Genetic Diversity and Selection Signal Analysis of Hu Sheep Based on SNP50K BeadChip
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Ethical Approval
2.2. DNA Extraction, Genotyping and Quality Control
2.3. Genetic Diversity
2.3.1. Effective Population Size (Ne)
2.3.2. Heterozygosity Analysis
2.3.3. Polymorphism Information Content (PIC)
2.3.4. Polymorphic Marker Ratio (PN)
2.3.5. Minor Allele Frequency (MAF)
2.3.6. Effective Number of Allele
2.4. Kinship Analysis
2.4.1. Genetic Relationship Analysis Based on the G Matrix
2.4.2. Genetic Distance Analysis
2.5. Population Structure Analysis
2.5.1. Principal Component Analysis
2.5.2. Cluster Analysis and Pedigree Construction
2.6. Run of Homozygosity Detection
2.7. Selective Sweep Analysis
3. Results
3.1. Summary of Data
3.2. Genetic Diversity
3.3. Kinship Analysis
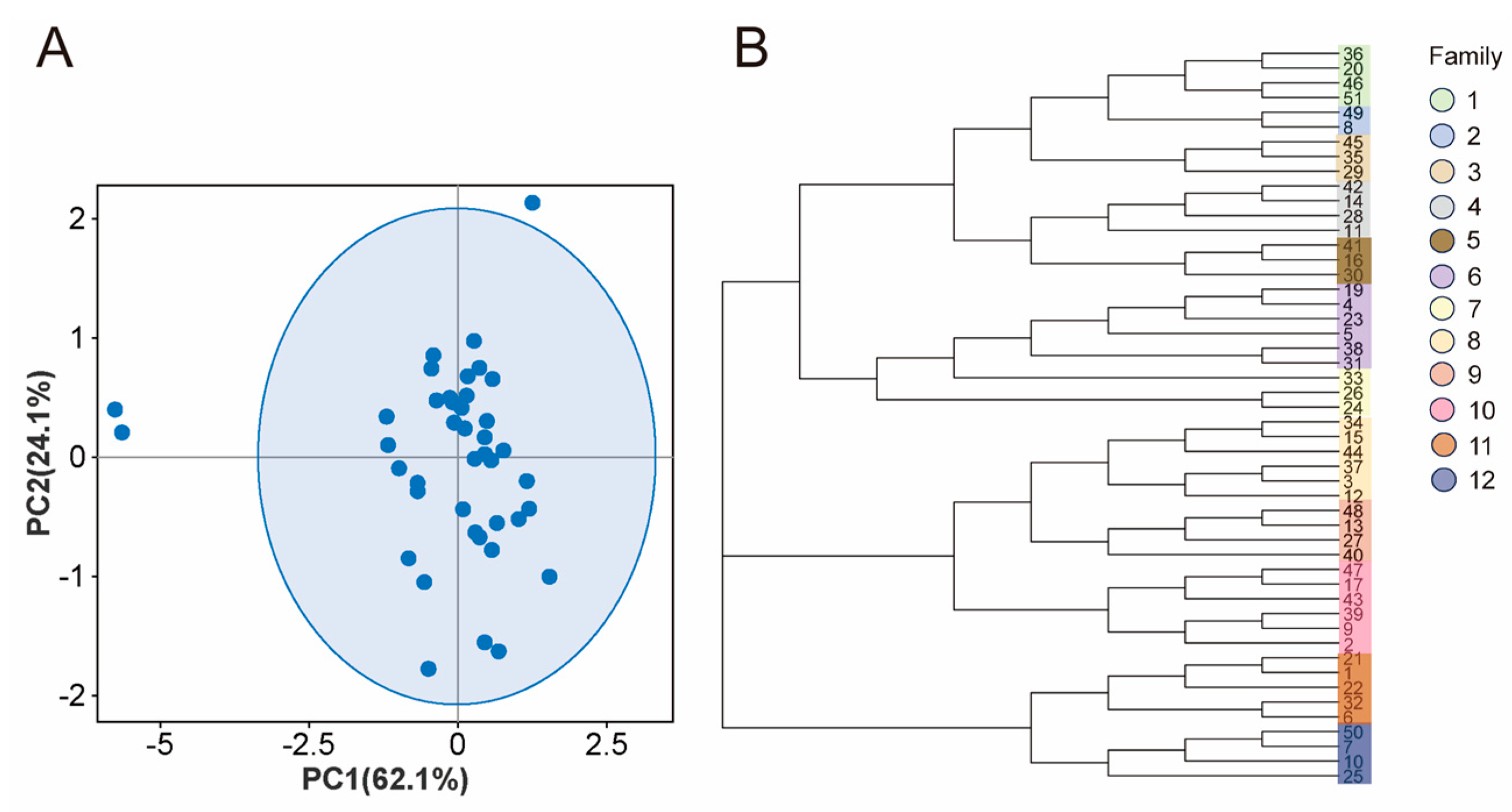
3.4. Population Structure Analysis
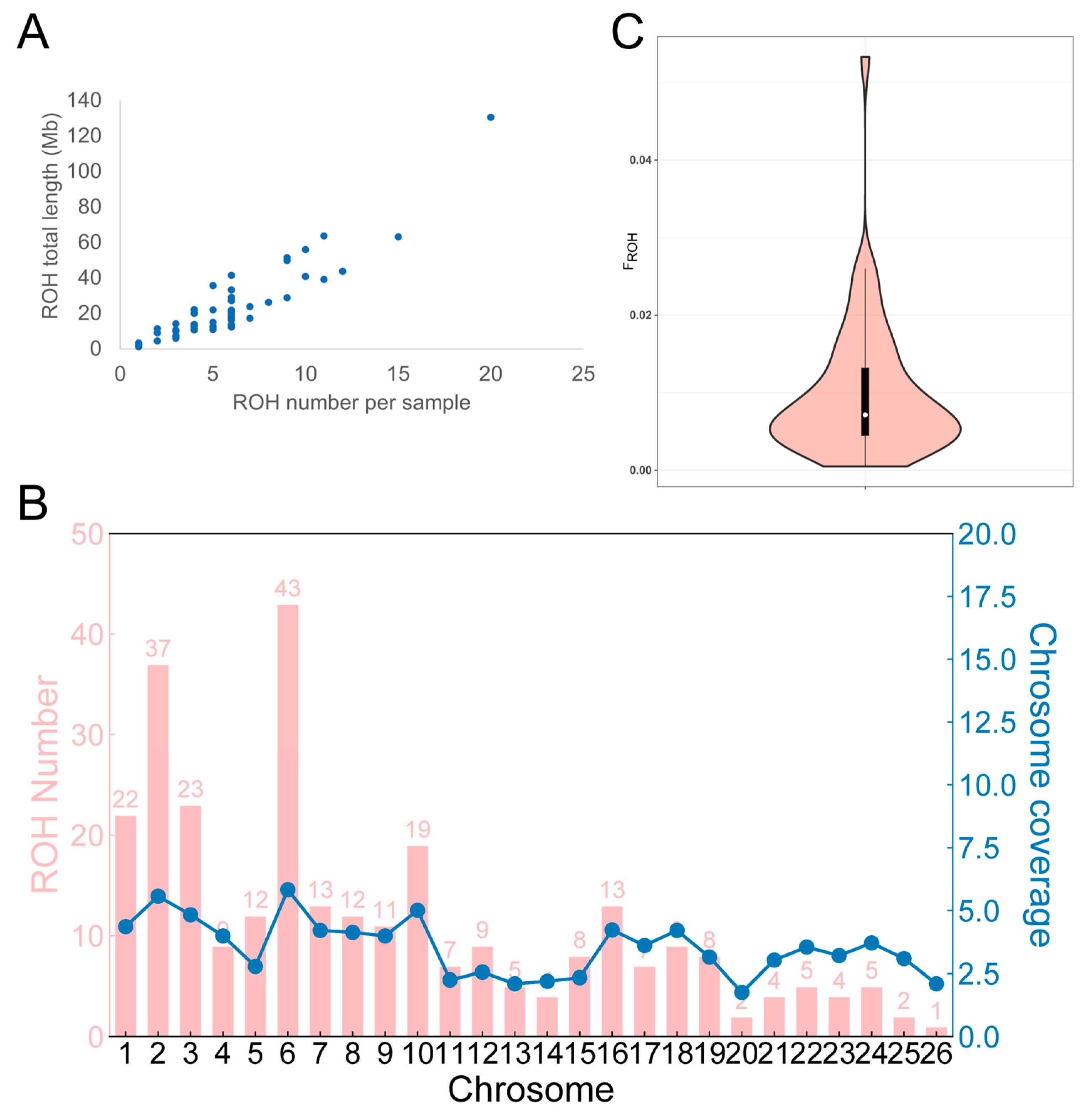
3.5. ROH Analysis
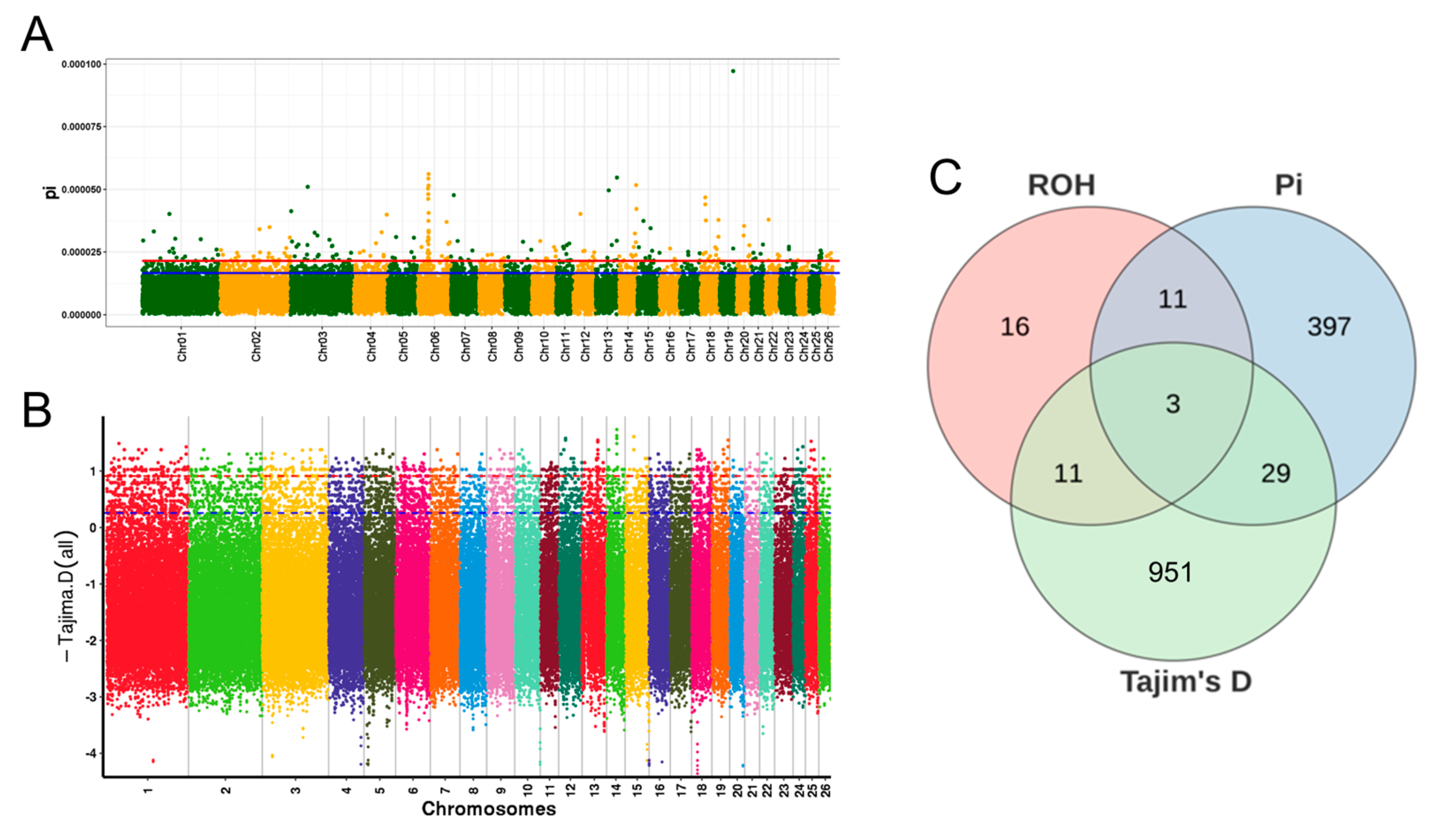
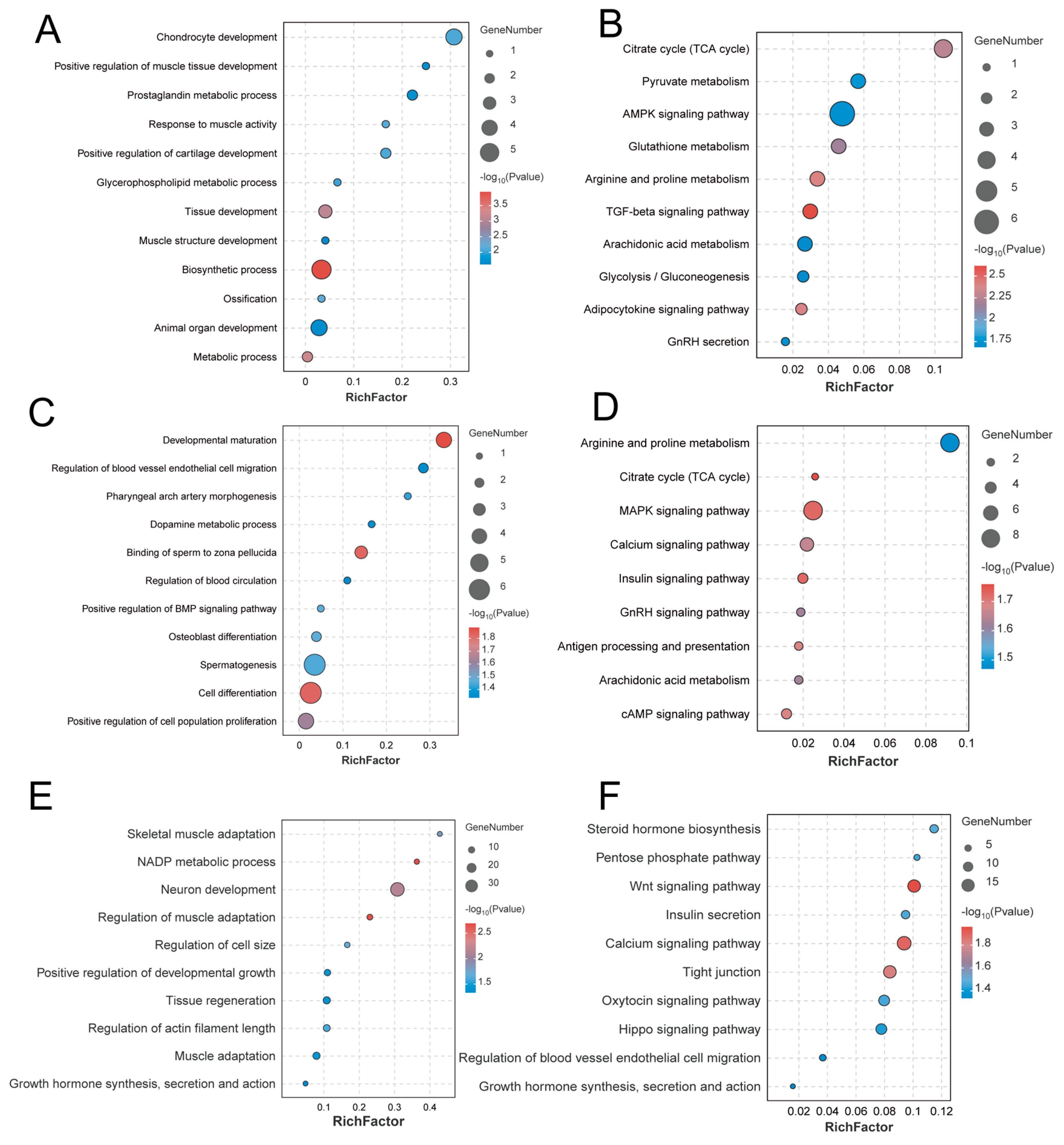
3.6. Candidate Gene Identification and Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yue, C.; Wang, J.; Shen, Y.; Zhang, J.; Liu, J.; Xiao, A.; Liu, Y.; Eer, H.; Zhang, Q.-E. Whole-genome DNA methylation profiling reveals epigenetic signatures in developing muscle in Tan and Hu sheep and their offspring. Front. Vet. Sci. 2023, 10, 1186040. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Wang, X.; Huang, C.; Yang, L.; Zhao, Q.; Chen, X.; Freitas-de-Melo, A.; Zhan, S.; Wang, L.; Dai, D.; et al. A genome-wide perspective on the diversity and selection signatures in indigenous goats using 53 K single nucleotide polymorphism array. Animal 2023, 17, 100706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lai, J.; Wang, X.; Li, M.; Zhang, Y.; Ji, C.; Chen, Q.; Lu, S. Genome-wide single nucleotide polymorphism (SNP) data reveal potential candidate genes for litter traits in a Yorkshire pig population. Arch. Anim. Breed. 2023, 66, 357–368. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, X.; Zhang, Y.; Zhao, Y.; Zhang, J.; Jia, Y.; Zhu, B.; Xu, L.; Zhang, L.; Gao, H.; et al. Genome-wide assessment of genetic diversity and population structure insights into admixture and introgression in Chinese indigenous cattle. BMC Genet. 2018, 19, 114. [Google Scholar] [CrossRef]
- Xu, S.S.; Ren, X.; Yang, G.L.; Xie, X.L.; Zhao, Y.X.; Zhang, M.; Shen, Z.Q.; Ren, Y.L.; Gao, L.; Shen, M.; et al. Genome-wide association analysis identifies the genetic basis of fat deposition in the tails of sheep (Ovis aries). Anim. Genet. 2017, 48, 560–569. [Google Scholar] [CrossRef]
- Sharma, V.; Zhou, M.; Wang, G.; Chen, M.; Pang, Q.; Jiang, S.; Zeng, J.; Du, D.; Yang, P.; Wu, W.; et al. Genetic diversity and population structure of sheep (Ovis aries) in Sichuan, China. PLoS ONE 2021, 16, e0257974. [Google Scholar] [CrossRef]
- Kumar, C.; Song, S.; Dewani, P.; Kumar, M.; Parkash, O.; Ma, Y.; Malhi, K.K.; Yang, N.; Mwacharo, J.M.; He, X.; et al. Population structure, genetic diversity and selection signatures within seven indigenous Pakistani goat populations. Anim. Genet. 2018, 49, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shengjiang·Nasier, Y.A.; Duman, S.L.; Qian, Y.; Cao, S.; Wang, W.; Meng, C.; Zhang, J.; Zhang, J. Evaluation of genetic 481 diversity and genetic structure in Kirgiz sheep population based on SNPs chip. Acta Vet. Zootech. Sin. 2023, 54, 572–583. [Google Scholar]
- Machová, K.; Marina, H.; Arranz, J.J.; Pelayo, R.; Rychtářová, J.; Milerski, M.; Vostrý, L.; Suárez-Vega, A. Genetic diversity of two native sheep breeds by genome-wide analysis of single nucleotide polymorphisms. Animal 2023, 17, 100690. [Google Scholar] [CrossRef]
- Ma, K.Y.; Li, D.P.; Qi, X.C.; Li, Q.; Wu, Y.; Song, J.J.; Zhang, Y.; Yang, H.; Li, T.T.; Ma, Y.J. Population structure, runs of homozygosity analysis and construction of single nucleotide polymorphism fingerprinting database of Longnan goat population. Food Energy Secur. 2023, 13, e517. [Google Scholar] [CrossRef]
- Ma, K.-Y.; Song, J.-J.; Li, D.-P.; Wu, Y.; Wang, C.-H.; Liu, Z.-L.; Li, T.-T.; Ma, Y.-J. Genomic structure analysis and construction of DNA fingerprint for four sheep populations. Animal 2024, 18, 101116. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Sun, W.; Guo, T.; Li, J.; Zhu, S.; Lu, Z.; Qiao, G.; Han, M.; Zhao, H.; Yang, B.; et al. Genome-Wide Selective Signatures Reveal Candidate Genes Associated with Hair Follicle Development and Wool Shedding in Sheep. Genes 2021, 12, 1924. [Google Scholar] [CrossRef]
- Gao, J.; Lyu, Y.; Zhang, D.; Reddi, K.K.; Sun, F.; Yi, J.; Liu, C.; Li, H.; Yao, H.; Dai, J.; et al. Genomic Characteristics and Selection Signatures in Indigenous Chongming White Goat (Capra hircus). Front. Genet. 2020, 11, 901. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Barbato, M.; Orozco-terWengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Silio, L.; Rodriguez, M.C.; Fernandez, A.; Barragan, C.; Benitez, R.; Ovilo, C.; Fernandez, A.I. Measuring inbreeding and inbreeding depression on pig growth from pedigree or SNP-derived metrics. J. Anim. Breed. Genet. 2013, 130, 349–360. [Google Scholar] [CrossRef]
- Hao, S.; Zhen, W.; Zhe, Z.; Qian, X.; Xu, Z.; Zhang, X.-Z.; Yang, H.-Z.; Zhu, M.-X.; Xue, M.; Liu, X.-H.; et al. Exploring the Current Situation of Conservation of Meishan Pigs Based on Genome Sequencing Data. J. Shanghai Jiaotong Univ. 2017, 35, 65–70. [Google Scholar]
- Botstein, D.R.; White, R.L.; Skolnick, M.H.; Davis, R.W. Construction of a Genetic Linkage Map in Man Using Restriction Fragment Length Polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.; Ma, Q.; Liu, S.; Ma, Y.; Jiang, L. Analysis of Family Structure and Paternity Test of Tan Sheep in Yanchi Area, China. Animals 2022, 12, 3099. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Signer-Hasler, H.; Burren, A.; Ammann, P.; Drgemller, C.; Flury, C. Runs of homozygosity and signatures of selection: A comparison among eight local Swiss sheep breeds. Anim. Genet. 2019, 50, 512–525. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Yue, Y.; Li, J.; Yang, B.; Chen, B.; Liu, J.; Lu, Z. Transcriptomics and metabolomics reveal improved performance of Hu sheep on hybridization with Southdown sheep. Food. Res. Int. 2023, 173, 113240. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Zhu, B.; Zhang, W.; Wang, Z.; Chen, Y.; Zhang, L.; Gao, X.; Gao, H.; Liu, G.E.; et al. Genome-wide scan reveals genetic divergence and diverse adaptive selection in Chinese local cattle. BMC Genom. 2019, 20, 494. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Y.Q.; Yan, Y.B.; Guo, H.Y.; Wang, Z.W. The genetic structure analysis and the comparative analysis of selection signals in Tezanghan sheep. Acta Vet. Zootech. Sin. 2024, 55, 2913–2926. [Google Scholar] [CrossRef]
- Zhang, R.B.; Zhou, D.H.; Zhou, L.S.; Gao, X.X.; Liu, N.; He, J.N. Analysis of genetic structure of conservation population in Jining gray goats based on 70K SNP chip. Acta Vet. Zootech. Sin. 2023, 54, 2836–2847. [Google Scholar] [CrossRef]
- Abied, A.; Bagadi, A.; Bordbar, F.; Pu, Y.B.; Augustino, S.M.A.; Xue, X.L.; Xing, F.; Gebreselassie, G.; Han, J.L.; Mwacharo, J.M.; et al. Genomic Diversity, Population Structure, and Signature of Selection in Five Chinese Native Sheep Breeds Adapted to Extreme Environments. Genes 2020, 11, 494. [Google Scholar] [CrossRef] [PubMed]
- An, Z.X.; Shi, L.G.; Hou, G.Y.; Zhou, H.L.; Xun, W.J. Genetic diversity and selection signatures in Hainan black goats revealed by whole-genome sequencing data. Animal 2024, 18, 101147. [Google Scholar] [CrossRef] [PubMed]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Neto, L.R.P.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. Plos Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Luigi-Sierra, M.G.; Fernandez, A.; Martinez, A.; Guan, D.; Delgado, J.V.; Alvarez, J.F.; Landi, V.; Such, F.X.; Jordana, J.; Saura, M.; et al. Genomic patterns of homozygosity and inbreeding depression in Murciano-Granadina goats. J. Anim. Sci. Biotechnol. 2022, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, S.; Ciani, E.; Sardina, M.T.; Sottile, G.; Pilla, F.; Portolano, B.; BiOvIta, C. Runs of homozygosity reveal genome-wide autozygosity in Italian sheep breeds. Anim. Genet. 2018, 49, 71–81. [Google Scholar] [CrossRef]
- Mokhber, M.; Moradi-Shahrbabak, M.; Sadeghi, M.; Moradi-Shahrbabak, H.; Stella, A.; Nicolzzi, E.; Rahmaninia, J.; Williams, J.L. A genome-wide scan for signatures of selection in Azeri and Khuzestani buffalo breeds. BMC Genom. 2018, 19, 449. [Google Scholar] [CrossRef]
- Li, Z.; Wei, S.; Li, H.; Wu, K.; Cai, Z.; Li, D.; Wei, W.; Li, Q.; Chen, J.; Liu, H.; et al. Genome-wide genetic structure and differentially selected regions among Landrace, Erhualian, and Meishan pigs using specific-locus amplified fragment sequencing. Sci. Rep. 2017, 7, 10063. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shi, L.; Li, Y.; Chen, L.; Garrick, D.; Wang, L.; Zhao, F. Estimates of genomic inbreeding and identification of candidate regions that differ between Chinese indigenous sheep breeds. J. Anim. Sci. Biotechnol. 2021, 12, 95. [Google Scholar] [CrossRef]
- Proto, G.G.; Mancin, E.; Sartori, C.; Mantovani, R. Unraveling inbreeding patterns and selection signals in Alpine Grey cattle. Animal 2024, 18, 101159. [Google Scholar] [CrossRef]
- Ghoreishifar, S.M.; Eriksson, S.; Johansson, A.M.; Khansefid, M.; Moghaddaszadeh-Ahrabi, S.; Parna, N.; Davoudi, P.; Javanmard, A. Signatures of selection reveal candidate genes involved in economic traits and cold acclimation in five Swedish cattle breeds. Genet. Sel. Evol. 2020, 52, 52. [Google Scholar] [CrossRef]
- Ramljak, J.; Spehar, M.; Ceranac, D.; Drzaic, V.; Pocrnic, I.; Barac, D.; Mioc, B.; Siric, I.; Barac, Z.; Ivankovic, A.; et al. Genomic Characterization of Local Croatian Sheep Breeds-Effective Population Size, Inbreeding & Signatures of Selection. Animals 2024, 14, 1928. [Google Scholar] [CrossRef]
- Li, X.; Fang, Y.; Chen, L.; Quan, H.; Wang, Y.; Ge, R.-S. Bone morphogenetic protein 4 inhibits rat stem/progenitor Leydig cell development and regeneration via SMAD-dependent and SMAD-independent signaling. Cell Death Dis. 2022, 13, 1039. [Google Scholar] [CrossRef]
- Ma, K.; Chen, N.; Wang, H.; Li, Q.; Shi, H.; Su, M.; Zhang, Y.; Ma, Y.; Li, T. The regulatory role of BMP4 in testicular Sertoli cells of Tibetan sheep. J. Anim. Sci. 2023, 101, skac393. [Google Scholar] [CrossRef]
- Abdurahman, A.; Du, X.; Yao, Y.; Sulaiman, Y.; Aniwashi, J.; Li, Q. Smad4 Feedback Enhances BMPR1B Transcription in Ovine Granulosa Cells. Int. J. Mol. Sci. 2019, 20, 2732. [Google Scholar] [CrossRef]
- Lv, F.-H.; Cao, Y.-H.; Liu, G.-J.; Luo, L.-Y.; Lu, R.; Liu, M.-J.; Li, W.-R.; Zhou, P.; Wang, X.-H.; Shen, M.; et al. Whole-Genome Resequencing of Worldwide Wild and Domestic Sheep Elucidates Genetic Diversity, Introgression, and Agronomically Important Loci. Mol. Biol. Evol. 2021, 39, msab353. [Google Scholar] [CrossRef]
- Yildirim, Y.; Ouriachi, T.; Woehlbier, U.; Ouahioune, W.; Balkan, M.; Malik, S.; Tolun, A. Linked homozygous BMPR1B and PDHA2 variants in a consanguineous family with complex digit malformation and male infertility. Eur. J. Hum. Genet. 2018, 26, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Rangaraj, N.; Shivaji, S. Activity of pyruvate dehydrogenase A (PDHA) in hamster spermatozoa correlates positively with hyperactivation and is associated with sperm capacitation. Biol. Reprod. 2006, 75, 767–777. [Google Scholar] [CrossRef]
- Leung, E.T.Y.; Lee, B.K.M.; Lee, C.-L.; Tian, X.; Lam, K.K.W.; Li, R.H.W.; Ng, E.H.Y.; Yeung, W.S.B.; Ou, J.-P.; Chiu, P.C.N. The role of spermatozoa-zona pellucida interaction in selecting fertilization-competent spermatozoa in humans. Front. Endocrinol. 2023, 14, 1135973. [Google Scholar] [CrossRef]
- Chen, S.-R.; Batool, A.; Wang, Y.-Q.; Hao, X.-X.; Chang, C.-S.; Cheng, C.Y.; Liu, Y.-X. The control of male fertility by spermatid-specific factors: Searching for contraceptive targets from spermatozoon’s head to tail. Cell Death Dis. 2016, 7, e2472. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, M.; Wu, Q.; Lu, H.; Lei, C.; Ahmed, Z.; Sun, J. Analysis of genetic diversity and selection characteristics using the whole genome sequencing data of five buffaloes, including Xilin buffalo, in Guangxi, China. Front. Genet. 2023, 13, 1084824. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Ouyang, J.; Liu, S.; Tang, H.; Xiong, Y.; Yan, X.; Chen, H. Genomic signatures reveal selection in Lingxian white goose. Poult. Sci. 2023, 102, 102269. [Google Scholar] [CrossRef] [PubMed]
- Tomar, A.K.; Rajak, S.K.; Aslam, M.M.K.; Chhikara, N.; Ojha, S.K.; Nayak, S.; Chhillar, S.; Kumaresan, A.; Yadav, S. Sub-fertility in crossbred bulls: Identification of proteomic alterations in spermatogenic cells using high throughput comparative proteomics approach. Theriogenology 2021, 169, 65–75. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Yao, W.; Luo, J.; Wu, J.; Zhang, F.; Li, C.; Gao, L.; Zhang, Y. Knockdown of PROX1 promotes milk fatty acid synthesis by targeting PPARGC1A in dairy goat mammary gland. Int. J. Biol. Macromol. 2024, 266, 131043. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Cai, B.; Kong, S.; Zhou, Z.; Zhang, J.; Zhang, X.; Nie, Q. PPARGC1A Is a Moderator of Skeletal Muscle Development Regulated by miR-193b-3p. Int. J. Mol. Sci. 2022, 23, 9575. [Google Scholar] [CrossRef] [PubMed]
- An, S.-Y.; Zhang, G.-M.; Liu, Z.-F.; Zhou, C.; Yang, P.-C.; Wang, F. MiR-1197-3p regulates testosterone secretion in goat Leydig cells via targeting PPARGC1A. Gene 2019, 710, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Su, P.; Wang, S.; Gu, Y.; Cao, X.; Lv, X.; Wang, S.; Getachew, T.; Mwacharo, J.M.; Haile, A.; et al. New Insight into the Role of the Leucine Aminopeptidase 3 (LAP3) in Cell Proliferation and Myogenic Differentiation in Sheep Embryonic Myoblasts. Genes 2022, 13, 1438. [Google Scholar] [CrossRef]
- Cai, K.; Liu, R.; Wei, L.; Wang, X.; Cui, H.; Luo, N.; Wen, J.; Chang, Y.; Zhao, G. Genome-wide association analysis identify candidate genes for feed efficiency and growth traits in Wenchang chickens. BMC Genom. 2024, 25, 645. [Google Scholar] [CrossRef]
- Huang, C.; Ge, F.; Ren, W.; Zhang, Y.; Wu, X.; Zhang, Q.; Ma, X.; Bao, P.; Guo, X.; Chu, M.; et al. Copy number variation of the HPGDS gene in the Ashidan yak and its associations with growth traits. Gene 2021, 772, 145382. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.G.; Yuan, Y.; Zhou, D.K.; Ma, Y.H.; Mahrous, K.F.; Wang, S.Z.; He, Y.M.; Duan, X.H.; Zhang, W.Y.; E, G. Genome-wide selection signal analysis of Australian Boer goat reveals artificial selection imprinting on candidate genes related to muscle development. Anim. Genet. 2021, 52, 550–555. [Google Scholar] [CrossRef]
- Ma, J.; Gao, X.; Li, J.; Gao, H.; Wang, Z.; Zhang, L.; Xu, L.; Gao, H.; Li, H.; Wang, Y.; et al. Assessing the Genetic Background and Selection Signatures of Huaxi Cattle Using High-Density SNP Array. Animals 2021, 11, 3469. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Cao, X.-K.; Peng, S.-J.; Wang, X.-G.; Li, Z.; Elnour, I.-E.; Huang, Y.-Z.; Lan, X.-Y.; Chen, H. Transcriptional regulation of the bovine FGFR1 gene facilitates myoblast proliferation under hypomethylation of the promoter. J. Cell. Physiol. 2020, 235, 8667–8678. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Fang, W.; Yuan, M.; Sun, H.; Wang, J. Transcriptome Analysis of Leg Muscles and the Effects of ALOX5 on Proliferation and Differentiation of Myoblasts in Haiyang Yellow Chickens. Genes 2023, 14, 1213. [Google Scholar] [CrossRef] [PubMed]


| Quality Control Standard | Number of SNPs |
|---|---|
| Total number of SNPs | 64,734 |
| SNP with MAF < 0.01 | 4123 |
| SNP with P < 10−6 of Hardy-Weinberg equilibrium | 105 |
| SNP with call rate < 0.90 | 1197 |
| SNPs on chromosome X | 1527 |
| SNPs on chromosome Y | 1251 |
| Insertion/Deletion | 9 |
| SNPs used after quality control | 56,522 |
| Index | Average Value |
|---|---|
| Effective population size (Ne) | 6.9 |
| Polymorphic information content (PIC) | 0.264 |
| Polymorphic marker ratio (PN) | 0.873 |
| Expected heterozygosity (He) | 0.356 |
| Observed heterozygosity (Ho) | 0.36 |
| Effective number of alleles | 1.563 |
| Minor allele frequency (MAF) | 0.253 |
| ROH Length (Mb) | ROH number | Percent (%) |
|---|---|---|
| 1–5 | 228 | 77.55 |
| 5–10 | 47 | 15.99 |
| 10–15 | 11 | 3.74 |
| 15–20 | 6 | 2.04 |
| >20 | 2 | 0.68 |
| Total | 294 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, K.; Song, J.; Li, D.; Li, T.; Ma, Y. Genetic Diversity and Selection Signal Analysis of Hu Sheep Based on SNP50K BeadChip. Animals 2024, 14, 2784. https://doi.org/10.3390/ani14192784
Ma K, Song J, Li D, Li T, Ma Y. Genetic Diversity and Selection Signal Analysis of Hu Sheep Based on SNP50K BeadChip. Animals. 2024; 14(19):2784. https://doi.org/10.3390/ani14192784
Chicago/Turabian StyleMa, Keyan, Juanjuan Song, Dengpan Li, Taotao Li, and Youji Ma. 2024. "Genetic Diversity and Selection Signal Analysis of Hu Sheep Based on SNP50K BeadChip" Animals 14, no. 19: 2784. https://doi.org/10.3390/ani14192784
APA StyleMa, K., Song, J., Li, D., Li, T., & Ma, Y. (2024). Genetic Diversity and Selection Signal Analysis of Hu Sheep Based on SNP50K BeadChip. Animals, 14(19), 2784. https://doi.org/10.3390/ani14192784






