoietin (EPO) and EPO receptor genes in familial erythrocytosis. Int. J. Lab. Hematol. 2019, 41, 162–167. [Google Scholar] [CrossRef]
- Beall, C.M.; Cavalleri, G.L.; Deng, L.; Elston, R.C.; Gao, Y.; Knight, J.; Li, C.; Li, J.C.; Liang, Y.; McCormack, M.; et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. USA 2010, 107, 11459–11464. [Google Scholar] [CrossRef]
- Zhou, H.; Hickford, J.G.; Fang, Q. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal. Biochem. 2006, 354, 159–161. [Google Scholar] [CrossRef]
- Lichtman, M.A.; Murphy, M.S.; Adamson, J.W. Detection of mutant hemoglobins with altered affinity for oxygen: A simplified technique. Ann. Intern. Med. 1976, 84, 517–520. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Stuart, J.A.; Aibueku, O.; Bagshaw, O.; Moradi, F. Hypoxia inducible factors as mediators of reactive oxygen/nitrogen species homeostasis in physiological normoxia. Med. Hypotheses 2019, 129, 109249. [Google Scholar] [CrossRef]
- Semenza, G.L. Perspectives on oxygen sensing. Cell 1999, 98, 281–284. [Google Scholar] [CrossRef]
- Li, F.; Qiao, Z.; Duan, Q.; Nevo, E. Adaptation of mammals to hypoxia. Anim. Models Exp. Med. 2021, 4, 311–318. [Google Scholar] [CrossRef]
- Schödel, J.; Grampp, S.; Maher, E.R.; Moch, H.; Ratcliffe, P.J.; Russo, P.; Mole, D.R. Hypoxia, Hypoxia-inducible Transcription Factors, and Renal Cancer. Eur. Urol. 2016, 69, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, N.R.; Semenza, G.L. Adaptive and Maladaptive Cardiorespiratory Responses to Continuous and Intermittent Hypoxia Mediated by Hypoxia-Inducible Factors 1 and 2. Physiol. Rev. 2012, 92, 967–1003. [Google Scholar] [CrossRef]
- Figlus, M.; Kaczorowska, B.; Jaskólski, D.J.; Kępczyński, Ł. Von Hippel-Lindau syndrome—A case report. Pol. Merkur. Lek. 2018, 44, 248–252. [Google Scholar]
- Yang, K.; Zhang, L.; Chen, W.; Cheng, J.; Zhao, X.; Zhang, Y.; Li, R.; Zhou, M.; Yao, Y.; Li, Y.; et al. Expression of EPO and related factors in the liver and kidney of plain and Tibetan sheep. Histol. Histopathol. 2023, 38, 1337–1347. [Google Scholar]
- Crait, J.R.; Prange, H.D.; Marshall, N.A.; Harlow, H.J.; Cotton, C.J.; Ben-David, M. High-altitude diving in river otters: Coping with combined hypoxic stresses. J. Exp. Biol. 2012, 215, 256–263. [Google Scholar] [CrossRef]
- Siri, W.E.; Van Dyke, D.C.; Winchell, H.S.; Pollycove, M.; Parker, H.G.; Cleveland, A.S. Early erythropoietin, blood, and physiological responses to severe hypoxia in man. J. Appl. Physiol. 1966, 21, 73–80. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, C.; Zhang, D.; Jiang, H.; Peng, S.; Liu, Y.; Zhao, K.; Wang, C.; Chen, L. Analysis of the erythropoietin of a Tibetan Plateau schizothoracine fish (Gymnocypris dobula) reveals enhanced cytoprotection function in hypoxic environments. BMC Evol. Biol. 2016, 16, 11. [Google Scholar] [CrossRef]
- Benavides, M.V.; Sonstegard, T.S.; Van Tassell, C. Genomic Regions Associated with Sheep Resistance to Gastrointestinal Nematodes. Trends Parasitol. 2016, 32, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Stembridge, M.; Williams, A.M.; Gasho, C.; Dawkins, T.G.; Drane, A.; Villafuerte, F.C.; Levine, B.D.; Shave, R.; Ainslie, P.N. The overlooked significance of plasma volume for successful adaptation to high altitude in Sherpa and Andean natives. Proc. Natl. Acad. Sci. USA 2019, 116, 16177–16179. [Google Scholar] [CrossRef] [PubMed]

| Gene | Exon | Primer Sequence (5′→3′) | Product Size (bp) | Annealing Temperature/°C | Purpose |
|---|---|---|---|---|---|
| EPO | 1 | F: TTGAAGTTTGGCCGGGAGAAGTG R: CCAAGCCAGCCTCACTCTCCTCC | 864 | 58 | PCR |
| EPO | 4 | F: GCTGTTTCTGTAAAGTGAGAGAAGGGC R: GCCAGTGGGGTCAAGAGGTCAG | 1008 | 57 | PCR |
| EPO | F: CTGTCTTTTCTGCTGTTTCCTCTG R: CTCCATCCTCTTCCAGGCATA | 207 | 60 | RT-qPCR | |
| β-actin | F: AGCCTTCCTTCCTGGGCATGGA R: GGACAGCACCGTGTTGGCGTAGA | 113 | 60 | RT-qPCR |
| SNP Site | Number | Genotype Frequency (Number) | Allele Frequency | |||
|---|---|---|---|---|---|---|
| g.855 A > C | 349 | AA | AC | CC | A | C |
| 0.8195 (280) | 0.1748 (61) | 0.0057 (2) | 0.9069 | 0.0931 | ||
| g.1985 T > G | 346 | GG | GT | TT | G | T |
| 0.3584 (124) | 0.5029 (174) | 0.1387 (48) | 0.6098 | 0.3902 | ||
| g.2115 G > C | 349 | CC | CG | GG | C | G |
| 0.6905 (241) | 0.2665 (93) | 0.0430 (15) | 0.8238 | 0.1762 | ||
| SNP Site | Ho | He | Ne | PIC | Hardy–Weinberg p-Value |
|---|---|---|---|---|---|
| g.855 A > C | 0.8312 | 0.1688 | 1.2031 | 0.1545 | p > 0.05 |
| g.1985 T > G | 0.5242 | 0.4758 | 1.9077 | 0.3625 | p > 0.05 |
| g.2115 G > C | 0.7096 | 0.2904 | 1.4092 | 0.2483 | p > 0.05 |
| SNP Site | Blood Gas | Genotype | ||
|---|---|---|---|---|
| g.855 A > C | AA (n = 286) | AC (n = 61) | CC (n = 2) | |
| pCO2 | 40.453 ± 0.643 | 41.742 ± 0.464 | 40.557 ± 0.372 | |
| pO2 | 38.618 ± 0.365 | 38.364 ± 0.546 | 38.037 ± 0.407 | |
| sO2 | 70.587 ± 0.823 | 70.561 ± 0.554 | 71.701 ± 0.865 | |
| p50 | 26.470 ± 0.783 | 26.669 ± 0.059 | 26.256 ± 0.697 | |
| g.1985 T > G | GG (n = 124) | GT (n = 172) | TT (n = 48) | |
| pCO2 | 40.236 ± 0.431 | 41.057 ± 0.758 | 40.531 ± 0.478 | |
| pO2 | 39.465 ± 0.668 | 38.380 ± 1.445 | 38.624 ± 0.930 | |
| sO2 | 70.493 ± 0.292 | 70.562 ± 0.281 | 70.537 ± 0.840 | |
| p50 | 26.764 ± 0.842 | 27.201 ± 0.329 | 26.684 ± 0.506 | |
| g.2115 G > C | CC (n = 241) | CG (n = 93) | GG (n = 15) | |
| pCO2 | 40.127 ± 0.473 | 40.757 ± 0.437 | 40.140 ± 0.356 | |
| pO2 | 38.365 ± 0.566 b | 38.064 ± 0.442 b | 43.404 ± 0.494 a | |
| sO2 | 70.642 ± 1.537 | 70.066 ± 0.457 | 70.791 ± 1.209 | |
| p50 | 27.537 ± 0.802 | 26.729 ± 0.593 | 26.371 ± 0.401 | |
| Haplotype | SNP1 | SNP2 | SNP3 | Frequency/% | Haplotype Combination | Haplotype Combination Frequency/% |
|---|---|---|---|---|---|---|
| H1(ACT) | A | C | T | 0.390 | H1H1 | 0.1521 |
| H2(ACG) | A | C | G | 0.344 | H1H2 | 0.1342 |
| H3(AGG) | A | G | G | 0.171 | H1H3 | 0.0667 |
| H4(CCG) | C | C | G | 0.096 | H1H4 | 0.0374 |
| H2H2 | 0.1183 | |||||
| H2H3 | 0.0588 | |||||
| H2H4 | 0.0330 |
| Haplotype Combination | pCO2 | pO2 | sO2 | p50 |
|---|---|---|---|---|
| H1H1 | 40.347 ± 0.640 | 38.725 ± 0.305 ab | 70.621 ± 0.773 | 26.484 ± 0.333 |
| H1H2 | 40.821 ± 1.063 | 37.923 ± 0.127 ab | 70.081 ± 0.493 | 26.880 ± 0.453 |
| H1H3 | 41.776 ± 0.902 | 38.276 ± 0.884 ab | 69.796 ± 0.545 | 26.633 ± 0.892 |
| H1H4 | 40.901 ± 0.258 | 38.190 ± 1.896 ab | 70.278 ± 0.984 | 26.043 ± 1.574 |
| H2H2 | 41.670 ± 0.515 | 36.551 ± 0.295 b | 70.683 ± 0.998 | 26.499 ± 0.339 |
| H2H3 | 39.754 ± 0.862 | 37.629 ± 0.653 b | 69.850 ± 0.739 | 25.971 ± 0.509 |
| H2H4 | 40.573 ± 0.163 | 40.304 ± 0.796 a | 70.801 ± 1.473 | 26.302 ± 0.331 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Y.; Xi, Q.; He, Z.; Sun, H.; Li, S. Expression and Variations in EPO Associated with Oxygen Metabolism in Tibetan Sheep. Animals 2024, 14, 535. https://doi.org/10.3390/ani14040535
Ren Y, Xi Q, He Z, Sun H, Li S. Expression and Variations in EPO Associated with Oxygen Metabolism in Tibetan Sheep. Animals. 2024; 14(4):535. https://doi.org/10.3390/ani14040535
Chicago/Turabian StyleRen, Yue, Qiming Xi, Zhaohua He, Hongxian Sun, and Shaobin Li. 2024. "Expression and Variations in EPO Associated with Oxygen Metabolism in Tibetan Sheep" Animals 14, no. 4: 535. https://doi.org/10.3390/ani14040535
APA StyleRen, Y., Xi, Q., He, Z., Sun, H., & Li, S. (2024). Expression and Variations in EPO Associated with Oxygen Metabolism in Tibetan Sheep. Animals, 14(4), 535. https://doi.org/10.3390/ani14040535





