Low Genetic Variability of the Tundra Vole in Lithuania
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
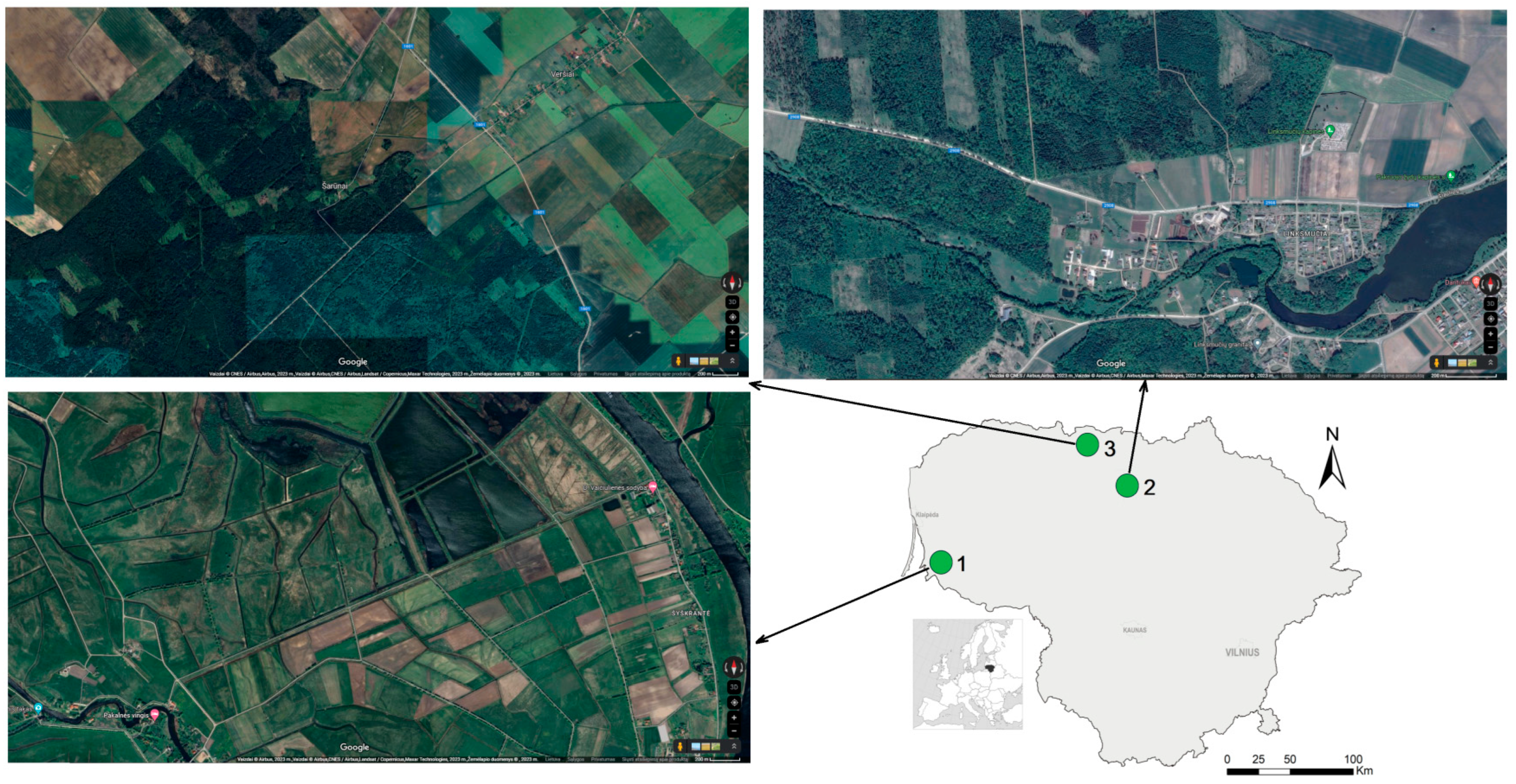
2.1. Small Mammal Sampling
2.2. DNA Isolation, PCR, and Sequencing
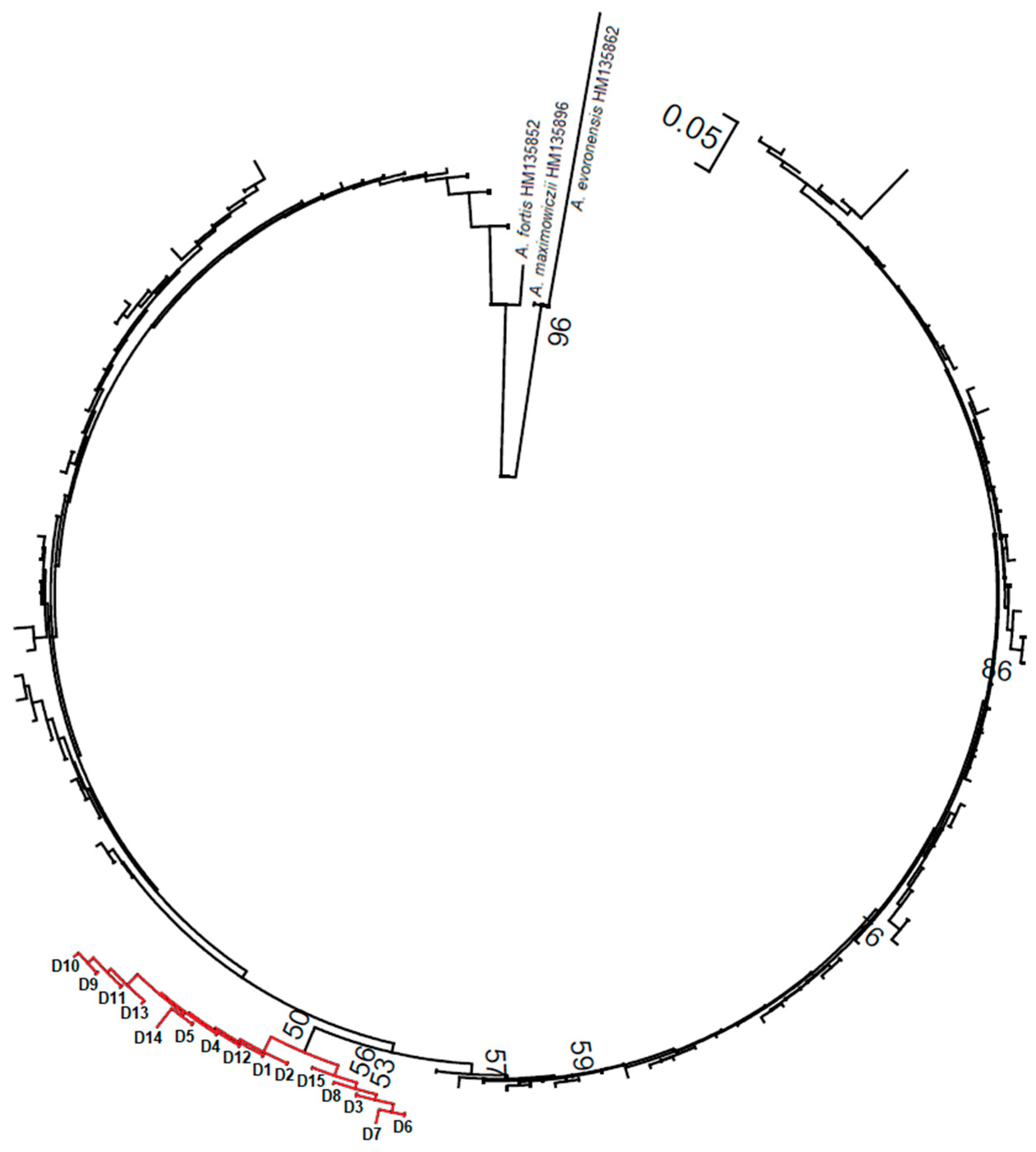
2.3. Phylogenetic Analyses
2.4. Population Genetic Analysis
3. Results
3.1. The Origin of Lithuanian Alexandromys oeconomus
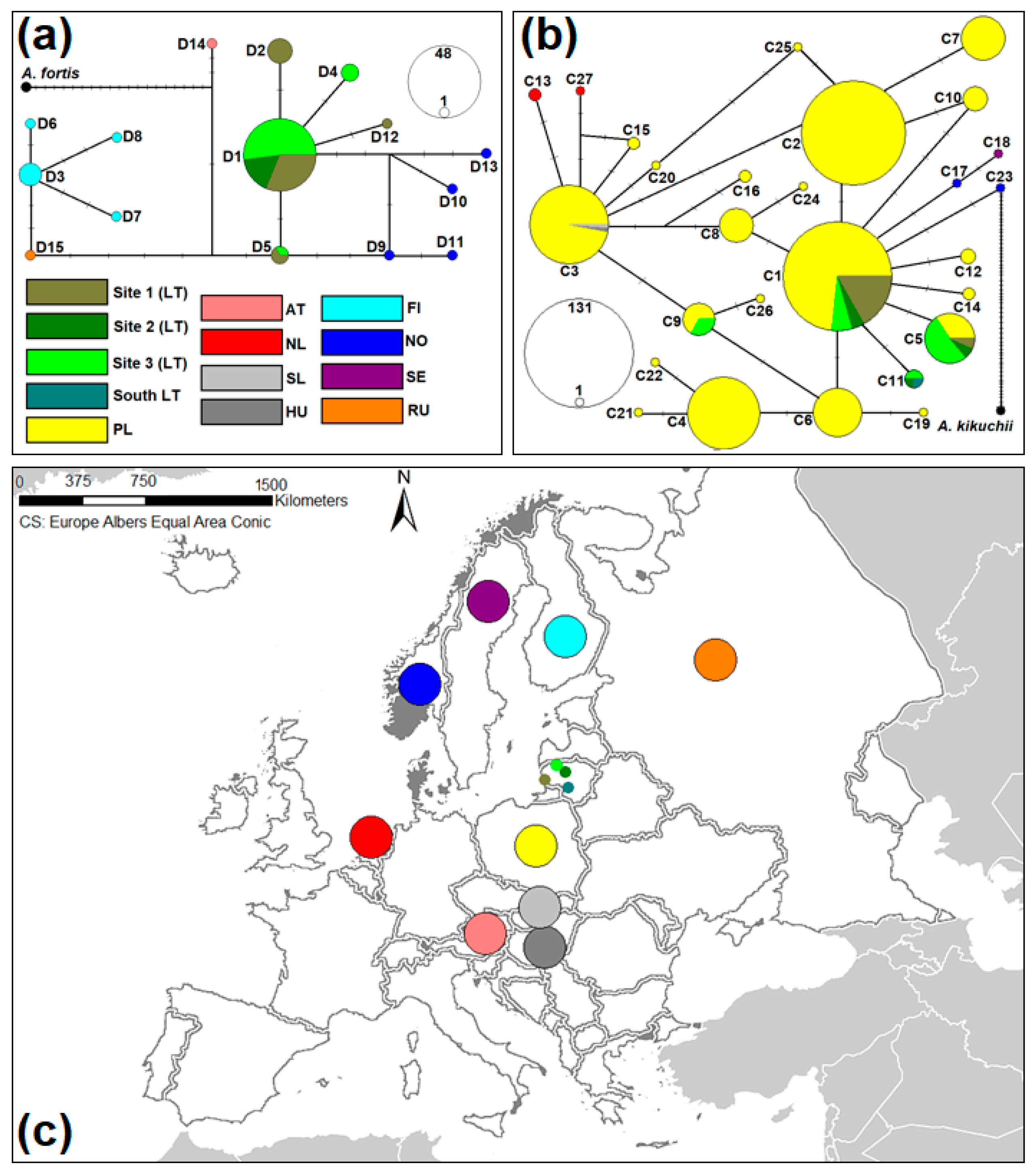
3.2. The Genetic Variability of A. oeconomus from Lithuania
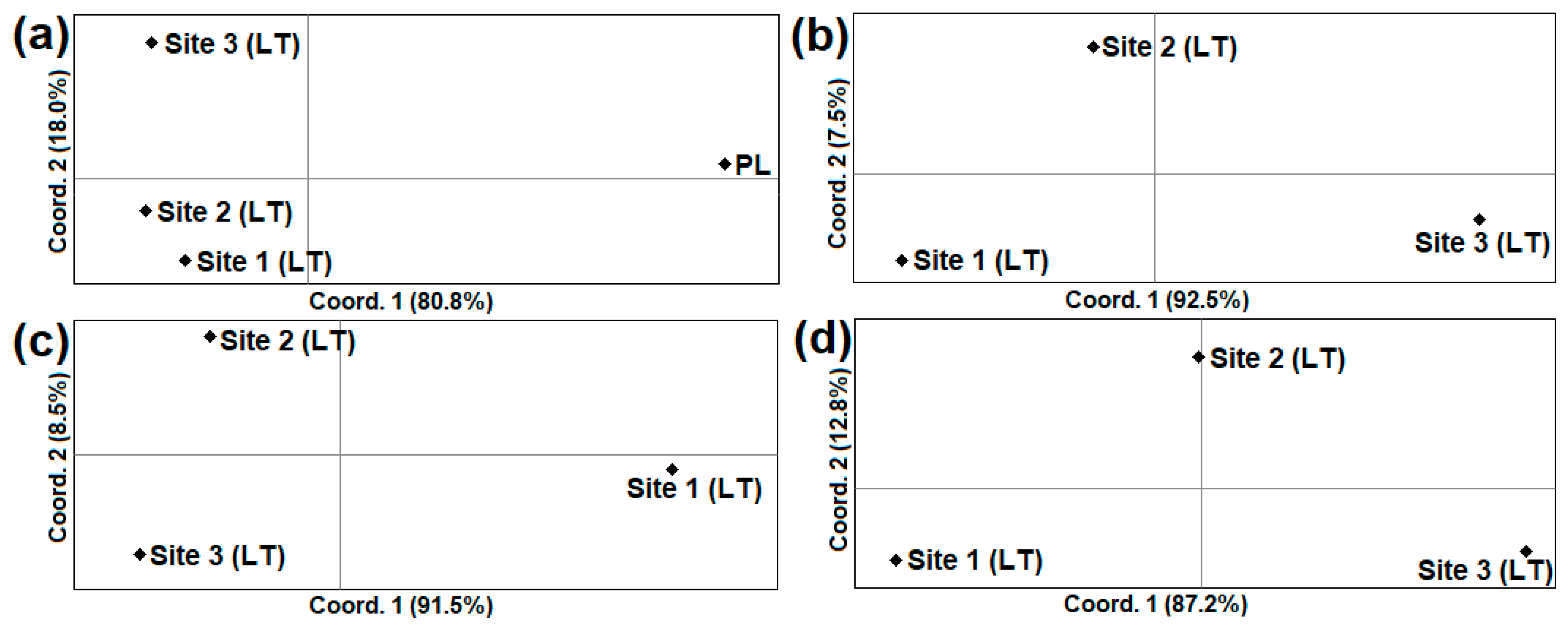
3.3. The Inter-Population Genetic Comparison of A. oeconomus from Lithuania
4. Discussion
4.1. Distribution of Alexandromys oeconomus
4.2. Low Genetic Variability of the Lithuanian Alexandromys oeconomus Population
4.3. The Isolation, Spread, and Genetic Diversity of Alexandromys oeconomus
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Haplotype | Country | GenBank Acc. No. | Frequency |
|---|---|---|---|
| Control region | |||
| D1 | Lithuania | OR806890 | 48 |
| D2 | Lithuania | OR806891 | 6 |
| D3 | Finland | AY305161–63, HM135801–02 | 5 |
| D4 | Lithuania | OR806892 | 3 |
| D5 | Lithuania | OR806893 | 3 |
| D6 | Finland | HM135799 | 1 |
| D7 | Finland | HM135798 | 1 |
| D8 | Finland | HM135800 | 1 |
| D9 | Norway | HM135796 | 1 |
| D10 | Norway | HM135797 | 1 |
| D11 | Norway | HM135795 | 1 |
| D12 | Lithuania | OR806894 | 1 |
| D13 | Norway | HM135929 | 1 |
| D14 | Austria | HM135930 | 1 |
| D15 | Russia | HM135928 | 1 |
| cytb | |||
| C1 | Poland | AY220010, KP684103, KP684111, KP684120 | 96 |
| C1 | Lithuania | OR806886 | 35 |
| C2 | Poland | KP684109 | 122 |
| C3 | Poland | AY220012, KP684101 | 68 |
| C3 | Hungary | AY220014 | 1 |
| C3 | Slovakia | AY220014 | 1 |
| C4 | Poland | GU987116, KP684104 | 60 |
| C5 | Lithuania | OR806887 | 19 |
| C5 | Poland | KP684105 | 10 |
| C6 | Poland | KP684112 | 26 |
| C7 | Poland | KP684107 | 22 |
| C8 | Poland | AY220013, KP684113 | 13 |
| C9 | Poland | KP684102 | 8 |
| C9 | Lithuania | OR806888 | 4 |
| C10 | Poland | KP684116 | 7 |
| C11 | Lithuania | AY220011, OR806889 | 4 |
| C12 | Poland | KP684114, KP684117 | 3 |
| C13 | Netherlands | AY220007 | 2 |
| C14 | Poland | KP684106 | 2 |
| C15 | Poland | KP684118 | 2 |
| C16 | Poland | KP684108 | 2 |
| C17 | Norway | AY220005 | 1 |
| C18 | Sweden | AY220003 | 1 |
| C19 | Poland | AY220008 | 1 |
| C20 | Poland | KP684119 | 1 |
| C21 | Poland | GU954319 | 1 |
| C22 | Poland | KP684121 | 1 |
| C23 | Norway | AY220004 | 1 |
| C24 | Poland | KP684110 | 1 |
| C25 | Poland | KP684115 | 1 |
| C26 | Poland | AY220009 | 1 |
| C27 | Netherlands | AY220006 | 1 |

References
- Garner, A.; Rachlow, J.L.; Hicks, J.F. Patterns of genetic diversity and its loss in mammalian populations. Conserv. Biol. 2005, 19, 1215–1221. [Google Scholar] [CrossRef]
- Bilton, D.T.; Mirol, P.M.; Mascheretti, S.; Fredga, K.; Zima, J.; Searle, J.B. Mediterranean Europe as an area of endemism for small mammals rather than a source for northwards postglacial colonization. Proc. R. Soc. Lond. B Biol. Sci. 1998, 265, 1219–1226. [Google Scholar] [CrossRef]
- Haynes, S.; Jaarola, M.; Searle, J.B. Phylogeography of the common vole (Microtus arvalis) with particular emphasis on the colonization of the Orkney archipelago. Mol. Ecol. 2003, 12, 951–956. [Google Scholar] [CrossRef]
- Deffontaine, V.; Libois, R.; Kotlík, P.; Sommer, R.; Nieberding, C.; Paradis, E.; Searle, J.B.; Michaux, J.R. Beyond the Mediterranean peninsulas: Evidence of central European glacial refugia for a temperate forest mammal species, the bank vole (Clethrionomys glareolus). Mol. Ecol. 2005, 14, 1727–1739. [Google Scholar] [CrossRef]
- Jaarola, M.; Searle, J.B. Phylogeography of field voles (Microtus agrestis) in Eurasia inferred from mitochondrial DNA sequences. Mol. Ecol. 2002, 11, 2613–2621. [Google Scholar] [CrossRef] [PubMed]
- Prakas, P.; Butkauskas, D.; Vaišvilienė, J.; Balčiauskienė, L.; Balčiauskas, L. The Spatial Pattern of the Two Genetic Lineages of the Field Vole in Lithuania. Diversity 2022, 14, 820. [Google Scholar] [CrossRef]
- Jaarola, M.; Martínková, N.; Gündüz, I.; Brunhoff, C.; Zima, J.; Nadachowski, A.; Amori, G.; Bulatova, N.S.; Chondropoulos, B.; Fraguedakis-Tsolis, S.; et al. Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2004, 33, 647–663. [Google Scholar] [CrossRef]
- Triant, D.A.; DeWoody, J.A. Accelerated Molecular Evolution in Microtus (Rodentia) as Assessed via Complete Mitochondrial Genome Sequences. Genetica 2006, 128, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, S.; Paupério, J.; Pavlova, S.V.; Alves, P.C.; Searle, J.B. The Microtus voles: Resolving the phylogeny of one of the most speciose mammalian genera using genomics. Mol. Phylogenet. Evol. 2018, 125, 85–92. [Google Scholar] [CrossRef]
- Łopucki, R.; Mróz, I.; Nowak-Życzyńska, Z.; Perlińska-Teresiak, M.; Owadowska-Cornil, E.; Klich, D. Genetic structure of the root vole Microtus oeconomus: Resistance of the habitat specialist to the natural fragmentation of preferred moist habitats. Genes 2022, 13, 434. [Google Scholar] [CrossRef]
- Haring, E.; Sheremetyeva, I.N.; Kryukov, A.P. Phylogeny of Palearctic vole species (genus Microtus, Rodentia) based on mitochondrial sequences. Mamm. Biol. 2011, 76, 258–267. [Google Scholar] [CrossRef]
- Alexandromys oeconomus . Available online: https://www.iucnredlist.org/species/13451/115113894 (accessed on 1 October 2023).
- ASM Mammal Diversity Database. Available online: https://www.mammaldiversity.org/ (accessed on 7 October 2023).
- Brunhoff, C.; Yoccoz, N.G.; Ims, R.A.; Jaarola, M. Glacial survival or late glacial colonization? Phylogeography of the root vole (Microtus oeconomus) in north-west Norway. J. Biogeogr. 2006, 33, 2136–2144. [Google Scholar] [CrossRef]
- Balčiauskas, L. New mammal species for Latvia, the root vole (Microtus oeconomus). Zool. Ecol. 2014, 24, 187–191. [Google Scholar] [CrossRef]
- Van de Zande, L.; Van Apeldoorn, R.C.; Blijdenstein, A.F.; De Jong, D.; Van Delden, W.; Bijlsma, R. Microsatellite analysis of population structure and genetic differentiation within and between populations of the root vole, Microtus oeconomus in the Netherlands. Mol. Ecol. 2000, 9, 1651–1656. [Google Scholar] [CrossRef] [PubMed]
- Hulejová Sládkovičová, V.; Dąbrowski, M.J.; Žiaka, D.; Miklós, P.; Gubányi, A.; La Haye, M.J.; Bekker, D.; Thissen, J.; Herzig-Straschil, B.; Kocian, Ľ.; et al. Genetic variability of the cold-tolerant Microtus oeconomus subspecies left behind retreating glaciers. Mamm. Biol. 2018, 88, 85–93. [Google Scholar] [CrossRef]
- Sládkovičová, V.; Žiak, D.; Miklós, P.; Gubányi, A.; Horváth, G. The history of genetic diversity and effective population size of an isolated Microtus oeconomus population on Kis Balaton. Mamm. Biol. 2022, 102, 87–98. [Google Scholar] [CrossRef]
- Brunhoff, C.; Galbreath, K.E.; Fedorov, V.B.; Cook, J.A.; Jaarola, M. Holarctic phylogeography of the root vole (Microtus oeconomus): Implications for late Quarternary biogeography of high latitudes. Mol. Ecol. 2003, 12, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Leijs, R.; van Apeldoorn, R.C.; Bijlsma, R. Low genetic differentiation in north-west European populations of the locally endangered root vole, Microtus oeconomus. Biol. Conserv. 1999, 87, 39–48. [Google Scholar] [CrossRef]
- Kelemen, K.A.; Urzi, F.; Buzan, E.; Horváth, G.F.; Tulis, F.; Baláž, I. Genetic variability and conservation of the endangered Pannonian root vole in fragmented habitats of an agricultural landscape. Nat. Conserv. 2021, 43, 167–191. [Google Scholar] [CrossRef]
- Dąbrowski, M.J.; Pomorski, J.J.; Gliwicz, J. Cytochrome b gene (cytb) sequence diversity in a Microtus oeconomus population from Bialowieza Primeval Forest. Acta Theriol. 2013, 58, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Pilot, M.; Dąbrowski, M.J.; Jancewicz, E.; Schtickzelle, N.; Gliwicz, J. Temporally stable genetic variability and dynamic kinship structure in a fluctuating population of the root vole Microtus oeconomus. Mol. Ecol. 2010, 19, 2800–2812. [Google Scholar] [CrossRef]
- Pereverzeva, V.V.; Dokuchaev, N.E.; Primak, A.A.; Dubinin, E.A. Variability of the Cytochrome b Gene of mtDNA of the Tundra Vole (Alexandromys oeconomus Pallas, 1776) in Northeast Asia and Alaska. Biol. Bull. Rev. 2023, 13, 336–351. [Google Scholar] [CrossRef]
- Likevičienė, N. Lietuvos TSR Peliniai Graužikai. Ph.D. Thesis, Vilnius University, Vilnius, Lithuania, 1959. [Google Scholar]
- Prūsaitė, J. Fauna of Lithuania. Mammals; Mokslas: Vilnius, Lithuania, 1988; pp. 143–144. [Google Scholar]
- Jancewicz, E.; Falkowska, E.; Ratkiewicz, M. mtDNA evidence for a local northern latitude Pleistocene refugium for the root vole (Microtus oeconomus, Arvicolinae, Rodentia) from Eastern Poland. J. Zool. Syst. Evol. Res. 2015, 53, 331–339. [Google Scholar] [CrossRef]
- Jancewicz, E.; Falkowska, E. The effect of Pleistocene glacial morphogenesis on the genetic structure of the humid-and cold-tolerant root vole Microtus oeconomus (Rodentia, Cricetidae) in Poland, central Europe. Quat. Res. 2020, 93, 225–242. [Google Scholar] [CrossRef]
- Balčiauskas, L.; Balčiauskienė, L.; Baltrūnaitė, L. Root vole, Microtus oeconomus, in Lithuania: Changes in the distribution range. Folia Zool. 2010, 59, 267–277. [Google Scholar] [CrossRef]
- Balčiauskas, L.; Balčiauskienė, L. Long-term changes in a small mammal community in a temperate zone meadow subject to seasonal floods and habitat transformation. Integr. Zool. 2022, 17, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Alejūnas, P.; Stirkė, V. Small mammals in northern Lithuania: Species diversity and abundance. Ekologija 2010, 56, 110–115. [Google Scholar] [CrossRef]
- Čepukienė, A.; Jasiulionis, M. Small mammal community changes during forest succession (Pakruojis district, north Lithuania). Zool. Ecol. 2012, 22, 144–149. [Google Scholar] [CrossRef]
- Balčiauskas, L.; Stirkė, V.; Balčiauskienė, L. Rodent fertility in commercial orchards in relation to body mass and body condition. Agric. Ecosyst. Environ. 2022, 329, 107886. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Iwasa, M.A.; Kostenko, V.A.; Frisman, L.V.; Kartavtseva, I.V. Phylogeography of the root vole Microtus oeconomus in Russian far east: A special reference to comparison between Holarctic and Palaearctic voles. Mammal Study 2009, 34, 123–130. [Google Scholar] [CrossRef]
- Bannikova, A.A.; Lebedev, V.S.; Lissovsky, A.A.; Matrosova, V.; Abramson, N.I.; Obolenskaya, E.V.; Tesakov, A.S. Molecular phylogeny and evolution of the Asian lineage of vole genus Microtus (Rodentia: Arvicolinae) inferred from mitochondrial cytochrome b sequence. Biol. J. Linn. Soc. Lond. 2010, 99, 595–613. [Google Scholar] [CrossRef]
- Fink, S.; Fischer, M.C.; Excoffier, L.; Heckel, G. Genomic scans support repetitive continental colonization events during the rapid radiation of voles (Rodentia: Microtus): The utility of AFLPs versus mitochondrial and nuclear sequence markers. Syst. Biol. 2010, 59, 548–572. [Google Scholar] [CrossRef]
- Liu, S.; Jin, W.; Liu, Y.; Murphy, R.W.; Lv, B.; Hao, H.; Liao, R.; Sun, Z.; Tang, M.; Chen, W.; et al. Taxonomic position of Chinese voles of the tribe Arvicolini and the description of 2 new species from Xizang, China. J. Mammal. 2017, 98, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Lissovsky, A.A.; Petrova, T.V.; Yatsentyuk, S.P.; Golenishchev, F.N.; Putincev, N.I.; Kartavtseva, I.V.; Sheremetyeva, N.I.; Abramson, N.I. Multilocus phylogeny and taxonomy of East Asian voles Alexandromys (Rodentia, Arvicolinae). Zool. Scr. 2017, 47, 9–20. [Google Scholar] [CrossRef]
- Galbreath, K.E.; Cook, J.A. Genetic consequences of Pleistocene glaciations for the tundra vole (Microtus oeconomus) in Beringia. Mol. Ecol. 2004, 13, 135–148. [Google Scholar] [CrossRef]
- Villesen, P. FaBox: An online toolbox for fasta sequences. Mol. Ecol. Notes 2007, 7, 965–968. [Google Scholar] [CrossRef]
- Kumar, S. Patterns of nucleotide substitution in mitochondrial protein coding genes of vertebrates. Genetics 1996, 143, 537–548. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (Version 3.0): An Integrated Software Package for Population Genetics Data Analysis. Evol. Bioinform. 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Falkowska, E.; Jancewicz, E. Landscape diversity vs. population resilience of a wetland species near the limits of its range (the root vole Microtus oeconomus in Poland)–Implications for species conservation. Catena 2022, 211, 105947. [Google Scholar] [CrossRef]
- Tast, J. The root vole, Microtus oeconomus (Pallas), as an inhabitant of seasonally flooded land. Ann. Zool. Fenn. 1966, 3, 127–171. Available online: https://www.jstor.org/stable/23731273 (accessed on 3 November 2023).
- Stirkė, V.; Balčiauskas, L.; Balčiauskienė, L. Common vole as a focal small mammal species in orchards of the Northern Zone. Diversity 2021, 13, 134. [Google Scholar] [CrossRef]
- Kalivodová, M.; Kanka, R.; Miklós, P.; Sládkovičová, V.H.; Žiak, D. Importance of wetland refugia in agricultural landscape provided based on the community characteristics of small terrestrial mammals. Ekológia 2018, 37, 358–368. [Google Scholar] [CrossRef]
- Neby, M.; Kamenova, S.; Devineau, O.; Ims, R.A.; Soininen, E.M. Issues of under-representation in quantitative DNA metabarcoding weaken the inference about diet of the tundra vole Microtus oeconomus. PeerJ 2021, 9, e11936. [Google Scholar] [CrossRef] [PubMed]
- Steen, H. Low survival of long distance dispersers of the root vole (Microtus oeconomus). Ann. Zool. Fenn. 1994, 31, 271–274. Available online: https://www.jstor.org/stable/23735382 (accessed on 5 November 2023).
- Gubányi, A.; Dudich, A.; Stollmann, A.; Ambros, M. Distribution and conservation management of the root vole (Microtus oeconomus) populations along the Danube in Central Europe (Rodentia: Arvicolinae). Lynx 2009, 40, 29–42. [Google Scholar]
- Steketee, J.K.; Rocha, A.V.; Gough, L.; Griffin, K.L.; Klupar, I.; An, R.; Williamson, N.; Rowe, R.J. Small herbivores with big impacts: Tundra voles (Microtus oeconomus) alter post-fire ecosystem dynamics. Ecology 2022, 103, e3689. [Google Scholar] [CrossRef]
- Wood, D.A.; Rose, J.P.; Halstead, B.J.; Stoelting, R.E.; Swaim, K.E.; Vandergast, A.G. Combining genetic and demographic monitoring better informs conservation of an endangered urban snake. PLoS ONE 2020, 15, e0231744. [Google Scholar] [CrossRef]
- Mathur, S.; Tomeček, J.M.; Tarango-Arámbula, L.A.; Perez, R.M.; DeWoody, J.A. An evolutionary perspective on genetic load in small, isolated populations as informed by whole genome resequencing and forward-time simulations. Evolution 2023, 77, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Thissen, J.B.; Bekker, D.L.; Spreitzer, K.; Herzig-Straschil, B. The distribution of the Pannonic root vole (Microtus oeconomus mehelyi Ehik, 1928) in Austria. Lutra 2015, 58, 3–22. [Google Scholar]
- Domínguez, J.C.; Alda, F.; Calero-Riestra, M.; Olea, P.P.; Martínez-Padilla, J.; Herranz, J.; Oñate, J.J.; Santamaría, A.; Viñuela, J.; García, J.T. Genetic footprints of a rapid and large-scale range expansion: The case of cyclic common vole in Spain. Heredity 2023, 130, 381–393. [Google Scholar] [CrossRef]
- García, J.T.; Domínguez-Villaseñor, J.; Alda, F.; Calero-Riestra, M.; Olea, P.P.; Fargallo, J.A.; Martínez-Padilla, J.; Herranz, J.; Oñate, J.J.; Santamaría, A.; et al. A complex scenario of glacial survival in Mediterranean and continental refugia of a temperate continental vole species (Microtus arvalis) in Europe. J. Zool. Syst. Evolut. Res. 2020, 58, 459–474. [Google Scholar] [CrossRef]

| Sample | n | S | h | K | Hd ± SD | π ± SD | Tajima D |
|---|---|---|---|---|---|---|---|
| cytb | |||||||
| Lithuania | 62 | 4 | 4 | 0.80011 | 0.589 ± 0.047 | 0.00115 ± 0.00017 | −0.13146 |
| Lithuania, present study | 61 | 4 | 4 | 0.78033 | 0.577 ± 0.047 | 0.00112 ± 0.00017 | −0.18937 |
| Lithuania, Site 1 | 24 | 1 | 2 | 0.15942 | 0.159 ± 0.094 | 0.00023 ± 0.00014 | −0.68111 |
| Lithuania, Site 2 | 8 | 2 | 3 | 0.67857 | 0.607 ± 0.164 | 0.00097 ± 0.00032 | −0.44794 |
| Lithuania, Site 3 | 29 | 4 | 4 | 1.14286 | 0.655 ± 0.065 | 0.00164 ± 0.00029 | 0.31626 |
| Poland | 448 | 19 | 21 | 2.80442 | 0.833 ± 0.008 | 0.00402 ± 0.00009 | −0.03417 |
| Central Europe 1 | 518 | 26 | 27 | 2.72261 | 0.842 ± 0.008 | 0.00391 ± 0.00008 | −0.80183 |
| Northern Europe 2 | 44 | 38 | 24 | 5.68499 | 0.958 ± 0.014 | 0.00816 ± 0.00071 | −1.20614 |
| Overall | 562 | 60 | 51 | 4.64387 | 0.865 ± 0.007 | 0.00666 ± 0.00006 | −1.32441 |
| control region | |||||||
| Lithuania | 61 | 4 | 5 | 0.40328 | 0.372 ± 0.076 | 0.00107 ± 0.00024 | −1.14912 |
| Lithuania, Site 1 | 24 | 3 | 4 | 0.63406 | 0.562 ± 0.092 | 0.00168 ± 0.00035 | −0.53008 |
| Lithuania, Site 2 | 8 | 0 | 1 | – | – | 0 | – |
| Lithuania, Site 3 | 29 | 2 | 3 | 0.26108 | 0.254 ± 0.100 | 0.00069 ± 0.00028 | −1.00859 |
| Northern Europe 3 | 14 | 19 | 10 | 5.86813 | 0.890 ± 0.081 | 0.01552 ± 0.00252 | −0.07419 |
| Overall | 75 | 22 | 15 | 2.52252 | 0.582 ± 0.007 | 0.00667 ± 0.00126 | −1.33757 |
| cytb + control region | |||||||
| Lithuania | 61 | 8 | 8 | 1.18361 | 0.754 ± 0.039 | 0.00106 ± 0.00012 | −0.80927 |
| Lithuania, Site 1 | 24 | 4 | 5 | 0.79348 | 0.656 ± 0.083 | 0.00071 ± 0.00013 | −0.70896 |
| Lithuania, Site 2 | 8 | 2 | 3 | 0.67857 | 0.607 ± 0.164 | 0.00061 ± 0.00020 | −0.44794 |
| Lithuania, Site 3 | 29 | 6 | 6 | 1.40394 | 0.761 ± 0.054 | 0.00126 ± 0.00019 | −0.23334 |
| Site 1 (LT) | Site 2 (LT) | Site 3 (LT) | Poland | Northern Europe | |
|---|---|---|---|---|---|
| Site1 (LT) | 0.05900 | 0.12513 ** | – | 0.63653 ** | |
| Site 2 (LT) | 0.06606 | –0.02346 | – | 0.52409 ** | |
| Site 3 (LT) | 0.22077 ** | 0.02509 | – | 0.68879 ** | |
| Poland | 0.18365 ** | 0.16660 * | 0.20425 ** | - | |
| Northern Europe | 0.77818 ** | 0.72406 ** | 0.77798 ** | 0.82620 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prakas, P.; Butkauskas, D.; Balčiauskienė, L.; Balčiauskas, L. Low Genetic Variability of the Tundra Vole in Lithuania. Animals 2024, 14, 270. https://doi.org/10.3390/ani14020270
Prakas P, Butkauskas D, Balčiauskienė L, Balčiauskas L. Low Genetic Variability of the Tundra Vole in Lithuania. Animals. 2024; 14(2):270. https://doi.org/10.3390/ani14020270
Chicago/Turabian StylePrakas, Petras, Dalius Butkauskas, Laima Balčiauskienė, and Linas Balčiauskas. 2024. "Low Genetic Variability of the Tundra Vole in Lithuania" Animals 14, no. 2: 270. https://doi.org/10.3390/ani14020270
APA StylePrakas, P., Butkauskas, D., Balčiauskienė, L., & Balčiauskas, L. (2024). Low Genetic Variability of the Tundra Vole in Lithuania. Animals, 14(2), 270. https://doi.org/10.3390/ani14020270








