Mucosal Exosome Proteomics of Hybrid Grouper Epinephelus fuscoguttatus♀ × E. lanceolatus♂ Infected by Pseudomonas plecoglossicida
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Exosome Isolation
2.2. Exosomes Characterization
2.2.1. Morphology Characterization Using Transmission Electron Microscopy (TEM)
2.2.2. Particle Size Measurement Using Nanoparticle Tracking Analysis (NTA)
2.2.3. Western Blot Analysis
2.3. Exosomal Protein Analysis
2.3.1. Total Protein Extraction and Protein Quality Test
2.3.2. Trypsin Digestion and Fraction Separation
2.3.3. LC-MS/MS Analysis
2.3.4. Proteomics Bioinformatics Analysis
2.4. Biomarkers Screen and Validation
3. Results
3.1. Mucous Exosomes Characterization
3.2. Quality Control of Proteomic Sequencing
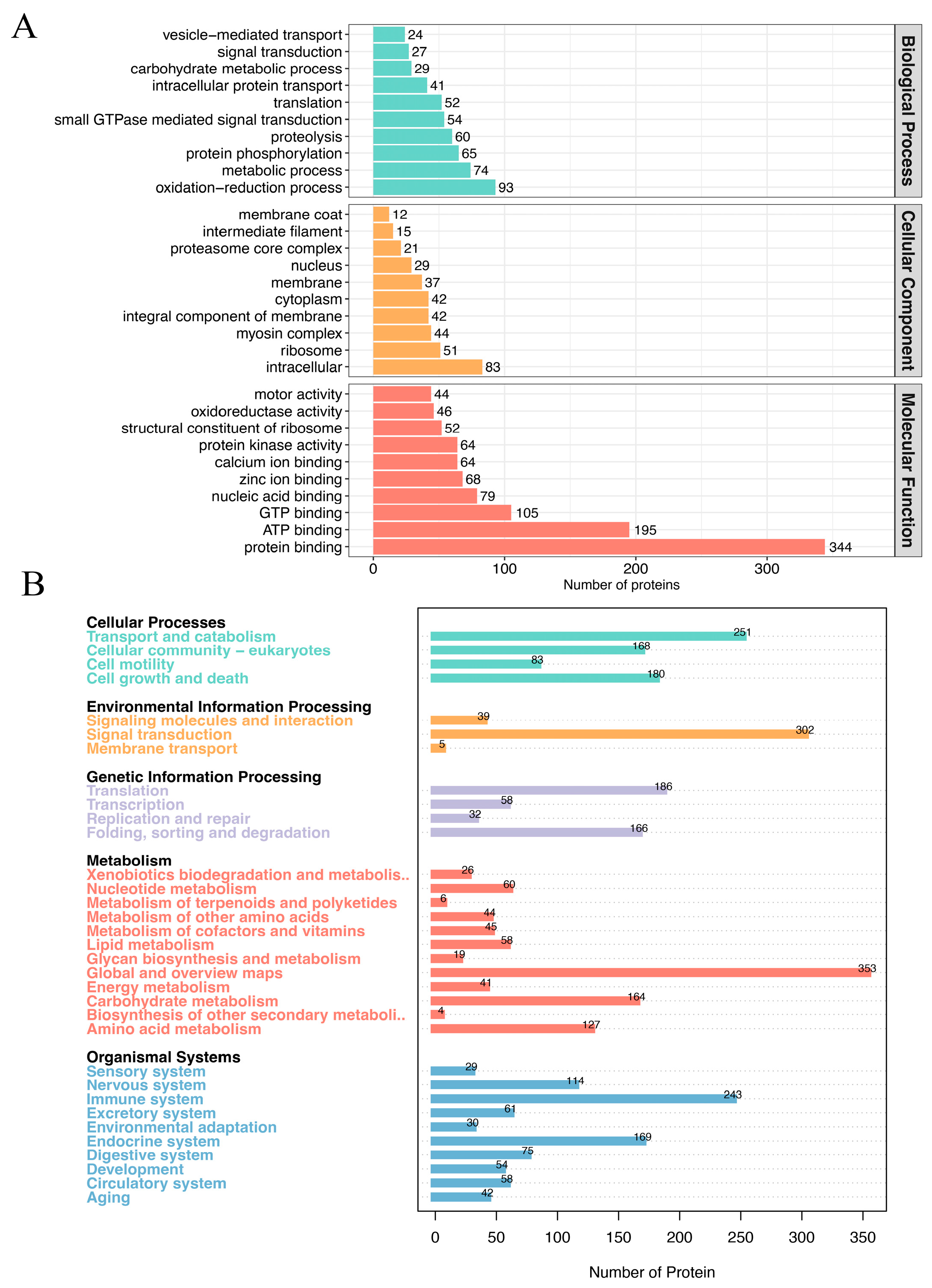
3.3. Functional Annotation of Protein Cargos Inside the Mucus Exosomes
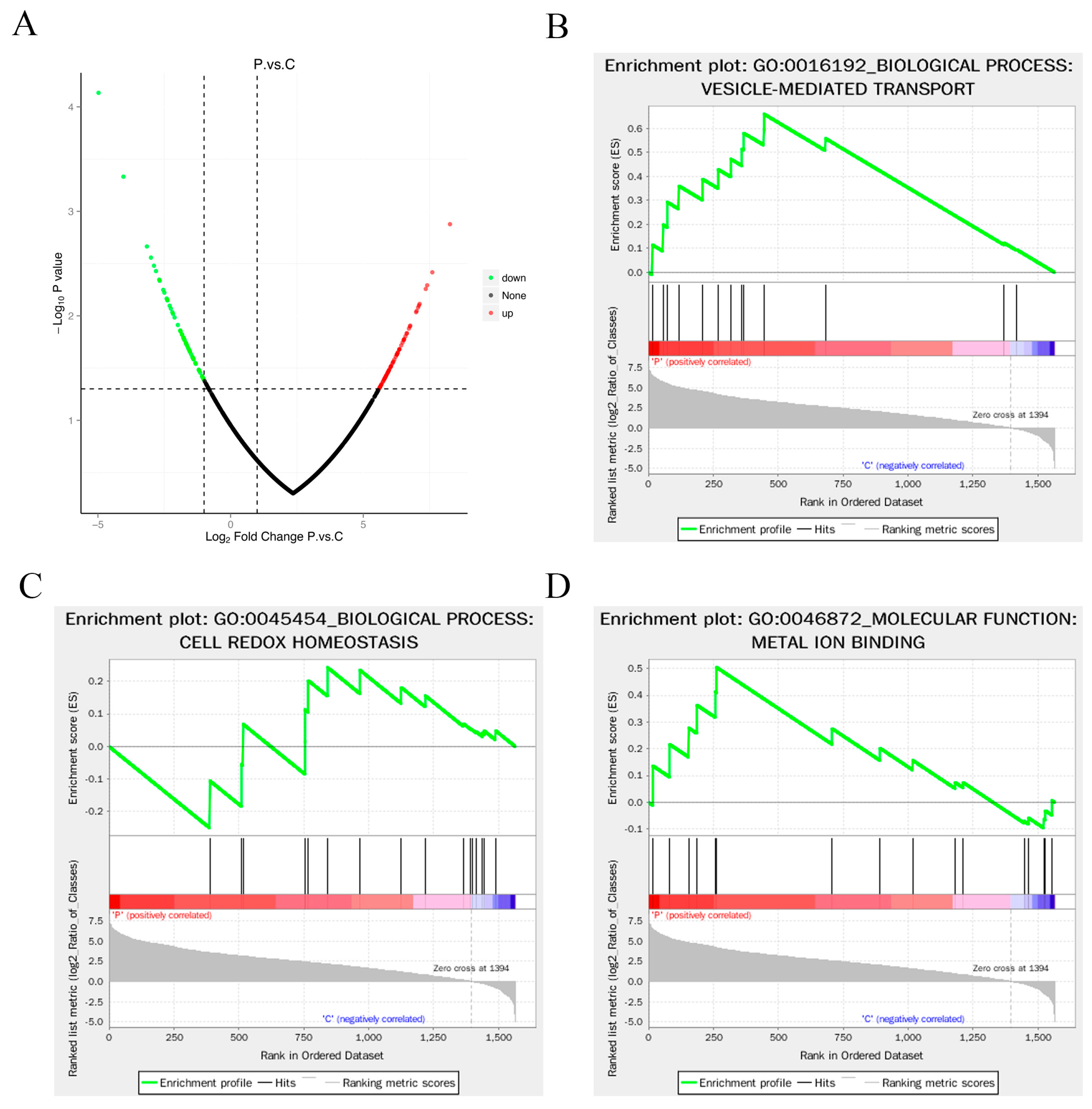
3.4. Enrichment Analysis of Differentially Expressed Proteins
3.5. Subcellular Localization Analysis of Differentially Expressed Proteins
3.6. Differentially Expressed Proteins Interaction Analysis
3.7. Validation of DEP Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, L.; Zhao, L.; Liu, W.; Xu, X.; Su, Y.; Qin, Y.; Yan, Q. Dual RNA-Seq Unveils Pseudomonas plecoglossicida htpG Gene Functions During Host-Pathogen Interactions with Epinephelus coioides. Front. Immunol. 2019, 10, 984. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-H.; Xiong, J.-B.; Ding, F.-F.; Chen, J. Immune and gut bacterial successions of large yellow croaker (Larimichthys crocea) during Pseudomonas plecoglossicida infection. Fish Shellfish Immunol. 2020, 99, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.T.; Zhou, S.M.; An, S.W.; Chen, L.; Wang, G.L. Visceral granulomas in farmed large yellow croaker, Larimichthys crocea (Richardson), caused by a bacterial pathogen, Pseudomonas plecoglossicida. J. Fish Diseases 2014, 37, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Li, M.; Chen, J. Draft Genome Sequence of Pseudomonas plecoglossicida Strain NB2011, the Causative Agent of White Nodules in Large Yellow Croaker (Larimichthys crocea). Genome Announc. 2013, 1, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Nie, P.; Zhao, L.; Huang, L.; Qin, Y.; Xu, X.; Zhan, J.; Yan, Q. Dual RNA-Seq Unveils the Role of the Pseudomonas plecoglossicida fliA Gene in Pathogen-Host Interaction with Larimichthys crocea. Microorganisms 2019, 7, 443. [Google Scholar] [CrossRef]
- Tang, Y.; Sun, Y.; Zhao, L.; Xu, X.; Huang, L.; Qin, Y.; Su, Y.; Yi, G.; Yan, Q. Mechanistic insight into the roles of Pseudomonas plecoglossicida clpV gene in host-pathogen interactions with Larimichthys crocea by dual RNA-seq. Fish Shellfish Immunol. 2019, 93, 344–353. [Google Scholar] [CrossRef]
- Nishimori, E.; Kita-Tsukamoto, K.; Wakabayashi, H. Pseudomonas plecoglossicida sp. nov., the causative agent of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 1, 83–89. [Google Scholar] [CrossRef]
- Akayli, T.; Çanak, Ö.; Başaran, B. A New Pseudomonas Species Observed in Cultured Young Rainbow Trout (Oncorhynchus mykis Walbaum, 1792): Pseudomonas plecoglossicida. Biyol. Bilim. Araştırma Dergisi. 2011, 4, 107–111. [Google Scholar]
- Sun, Y.; Luo, G.; Zhao, L.; Huang, L.; Qin, Y.; Su, Y.; Yan, Q. Integration of RNAi and RNA-seq Reveals the Immune Responses of Epinephelus coioides to sigX Gene of Pseudomonas plecoglossicida. Front. Immunol. 2018, 9, 1624. [Google Scholar] [CrossRef]
- Tao, Z.; Zhou, T.; Zhou, S.; Wang, G. Temperature-regulated expression of type VI secretion systems in fish pathogen Pseudomonas plecoglossicida revealed by comparative secretome analysis. FEMS Microbiol. Lett. 2016, 363, fnw261. [Google Scholar] [CrossRef]
- Tao, Z.; Xu, Y.; Zhou, S.; Qian, D.; Liu, M.; Li, W.; Xu, W.; Yan, X. Acquisition of a type VI secretion system is critical for Pseudomonas plecoglossicida induced granulomas in fish internal organs. Aquaculture 2020, 516, 734629. [Google Scholar] [CrossRef]
- Miller, B.S.; Bezinge, L.; Gliddon, H.D.; Huang, D.; Dold, G.; Gray, E.R.; Heaney, J.; Dobson, P.J.; Nastouli, E.; Morton, J.J.L.; et al. Spin-enhanced nanodiamond biosensing for ultrasensitive diagnostics. Nature 2020, 587, 588–593. [Google Scholar] [CrossRef]
- Cherkaoui, D.; Mesquita, S.G.; Huang, D.; Lugli, E.B.; Webster, B.L.; McKendry, R.A. CRISPR-assisted test for Schistosoma haematobium. Sci. Rep. 2023, 13, 4990. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Jia, L.; He, X.; Zhang, B. Proteomics of mucosal exosomes of Cynoglossus semilaevis altered when infected by Vibrio harveyi. Dev. Comp. Immunol. 2021, 119, 104045. [Google Scholar] [CrossRef]
- Zhao, N.; Jia, L.; Wang, Q.; Deng, Q.; Ru, X.; Zhu, C.; Zhang, B. The feasibility of skin mucus replacing exosome as a pool for bacteria-infected markers development via comparative proteomic screening in teleost. Fish Shellfish Immunol. 2023, 132, 108483. [Google Scholar] [CrossRef]
- Xiu, L.; Wu, Y.; Lin, G.; Zhang, Y.; Huang, L. Bacterial membrane vesicles: Orchestrators of interkingdom interactions in microbial communities for environmental adaptation and pathogenic dynamics. Front. Immunol. 2024, 15, 1371317. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Cheruvanky, A.; Zhou, H.; Pisitkun, T.; Kopp, J.B.; Knepper, M.A.; Yuen, P.S.T.; Star, R.A. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Ren. Physiol. 2007, 292, F1657–F1661. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduction and Targeted Therapy. Signal Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Genschmer, K.R.; Russell, D.W.; Lal, C.; Szul, T.; Bratcher, P.E.; Noerager, B.D.; Abdul Roda, M.; Xu, X.; Rezonzew, G.; Viera, L.; et al. Activated PMN Exosomes: Pathogenic Entities Causing Matrix Destruction and Disease in the Lung. Cell 2019, 176, 113–126. [Google Scholar] [CrossRef]
- LeBleu, V.S.; Kalluri, R. Exosomes as a Multicomponent Biomarker Platform in Cancer. Trends Cancer 2020, 6, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Li, I.; Nabet, B.Y. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. Cancer 2019, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- D’Agnelli, S.; Gerra, M.C.; Bignami, E.; Arendt-Nielsen, L. Exosomes as a new pain biomarker opportunity. Mol. Pain 2020, 16, 1744806920957800. [Google Scholar] [CrossRef] [PubMed]
- Tavasolian, F.; Moghaddam, A.S.; Rohani, F.; Abdollahi, E.; Janzamin, E.; Momtazi-Borojeni, A.A.; Moallem, S.A.; Jamialahmadi, T.; Sahebkar, A. Exosomes: Effectual players in rheumatoid arthritis. Autoimmun. Rev. 2020, 19, 102511. [Google Scholar] [CrossRef]
- Salomon, C.; Guanzon, D.; Scholz-Romero, K.; Longo, S.; Correa, P.; Illanes, S.E.; Rice, G.E. Placental Exosomes as Early Biomarker of Preeclampsia: Potential Role of Exosomal MicroRNAs Across Gestation. J. Clin. Endocrinol. Metab. 2017, 102, 3182–3194. [Google Scholar] [CrossRef]
- Přikryl, P.; Satrapová, V.; Frýdlová, J.; Hrušková, Z.; Zima, T.; Tesar, V.; Vokurka, M. Mass spectrometry-based proteomic exploration of the small urinary extracellular vesicles in ANCA-associated vasculitis in comparison with total urine. J. Proteom. 2020, 233, 104067. [Google Scholar] [CrossRef]
- Iliev, D.B.; Jørgensen, S.M.; Rode, M.; Krasnov, A.; Harneshaug, I.; Jørgensen, J.B. CpG-induced secretion of MHCIIβ and exosomes from salmon (Salmo salar) APCs. Dev. Comp. Immunol. 2010, 34, 29–41. [Google Scholar] [CrossRef]
- Sun, Z.; Hao, T.; Tian, J. Identification of exosomes and its signature miRNAs of male and female Cynoglossus semilaevis. Sci. Rep. 2017, 7, 860. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef]
- Zhao, N.; Zhang, B.; Xu, Z.-Q.; Jia, L.; Li, M.; He, X.; Bao, B. Detecting Cynoglossus semilaevis infected with Vibrio harveyi using micro RNAs from mucous exosomes. Mol. Immunol. 2020, 128, 268–276. [Google Scholar] [CrossRef]
- Magnadóttir, B.; Kraev, I.; Guðmundsdóttir, S.; Dodds, A.W.; Lange, S. Extracellular vesicles from cod (Gadus morhua L.) mucus contain innate immune factors and deiminated protein cargo. Dev. Comp. Immunol. 2019, 99, 103397. [Google Scholar] [CrossRef] [PubMed]
- Criscitiello, M.F.; Kraev, I.; Lange, S. Deiminated proteins in extracellular vesicles and plasma of nurse shark (Ginglymostoma cirratum)—Novel insights into shark immunity. Fish Shellfish Immunol. 2019, 92, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Xu, W.; Zhao, L.; Xu, X.; Luo, Z.; Huang, L.; Yan, Q. Protection against Pseudomonas plecoglossicida in Epinephelus coioides immunized with a cspA1-knock-down live attenuated vaccine. Fish Shellfish Immunol. 2019, 89, 498–504. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Kowal, E.J.K.; Ter-Ovanesyan, D.; Regev, A.; Church, G.M. Extracellular Vesicle Isolation and Analysis by Western Blotting. In Extracellular Vesicles: Methods and Protocols; Kuo, W.P., Jia, S., Eds.; Extracellular Vesicles. Methods in Molecular Biology, vol 1660; Humana Press: New York, NY, USA, 2017; pp. 143–152. [Google Scholar]
- Choi, D.; Go, G.; Kim, D.-K.; Lee, J.; Park, S.-M.; Di Vizio, D.; Gho, Y.S. Quantitative proteomic analysis of trypsin-treated extracellular vesicles to identify the real-vesicular proteins. J. Extracell Vesicles 2020, 9, 1757209. [Google Scholar] [CrossRef]
- Bandu, R.; Oh, J.W.; Kim, K.P. Mass spectrometry-based proteome profiling of extracellular vesicles and their roles in cancer biology. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Medina-Gali, R.; Belló-Pérez, M.; Ciordia, S.; Mena, M.C.; Coll, J.; Novoa, B.; Ortega-Villaizán, M.d.M.; Perez, L. Plasma proteomic analysis of zebrafish following spring viremia of carp virus infection. Fish Shellfish Immunol. 2019, 86, 892–899. [Google Scholar] [CrossRef]
- Chen, R.; Mao, Y.; Wang, J.; Liu, M.; Qiao, Y.; Zheng, L.; Su, Y.; Ke, Q.; Zheng, W. Molecular mechanisms of an antimicrobial peptide piscidin (Lc-pis) in a parasitic protozoan, Cryptocaryon irritans. BMC Genom. 2018, 19, 192. [Google Scholar] [CrossRef]
- Benhamed, S.; Guardiola, F.A.; Mars, M.; Esteban, M.Á. Pathogen bacteria adhesion to skin mucus of fishes. Vet. Microbiol. 2014, 171, 1–12. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Krumeich, S.; Raposo, G.; Théry, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell Vesicles 2012, 1, 18397. [Google Scholar] [CrossRef]
- Martínez-González, E.; Brochado-Kith, Ó.; Gómez-Sanz, A.; Martín-Carbonero, L.; Jimenez-Sousa, M.Á.; Martínez-Román, P.; Resino, S.; Briz, V.; Fernández-Rodríguez, A. Comparison of methods and characterization of small RNAs from plasma extracellular vesicles of HIV/HCV coinfected patients. Sci. Rep. 2020, 10, 11140. [Google Scholar] [CrossRef] [PubMed]
- Diaz, G.; Bridges, C.; Lucas, M.; Cheng, Y.; Schorey, J.; Dobos, K.; Kruh-Garcia, N. Protein Digestion, Ultrafiltration, and Size Exclusion Chromatography to Optimize the Isolation of Exosomes from Human Blood Plasma and Serum. J. Vis. Experiments 2018, 134, 57467. [Google Scholar] [CrossRef]
- Street, J.M.; Koritzinsky, E.H.; Glispie, D.M.; Star, R.A.; Yuen, P.S.T. Chapter Three—Urine Exosomes: An Emerging Trove of Biomarkers. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 103–122. [Google Scholar]
- Lu, R.; Zhang, W.; Zhang, Y.; Yang, L.; Wang, J.; Meng, X.; Nie, G. Isolation and identification of hepatocellular exosomes and their effects on the expression of miR-122/33 and immune-related genes in grass carp(Ctenopharyngodon idella). J. Fish. China 2020, 44, 1–10. [Google Scholar]
- He, R.; Zuo, Y.; Zhao, L.; Ma, Y.; Yan, Q.; Huang, L. Copper stress by nutritional immunity activates the CusS-CusR two-component system that contributes to Vibrio alginolyticus anti-host response but affects virulence-related properties. Aquaculture 2021, 532, 736012. [Google Scholar] [CrossRef]
- Huang, L.; Zuo, Y.; Qin, Y.; Zhao, L.; Lin, M.; Yan, Q. The Zinc Nutritional Immunity of Epinephelus coioides Contributes to the Importance of znuC During Pseudomonas plecoglossicida. Infection 2021, 12, 678699. [Google Scholar] [CrossRef]
- Wang, J.; Li, Q.; Huang, L. Effect of the zinc transporter ZupT on the virulence mechanisms of mesophilic Aeromonas salmonicida SRW-OG1. Anim. Res. One Health 2023, 1, 30–42. [Google Scholar] [CrossRef]
- Wang, L.; Shao, C.; Xu, W.; Zhou, Q.; Wang, N.; Chen, S. Proteome profiling reveals immune responses in Japanese flounder (Paralichthys olivaceus) infected with Edwardsiella tarda by iTRAQ analysis. Fish Shellfish Immunol. 2017, 66, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Martínez, D.; Oyarzún, R.; Vargas-Lagos, C.; Pontigo, J.P.; Soto-Dávila, M.; Saravia, J.; Romero, A.; Núñez, J.J.; Yáñez, A.J.; Vargas-Chacoff, L. Identification, characterization and modulation of ferritin-H in the sub-Antarctic Notothenioid Eleginops maclovinus challenged with Piscirickettsia salmonis. Dev. Comp. Immunol. 2017, 73, 88–96. [Google Scholar] [CrossRef]
- Long, M.; Zhao, J.; Li, T.; Tafalla, C.; Zhang, Q.; Wang, X.; Gong, X.; Shen, Z.; Li, A. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J. Proteom. 2015, 122, 41–54. [Google Scholar] [CrossRef]
- Yaacob, E.N.; De Geest, B.G.; Goethals, J.; Bajek, A.; Dierckens, K.; Bossier, P.; Vanrompay, D. Recombinant ferritin-H induces immunosuppression in European sea bass larvae (Dicentrarchus labrax) rather than immunostimulation and protection against a Vibrio anguillarum infection. Vet. Immunol. Immunopathol. 2018, 204, 19–27. [Google Scholar] [CrossRef]
- Wu, C.-X.; Liu, Z.-F. Proteomic Profiling of Sweat Exosome Suggests its Involvement in Skin Immunity. J. Investig. Dermatol. 2018, 138, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef] [PubMed]
- Valero, Y.; Arizcun, M.; Esteban, M.Á.; Cuesta, A.; Chaves-Pozo, E. Transcription of histones H1 and H2B is regulated by several immune stimuli in gilthead seabream and European sea bass. Fish Shellfish Immunol. 2016, 57, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Gu, H.J.; Huang, H.Q.; Wang, H.Y.; Xia, Z.H.; Hu, Y.H. Characterization, expression, and antimicrobial activity of histones from Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2020, 96, 235–244. [Google Scholar] [CrossRef]





| Primers | Sequences (5′-3′) | Reference |
|---|---|---|
| β-actin-F | GTGCTGTCTTTCCCTCCATC | [39] |
| β-actin-R | CTCTTGCTCTGGGCTTCATC | |
| Grouper-Annexin A5-F | GAGGCAGGAGATCAAGACCG | |
| Grouper-Annexin A5-R | GAGCCACGATCAGGGTTTCA | |
| Grouper-E 3 subunit M-F | TGTCCGTGCTCTCAAAGACC | |
| Grouper-E 3 subunit M-R | TGCTAGCTTTCCGCTCACAA |
| Protein | Description | log2FC | p Value | Subcellular Localization | |
|---|---|---|---|---|---|
| 1 | Protein_21745 | Plasma kallikrein | 8.26 | 0.001 | extracell protein |
| 2 | Protein_8061 | Annexin A5 | 7.60 | 0.003 | extracell protein |
| 3 | Protein_2146 | Eukaryotic translation initiation factor 3 subunit M | 7.40 | 0.005 | -- |
| 4 | Protein_11510 | S-methyl-5-thioadenosine phosphorylase | 7.35 | 0.005 | cytoplasm protein |
| 5 | Protein_7712 | Collagen alpha-1(XIV) chain | 7.12 | 0.007 | extracell protein |
| 6 | Protein_8704 | Catenin alpha-1 | 7.09 | 0.007 | plasma membrane protein |
| 7 | Protein_10555 | 60S ribosomal protein L17 | 7.06 | 0.008 | -- |
| 8 | Protein_21908 | 26S proteasome regulatory subunit 6A | 7.01 | 0.008 | nucleus protein |
| 9 | Protein_509 | Actin, muscle | 6.99 | 0.009 | centrosome protein |
| 10 | Protein_3526 | Annexin A13 | 6.77 | 0.012 | plasma membrane protein |
| 11 | Protein_21718 | FACT complex subunit SSRP1 | 6.75 | 0.012 | nucleus protein |
| 12 | Protein_4455 | Major vault protein (Fragment) | 6.72 | 0.013 | nucleus protein |
| 13 | Protein_4749 | Calpain-5 | 6.64 | 0.014 | cytoplasm protein |
| 14 | Protein_3222 | Nck-associated protein 1 | 6.63 | 0.014 | -- |
| 15 | Protein_921 | Extended synaptotagmin-1 | 6.58 | 0.015 | microsome protein |
| 16 | Protein_1062 | Coatomer subunit beta | 6.54 | 0.016 | plasma membrane protein |
| 17 | Protein_4013 | Sodium/potassium-transporting ATPase subunit alpha-3 | 6.53 | 0.016 | nucleus protein |
| 18 | Protein_20191 | Myosin-10 | 6.52 | 0.017 | nucleus protein |
| 19 | Protein_5803 | Desmoplakin | 6.48 | 0.018 | -- |
| 20 | Protein_6704 | Synaptotagmin-like protein 4 | 6.42 | 0.019 | cytoplasm protein |
| 21 | Protein_1348 | Nectin-2 | −1.03 | 0.040 | plasma membrane protein |
| 22 | Protein_3796 | COMM domain-containing protein 6 | −1.06 | 0.039 | nucleus protein |
| 23 | Protein_1699 | Galactose-specific lectin nattectin | −1.07 | 0.038 | plasma membrane protein |
| 24 | Protein_19641 | ATP-dependent translocase ABCB1 | −1.07 | 0.038 | mitochondrion protein |
| 25 | Protein_10149 | Serine/threonine-protein kinase Nek9 | −1.14 | 0.035 | cytoplasm protein |
| 26 | Protein_22073 | RNA-binding protein 8A | −1.18 | 0.033 | nucleus protein |
| 27 | Protein_13899 | Transmembrane emp24 domain-containing protein 1 | −1.19 | 0.033 | Golgi apparatus protein |
| 28 | Protein_6507 | Aldehyde dehydrogenase family 3 member B1 | −1.20 | 0.033 | endoplasmic reticulum protein |
| 29 | Protein_20110 | N-alpha-acetyltransferase 15, NatA auxiliary subunit | −1.31 | 0.029 | cytoskeleton protein |
| 30 | Protein_4676 | Low choriolytic enzyme | −1.33 | 0.028 | -- |
| 31 | Protein_11132 | Plectin | −1.40 | 0.026 | cytoskeleton protein |
| 32 | Protein_6024 | Plasminogen activator inhibitor 1 RNA-binding protein | −1.40 | 0.026 | cytoplasm protein |
| 33 | Protein_8938 | Cyclin-dependent kinase 4 inhibitor D | −1.42 | 0.025 | nucleus protein |
| 34 | Protein_3505 | Superoxide dismutase (Mn), mitochondrial | −1.43 | 0.025 | mitochondrion protein |
| 35 | Protein_10177 | Galactose-specific lectin nattectin | −1.44 | 0.025 | plasma membrane protein |
| 36 | Protein_10019 | Interleukin-6 receptor subunit beta | −1.49 | 0.023 | plasma membrane protein |
| 37 | Protein_2270 | Receptor-type tyrosine-protein phosphatase F | −1.53 | 0.022 | plasma membrane protein |
| 38 | Protein_20694 | NHL repeat-containing protein 3 | −1.54 | 0.022 | extracell protein |
| 39 | Protein_3470 | Carbonyl reductase (NADPH) 1 | −1.56 | 0.021 | cytoplasm protein |
| 40 | Protein_431 | E3 ubiquitin-protein ligase HECTD1 | −1.58 | 0.021 | endoplasmic reticulum protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.; Ma, X.; Zhong, S.; Guo, J.; Cheng, D.; Chen, X.; Huang, T.; Huang, L.; Qiao, Y.; Pengsakul, T. Mucosal Exosome Proteomics of Hybrid Grouper Epinephelus fuscoguttatus♀ × E. lanceolatus♂ Infected by Pseudomonas plecoglossicida. Animals 2024, 14, 3401. https://doi.org/10.3390/ani14233401
Yang D, Ma X, Zhong S, Guo J, Cheng D, Chen X, Huang T, Huang L, Qiao Y, Pengsakul T. Mucosal Exosome Proteomics of Hybrid Grouper Epinephelus fuscoguttatus♀ × E. lanceolatus♂ Infected by Pseudomonas plecoglossicida. Animals. 2024; 14(23):3401. https://doi.org/10.3390/ani14233401
Chicago/Turabian StyleYang, Dong, Xiaowan Ma, Shengping Zhong, Jiasen Guo, Dewei Cheng, Xuyang Chen, Teng Huang, Lixing Huang, Ying Qiao, and Theerakamol Pengsakul. 2024. "Mucosal Exosome Proteomics of Hybrid Grouper Epinephelus fuscoguttatus♀ × E. lanceolatus♂ Infected by Pseudomonas plecoglossicida" Animals 14, no. 23: 3401. https://doi.org/10.3390/ani14233401
APA StyleYang, D., Ma, X., Zhong, S., Guo, J., Cheng, D., Chen, X., Huang, T., Huang, L., Qiao, Y., & Pengsakul, T. (2024). Mucosal Exosome Proteomics of Hybrid Grouper Epinephelus fuscoguttatus♀ × E. lanceolatus♂ Infected by Pseudomonas plecoglossicida. Animals, 14(23), 3401. https://doi.org/10.3390/ani14233401








