Metagenomics Analysis Reveals the Composition and Functional Differences of Fecal Microbiota in Wild, Farm, and Released Chinese Three-Keeled Pond Turtles (Mauremys reevesii)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fecal Sampling
2.2. Bacterial DNA Extraction and Amplification
2.3. 16S rRNA Gene Sequencing Analysis
2.4. Metagenomics Sequencing Analysis
2.5. Statistical and Data Analysis
3. Results
3.1. Intestinal Microbial Community Structure, Composition, and Diversity
3.2. Metagenomics Analysis
3.3. KEGG Pathway Analysis
3.4. Antibiotic-Resistant Gene Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, I.; Liu, K.; Long, D.; Faisal, S.; Hilal, M.G.; Ali, I.; Huang, X.; Long, R. Ramadan Fasting Leads to Shifts in Human Gut Microbiota Structured by Dietary Composition. Front. Microbiol. 2021, 12, 642999. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, W.F. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998, 14, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
- Bengmark, S. Gut microbiota, immune development and function. Pharmacol. Res. 2013, 69, 87–113. [Google Scholar] [CrossRef] [PubMed]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Ezenwa, V.O.; Gerardo, N.M.; Inouye, D.W.; Medina, M.; Xavier, J.B. Animal behavior and the microbiome. Science 2012, 338, 198–199. [Google Scholar] [CrossRef]
- Alberdi, A.; Aizpurua, O.; Bohmann, K.; Zepeda-Mendoza, M.L.; Gilbert, M.T.P. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 2016, 31, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Deschasaux, M.; Bouter, K.E.; Prodan, A.; Levin, E.; Groen, A.K.; Herrema, H.; Tremaroli, V.; Bakker, G.J.; Attaye, I.; Pinto-Sietsma, S.J.; et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 2018, 24, 1526–1531. [Google Scholar] [CrossRef]
- Versalovic, J.; Relman, D. How bacterial communities expand functional repertoires. PLoS Biol. 2006, 4, e430. [Google Scholar] [CrossRef]
- Xia, J.H.; Lin, G.; Fu, G.H.; Wan, Z.Y.; Lee, M.; Wang, L.; Liu, X.J.; Yue, G.H. The intestinal microbiome of fish under starvation. BMC Genom. 2014, 15, 266. [Google Scholar] [CrossRef] [PubMed]
- Clayton, J.B.; Vangay, P.; Huang, H.; Ward, T.; Hillmann, B.M.; Al-Ghalith, G.A.; Travis, D.A.; Long, H.T.; Tuan, B.V.; Minh, V.V.; et al. Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. USA 2016, 113, 10376–10381. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Moeller, A.H.; Li, Y.; Mpoudi-Ngole, E.; Ahuka-Mundeke, S.; Lonsdorf, E.V.; Pusey, A.E.; Peeters, M.; Hahn, B.H.; Ochman, H. Rapid changes in the gut microbiome during human evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16431–16435. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends. Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.I.; Walker, D.W. Role of gut microbiota in aging-related health decline: Insights from invertebrate models. Cell. Mol. Life Sci. 2018, 75, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Bletz, M.C.; Goedbloed, D.J.; Sanchez, E.; Reinhardt, T.; Tebbe, C.C.; Bhuju, S.; Geffers, R.; Jarek, M.; Vences, M.; Steinfartz, S. Amphibian gut microbiota shifts differentially in community structure but converges on habitat-specific predicted functions. Nat. Commun. 2016, 15, 13699. [Google Scholar] [CrossRef] [PubMed]
- Eliades, S.J.; Brown, J.C.; Colston, T.J.; Fisher, R.N.; Niukula, J.B.; Gray, K.; Vadada, J.; Rasalato, S.; Siler, C.D. Gut microbial ecology of the Critically Endangered Fijian crested iguana (Brachylophus vitiensis): Effects of captivity status and host reintroduction on endogenous microbiomes. Ecol. Evol. 2021, 11, 4731–4743. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, T.; Li, Y.; Tang, Y.; Huang, Z. Gut microbiota are associated with sex and age of host: Evidence from semi-provisioned rhesus macaques in southwest Guangxi, China. Ecol. Evol. 2021, 11, 8096–8122. [Google Scholar] [CrossRef]
- Fong, J.J.; Sung, Y.H.; Ding, L. Comparative analysis of the fecal microbiota of wild and captive beal’s eyed turtle (Sacalia bealei) by 16S rRNA gene sequencing. Front. Microbiol. 2020, 1, 570890. [Google Scholar] [CrossRef]
- Qu, Y.F.; Wu, Y.Q.; Zhao, Y.T.; Lin, L.H.; Du, Y.; Li, P.; Li, H.; Ji, X. The invasive red-eared slider turtle is more successful than the native Chinese three-keeled pond turtle: Evidence from the gut microbiota. PeerJ 2020, 8, 271. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, H.; Nakao, A. Molecular cloning of full-length Dmrt1 cDNA of Reeves turtle (Chinemys reevesii). J. Vet. Med. Sci. 2008, 70, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Bu, R.; Ye, Z.; Shi, H. Hibernation in Reeves’ Turtles (Mauremys reevesii) in Qichun County, Hubei Province, China: Hibernation Beginning and End and Habitat Selection. Animals 2022, 12, 2411. [Google Scholar] [CrossRef] [PubMed]
- Lovich, J.; Yasukawa, Y.; Ota, H. Mauremys reevesii (Gray 1831)–Reeves’ turtle, Chinese three-keeled pond turtle. Conservation biology of freshwater turtles and tortoises: A compilation project of the IUCN/SSC tortoise and freshwater turtle specialist group. Chelonian Res. Monogr. 2011, 5, 10. [Google Scholar]
- Bu, R.; Ye, Z.; Shi, H. Habitat Selection and Home Range of Reeves’ Turtle (Mauremys reevesii) in Qichun County, Hubei Province, China. Animals 2023, 13, 1514. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, H.; Jing, Z.Z.; Zheng, W.; Luo, Y.R.; Chen, S.X.; Guo, F. Robust host source tracking building on the divergent and non-stochastic assembly of gut microbiomes in wild and farmed large yellow croaker. Microbiome 2022, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Besnier, F.; Ayllon, F.; Skaala, Ø.; Solberg, M.F.; Fjeldheim, P.T.; Anderson, K.; Knutar, S.; Glover, K.A. Introgression of domesticated salmon changes life history and phenology of a wild salmon population. Evol. Appl. 2022, 15, 853–864. [Google Scholar] [CrossRef]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132, e154944. [Google Scholar] [CrossRef]
- Guo, W.; Ren, K.; Ning, R.H.; Li, C.W.; Zhang, H.M.; Li, D.S.; Xu, L.; Sun, F.H.; Dai, M. Fecal microbiota transplantation provides new insight into wildlife conservation. Glob. Ecol. Conserv. 2020, 24, 01234. [Google Scholar] [CrossRef]
- Jiang, F.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Cai, Z.; Gao, H.; Chi, X.; Zhang, T. Comparative analysis of gut microbial composition and potential functions in captive forest and alpine musk deer. Appl. Microbiol. Biotechnol. 2022, 6, 1325–1339. [Google Scholar] [CrossRef]
- Ahasan, M.S.; Waltzek, T.B.; Huerlimann, R.; Ariel, E. Fecal bacterial communities of wild-captured and stranded green turtles (Chelonia mydas) on the Great Barrier Reef. FEMS Microbiol. Ecol. 2017, 93, fix139. [Google Scholar] [CrossRef]
- Wu, H.; Wu, F.T.; Zhou, Q.H.; Zhao, D.P. Comparative Analysis of Gut Microbiota in Captive and Wild Oriental White Storks: Implications for Conservation Biology. Front. Microbiol. 2021, 12, 649466. [Google Scholar] [CrossRef]
- Tang, S.; Li, Y.; Huang, C.; Yan, S.; Li, Y.; Chen, Z.; Wu, Z. Comparison of Gut Microbiota Diversity between Captive and Wild Tokay Gecko (Gekko gecko). Front Microbiol. 2022, 13, 897923. [Google Scholar] [CrossRef]
- Hiroshi, M.; Tamotsu, K.; Hiroaki, O.; Mitsuo, S.; Takumi, M.; Todd, D.T.; Atsushi, T.; Moriya, O.; Ken, K.; Ohno, H. Assessment of metagenomic workflows using a newly constructed human gut microbiome mock community. DNA Res. 2023, 30, 3. [Google Scholar] [CrossRef]
- Sun, H.Z.; Peng, K.L.; Xue, M.Y.; Liu, J.X. Metagenomics analysis revealed the distinctive ruminal microbiome and resistive profiles in dairy buffaloes. Anim. Microbiome 2021, 3, 44. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Fan, L.; Qiu, L.; Dong, X.; Wang, Q.; Hu, G.; Meng, S.; Li, D.; Chen, J. Metagenomics Analysis Reveals Compositional and Functional Differences in the Gut Microbiota of Red Swamp Crayfish, Procambarus clarkii, Grown on Two Different Culture Environments. Front. Microbiol. 2021, 12, 735190. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Liang, L.; Wang, Z.; Ai, P.; You, X.; Bian, C.; Shi, Q.; Dong, B. A Comparative Metagenomics Study on Gastrointestinal Microbiota in Amphibious Mudskippers and Other Vertebrate Animals. Animals 2019, 9, 660. [Google Scholar] [CrossRef]
- Kang, K.; Hu, Y.; Wu, S.; Shi, S. Comparative Metagenomic Analysis of Chicken Gut Microbial Community, Function, and Resistome to Evaluate Noninvasive and Cecal Sampling Resources. Animals 2021, 11, 1718. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, Y.; Ma, L.; Liu, J.; An, Q.; Zhang, C.; Yin, G.; Cao, Z.; Pan, H. Comparative Analyses of Antibiotic Resistance Genes in Jejunum Microbiota of Pigs in Different Areas. Front. Cell. Infect. Microbiol. 2022, 12, 887428. [Google Scholar] [CrossRef]
- Ning, Y.; Qi, J.; Dobbins, M.T.; Liang, X.; Jiang, G. Comparative Analysis of Microbial Community Structure and Function in the Gut of Wild and Captive Amur Tiger. Front. Microbiol. 2020, 11, 1665. [Google Scholar] [CrossRef]
- Chen, Y.; Xia, Z.; Li, H. Metagenomic comparison of gut communities between hawksbills (Eretmochelys imbricata) and green sea turtles (Chelonia mydas). Arch. Microbiol. 2022, 204, 450. [Google Scholar] [CrossRef]
- Wang, Y.; Qu, M.; Bi, Y.; Liu, W.J.; Ma, S.; Wan, B.; Hu, Y.; Zhu, B.; Zhang, G.; George, F.; et al. The multi-kingdom microbiome catalog of the chicken gastrointestinal tract. Biosaf. Health 2024, 06, 101–115. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Liu, F.; Cao, J.; Lv, N.; Zhu, B.; Zhang, G.; Gao, G.F. Integrated metagenomic and metatranscriptomic profiling reveals differentially expressed resistomes in human, chicken, and pig gut microbiomes. Env. Int. 2020, 138, 105649. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.; Hou, Z.; Yuan, J.; Liu, Y.; Qu, Y.; Liu, B. Taxonomic and functional metagenomic profiling of gastrointestinal tract microbiome of the farmed adult turbot (Scophthalmus maximus). FEMS. Microbiol. Ecol. 2013, 86, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. In Vienna: R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids. Res. 2010, 38, 132. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B. Community Ecology Package. 2013. Available online: http://cran.r-project.org (accessed on 28 December 2021).
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 8, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Backhed, F. The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Bea, K.; Christina, S.; Claude, L.; John, P.; Joël, D.; Paul, E.; Isabelle, M. Effects of isolation and confinement on gastrointestinal microbiota—A systematic review. Front. Nutr. 2023, 10, 1214016. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Li, C.Q.; Chen, S.Y.; Xiao, H. Metagenomic analysis reveals hidden links between gut microbes and habitat adaptation among cave and surface dwelling Sinocyclocheilus species. Zool. Res. 2023, 44, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xia, Y.; Sun, B. Linking the bacterial microbiome between gut and habitat soil of Tibetan macaque. (Macaca thibetana). Ecol. Evol. 2022, 12, e9227. [Google Scholar] [CrossRef]
- Gao, H.; Chi, X.; Qin, W.; Wang, L.; Song, P.; Cai, Z.; Zhang, J.; Zhang, T. Comparison of the gut microbiota composition between the wild and captive Tibetan wild ass (Equus kiang). J. Appl. Microbiol. 2019, 126, 1869–1878. [Google Scholar] [CrossRef]
- Tan, C.K.; Natrah, I.; Suyub, I.B.; Edward, M.J.; Kaman, N.; Samsudin, A.A. Comparative study of gut microbiota in wild and captive Malaysian Mahseer (Tor tambroides). Microbiologyopen 2019, 8, e00734. [Google Scholar] [CrossRef]
- Wang, W.; Zheng, S.; Sharshov, K.; Sun, H.; Yang, F.; Wang, X.; Li, L.; Xiao, Z. Metagenomic profiling of gut microbial communities in both wild and artificially reared Bar-headed goose (Anser indicus). Microbiologyopen 2017, 6, e00429. [Google Scholar] [CrossRef]
- Khan, I.; Huang, Z.; Liang, L.; Li, N.; Ali, Z.; Ding, L.; Hong, M.; Shi, H. Ammonia stress influences intestinal histomorphology, immune status and microbiota of Chinese striped-neck turtle (Mauremys sinensis). Ecotoxicol. Environ. Saf. 2021, 222, 112471. [Google Scholar] [CrossRef]
- Ding, L.; Huang, Z.; Lu, Y.; Liang, L.; Hong, M. Toxic effects of ammonia on intestinal health and microbiota in red-eared slider (Trachemys scripta elegans). Chemosphere 2021, 280, 130630. [Google Scholar] [CrossRef]
- Ahasan, M.S.; Waltzek, T.B.; Huerlimann, R.; Ariel, E. Comparative analysis of gut bacterial communities of green turtles (Chelonia mydas) pre-hospitalization and post-rehabilitation by high-throughput sequencing of bacterial 16S rRNA gene. Microbiol. Res. 2018, 207, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.L.; Dean, S.H.; Longo, A.V.; Rothermel, B.B.; Tuberville, T.D.; Zamudio, K.R. Kinship, inbreeding and fine-scale spatial structure influence gut microbiota in a hindgut-fermenting tortoise. Mol. Ecol. 2015, 24, 2521–2536. [Google Scholar] [CrossRef]
- Arizza, V.; Vecchioni, L.; Caracappa, S.; Sciurba, G.; Berlinghieri, F.; Gentile, A.; Persichetti, M.F.; Arculeo, M.; Alduina, R. New insights into the gut microbiome in logger head sea turtles Caretta caretta stranded on the Mediterranean coast. PLoS ONE 2019, 4, e0220329. [Google Scholar] [CrossRef]
- Fugate, H.M.; Kapfer, J.M.; McLaughlin, R.W. Analysis of the microbiota in the fecal material of painted turtles (Chrysemys picta). Curr. Microbiol. 2020, 77, 11–14. [Google Scholar] [CrossRef]
- Fan, L.; Li, Q.X. Characteristics of intestinal microbiota in the Pacific white shrimp Litopenaeus vannamei differing growth performances in the marine cultured environment. Aquaculture 2019, 505, 450–461. [Google Scholar] [CrossRef]
- Bloodgood, J.C.G.; Hernandez, S.M.; Isaiah, A.; Suchodolski, J.S.; Hoopes, L.A.; Thompson, P.M.; Waltzek, T.B.; Norton, T.M. The effect of diet on the gastrointestinal microbiome of juvenile rehabilitating green turtles (Chelonia mydas). PLoS ONE 2020, 15, e0227060. [Google Scholar] [CrossRef]
- Hooper, L.V.; Stappenbeck, T.S.; Hong, C.V.; Gordon, J.I. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nature immunology. Nat. Immunol. 2003, 4, 269–273. [Google Scholar] [CrossRef]
- Bereded, N.K.; Abebe, G.B.; Fanta, S.W.; Curto, M.; Waidbacher, H.; Meimberg, H.; Domig, K.J. The impact of sampling season and catching site (wild and aquaculture) on gut microbiota composition and diversity of Nile tilapia (Oreochromis niloticus). Biology 2021, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Terova, G.; Díaz, N.; Rimoldi, S.; Ceccotti, C.; Gliozheni, E.; Piferrer, F. Effects of Sodium Butyrate Treatment on Histone Modifications and the Expression of Genes Related to Epigenetic Regulatory Mechanisms and Immune Response in European Sea Bass (Dicentrarchus Labrax) Fed a Plant-Based Diet. PLoS ONE 2016, 11, e0160332. [Google Scholar] [CrossRef]
- Vandamme, P.; Bernardet, J.F.; Segers, P.; Kersters, K.; Holmes, B. New Perspectives in the Classification of the Flavobacteria: Description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. Rev. Int. J. Syst. Evol. Microbiol. 1994, 44, 827–831. [Google Scholar] [CrossRef]
- Son, Y.; Min, J.; Park, W. Chryseobacterium faecale sp. nov., isolated from camel feces. Int. J. Syst. Evol. Microbiol. 2022, 72, 005405. [Google Scholar] [CrossRef]
- Loch, T.P.; Faisal, M. Chryseobacterium aahli sp. nov., isolated from lake trout (Salvelinus namaycush) and brown trout (Salmo trutta), and emended descriptions of Chryseobacterium ginsenosidimutans and Chryseobacterium gregarium. Int. J. Syst. Evol. Microbiol. 2014, 64, 1573–1579. [Google Scholar] [CrossRef]
- Sharma, P.; Gupta, S.K.; Diene, S.M.; Rolain, J.M. Whole-genome sequence of Chryseobacterium oranimense, a colistin-resistant bacterium isolated from a cystic fibrosis patient in France. Antimicrob. Agents. Chemother. 2015, 59, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Ilardi, P.; Abad, J.; Rintamäki, P.; Bernardet, J.F.; Avendaño-Herrera, R. Phenotypic, serological and molecular evidence of Chryseobacterium piscicola in farmed Atlantic salmon, Salmo salar L., in Finland. J. Fish. Dis. 2010, 33, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Shahi, N.; Sharma, P.; Pandey, J.; Bisht, I.; Mallik, S.K. Characterization and pathogenicity study of Chryseobacterium scophthalmum recovered from gill lesions of diseased golden mahseer, Tor putitora (Hamilton, 1822) in India. Aquaculture 2018, 485, 81–92. [Google Scholar] [CrossRef]
- Kim, T.I.; Ki, K.S.; Lim, D.H.; Vijayakumar, M.; Park, S.M.; Choi, S.H.; Kim, K.Y.; Im, S.K.; Park, B.Y. Novel Acinetobacter parvus HANDI 309 microbial biomass for the production of N-acetyl-β-d-glucosamine (GlcNAc) using swollen chitin substrate in submerged fermentation. Biotechnol. Biofuels 2017, 10, 59. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef]
- Huang, Q.; Sham, R.C.; Deng, Y.; Mao, Y.; Wang, C.; Zhang, T.; Leung, K.-Y. Diversity of gut microbiomes in marine fishes is shaped by host-related factors. Mol. Ecol. 2020, 29, 5019–5034. [Google Scholar] [CrossRef]
- Tsuchiya, C.; Sakata, T.; Sugita, H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 2018, 46, 43–48. [Google Scholar] [CrossRef]
- Wang, A.; Zhang, Z.; Ding, Q.; Yang, Y.; Bindelle, J.; Ran, C.; Zhou, Z. Intestinal Cetobacterium and acetate modify glucose homeostasis via parasympathetic activation in zebrafish. Gut Microbes 2021, 13, 1–15. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Y.; Yang, Y.; Wang, A.; Sharshov, K.; Li, Y.; Cao, M.; Mao, P.; Li, L. Comparative analyses of the gut microbiota among three different wild geese species in the genus Anser. J. Basic. Microbiol. 2018, 58, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yao, Y.; Li, D.; Xu, H.; Wu, J.; Wen, A.; Xie, M.; Ni, Q.; Zhang, M.; Peng, G.; et al. Characterization of the gut microbiota in six geographical populations of Chinese rhesus macaques (Macaca mulatta), implying an adaptation to high-altitude environment. Microb. Ecol. 2018, 76, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Benton, T.G.; Bieg, C.; Harwatt, H.; Pudasaini, R.; Wellesley, L. Food system impacts on biodiversity loss. In Three Levers for Food System Transformation in Support of Nature; Chatham House: London, UK, 2021; Volume 3, pp. 2–3. [Google Scholar]
- Petrin, S.; Patuzzi, I.; Di Cesare, A.; Tiengo, A.; Sette, G.; Biancotto, G.; Corno, G.; Drigo, M.; Losasso, C.; Cibin, V. Evaluation and quantification of antimicrobial residues and antimicrobial resistance genes in two Italian swine farms. Environ. Pollut. 2019, 255, 113183. [Google Scholar] [CrossRef] [PubMed]
- Garrod, L.P.; O’grady, F. Antibiotic and Chemotherapy, 3rd ed.; CABI: Wallingford, UK, 1971. [Google Scholar]
- Seribelli, A.A.; da Silva, P.; da Cruz, M.F.; de Almeida, F.; Frazão, M.R.; Medeiros, M.I.C.; Rodrigues, D.-P.; Kich, J.D.; de Jesus Benevides, L.; Soares, S.C.; et al. Insights about the epidemiology of Salmonella Typhimurium isolates from different sources in Brazil using comparative genomics. Gut. Pathog. 2021, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015, 9, 2490–2502. [Google Scholar] [CrossRef] [PubMed]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016, 7, 1881. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.Q.; Colquhoun, D.J.; Nikuli, H.L.; Sørum, H. Prevalence of antibiotic resistance genes in the bacterial flora of integrated fish farming environments of Pakistan and Tanzania. Environ. Sci. Technol. 2012, 46, 8672–8679. [Google Scholar] [CrossRef]
- Maxwell, A. DNA gyrase as a drug target. Trends. Microbiol. 1997, 5, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Salwan, R.; Sharma, V. Bioactive compounds of Streptomyces: Biosynthesis to applications. Stud. Nat. Prod. Chem. 2020, 64, 467–491. [Google Scholar] [CrossRef]
- Koul, A.; Arnoult, E.; Lounis, N.; Guillemont, J.; Andries, K. The challenge of new drug discovery for tuberculosis. Nature 2011, 469, 483–490. [Google Scholar] [CrossRef]
- Shirude, P.S.; Hameed, S. Nonfluoroquinolone-based inhibitors of mycobacterial type II topoisomerase as potential therapeutic agents for TB. Annu. Rep. Med. Chem. 2012, 47, 319–330. [Google Scholar] [CrossRef]
- Chopra, S.; Matsuyama, K.; Tran, T.; Malerich, J.P.; Wan, B.; Franzblau, S.G.; Lun, S.; Guo, H.; Maiga, M.C.; Bishai, W.R.; et al. Evaluation of gyrase B as a drug target in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2012, 67, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Sugino, A.; Bott, K. Bacillus subtilis deoxyribonucleic acid gyrase. J. Bacteriol. 1980, 141, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Young, F.; Wilson, G. Chromosomal Map of Bacillus subtilis; Spores VI; American Society for Microbiology: Washington, DC, USA, 1975; pp. 596–614. [Google Scholar]


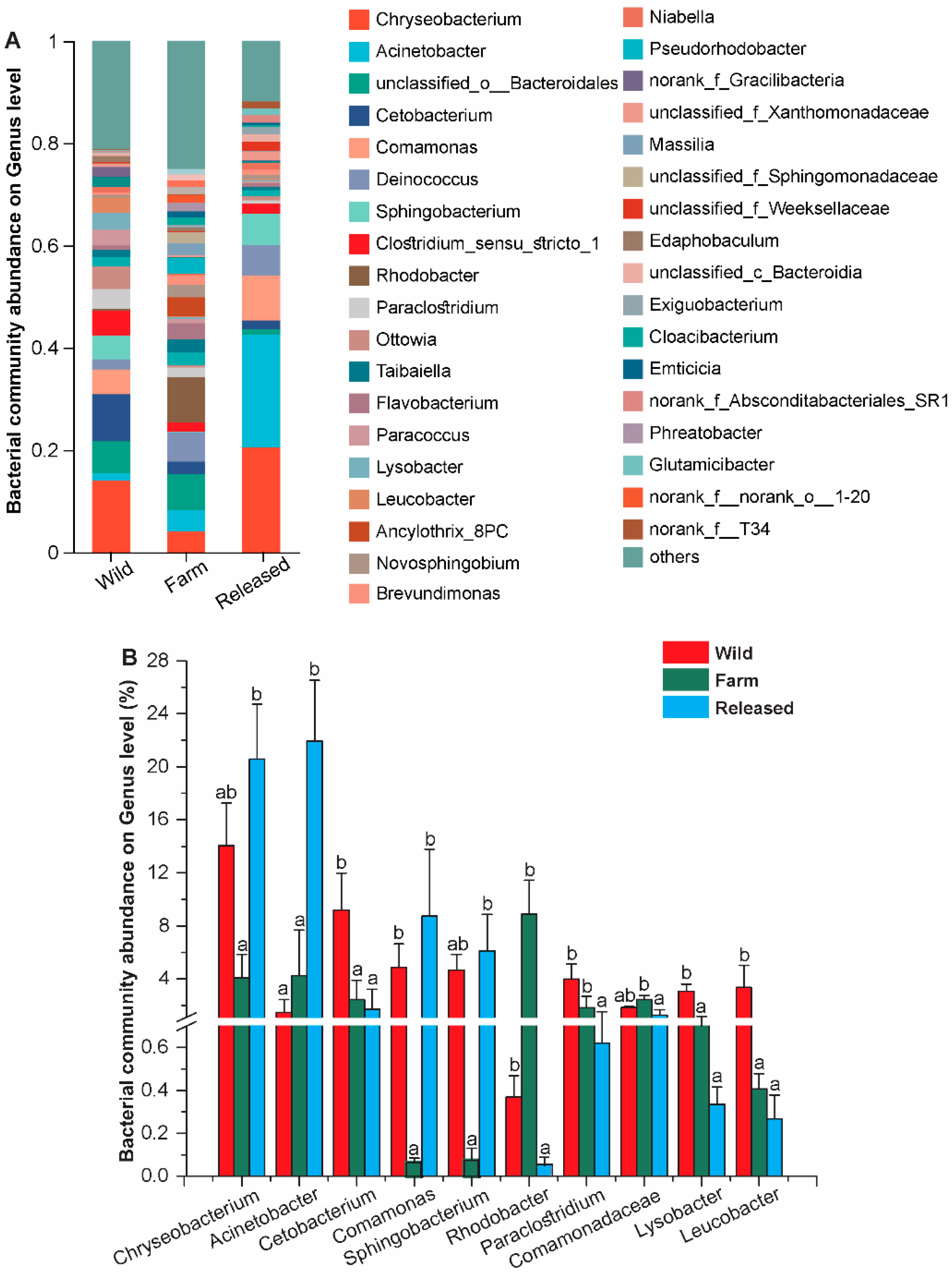
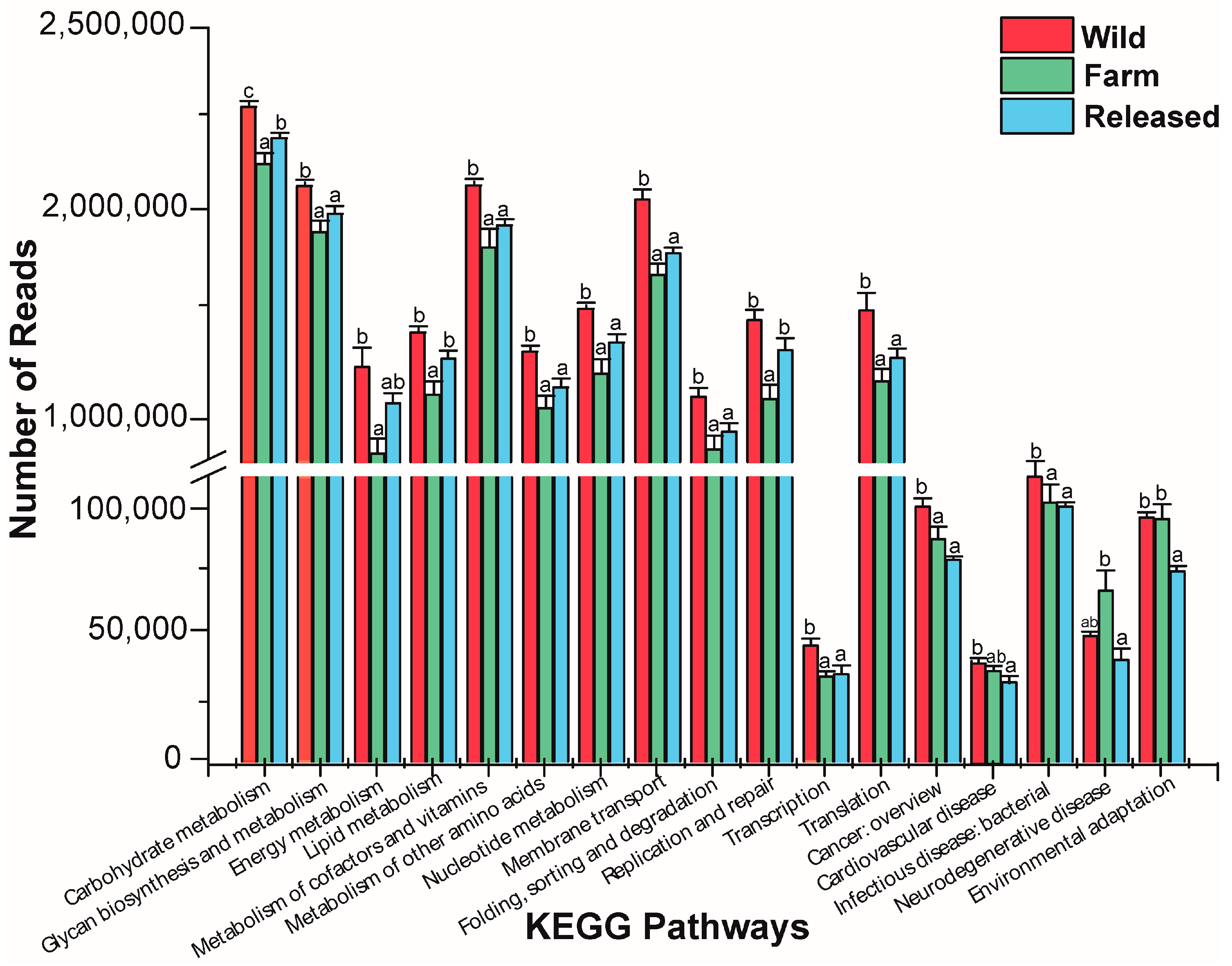


| Habitats | Richness and Diversity Estimators | ||||
|---|---|---|---|---|---|
| OTUs | ACE | Chao1 | Shannon | Simpson | |
| Wild | 247.50 ± 14.34 a | 337.84 ± 13.7 a | 353.87 ± 16.65 a | 3.7504 ± 0.16 a | 0.05 ± 0.00 ab |
| Farm | 408.80 ± 5.82 b | 539.06 ± 7.53 c | 559.51 ± 7.53 b | 4.2813 ± 0.08 b | 0.03 ± 0.00 a |
| Released | 282.60 ± 9.57 a | 379.93 ± 13.98 b | 393.80 ± 18.38 a | 3.5435 ± 0.19 a | 0.07 ± 0.01 b |
| ARGs | Wild | Farm | Released |
|---|---|---|---|
| macB | 6.65 ± 0.21 ab | 6.94 ± 0.17 b | 6.08 ± 0.28 a |
| tetA(58) | 5.01 ± 0.24 b | 3.74 ± 0.09 a | 4.65 ± 0.52 ab |
| bcrA | 3.14 ± 0.11 | 2.73 ± 0.04 | 3.03 ± 0.25 |
| msbA | 2.63 ± 0.12 | 2.48 ± 0.08 | 2.46 ± 0.19 |
| oleC | 2.29 ± 0.19 | 2.76 ± 0.09 | 2.14 ± 0.28 |
| mtrA | 2.01 ± 0.13 | 2.61 ± 0.18 | 2.32 ± 0.28 |
| novA | 2.06 ± 0.30 | 2.51 ± 0.14 | 2.31 ± 0.23 |
| TaeA | 1.60 ± 0.01 a | 1.91 ± 0.03 b | 1.82 ± 0.03 b |
| evgS | 1.31 ± 0.12 a | 2.38 ± 0.08 b | 1.62 ± 0.20 a |
| srpYmcr | 1.47 ± 0.05 | 1.43 ± 0.06 | 1.55 ± 0.08 |
| Others | 71.80 ± 0.91 | 70.47 ± 0.66 | 71.96 ± 1.08 |
| Class | Wild | Farm | Released |
|---|---|---|---|
| Macrolide antibiotic | 407,268 ± 10,238 b | 319,132 ± 11,347 a | 320,378 ± 6298 a |
| Aminocoumarin antibiotic | 209,156 ± 7231 b | 144,748 ± 8112 a | 173,676 ± 4845 ab |
| Tetracycline antibiotic | 217,246 ± 7573 b | 146,014 ± 6704 a | 180,496 ± 5449 ab |
| Peptide antibiotic | 145,318 ± 2303 c | 88,592 ± 397 a | 120,184 ± 2470 b |
| Nitroimidazole antibiotic | 92,770 ± 2720 b | 58,586 ± 2643 a | 68,382 ± 1379 a |
| Rifamycin antibiotic | 73,508 ± 2612 b | 51,152 ± 19,550 a | 53,920 ± 1805 a |
| Acridine dye | 32,406 ± 2425 b | 18,526 ± 9649 a | 27,502 ± 1128 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, I.; Bu, R.; Ali, Z.; Iqbal, M.S.; Shi, H.; Ding, L.; Hong, M. Metagenomics Analysis Reveals the Composition and Functional Differences of Fecal Microbiota in Wild, Farm, and Released Chinese Three-Keeled Pond Turtles (Mauremys reevesii). Animals 2024, 14, 1750. https://doi.org/10.3390/ani14121750
Khan I, Bu R, Ali Z, Iqbal MS, Shi H, Ding L, Hong M. Metagenomics Analysis Reveals the Composition and Functional Differences of Fecal Microbiota in Wild, Farm, and Released Chinese Three-Keeled Pond Turtles (Mauremys reevesii). Animals. 2024; 14(12):1750. https://doi.org/10.3390/ani14121750
Chicago/Turabian StyleKhan, Ijaz, Rongping Bu, Zeeshan Ali, Muhammad Shahid Iqbal, Haitao Shi, Li Ding, and Meiling Hong. 2024. "Metagenomics Analysis Reveals the Composition and Functional Differences of Fecal Microbiota in Wild, Farm, and Released Chinese Three-Keeled Pond Turtles (Mauremys reevesii)" Animals 14, no. 12: 1750. https://doi.org/10.3390/ani14121750
APA StyleKhan, I., Bu, R., Ali, Z., Iqbal, M. S., Shi, H., Ding, L., & Hong, M. (2024). Metagenomics Analysis Reveals the Composition and Functional Differences of Fecal Microbiota in Wild, Farm, and Released Chinese Three-Keeled Pond Turtles (Mauremys reevesii). Animals, 14(12), 1750. https://doi.org/10.3390/ani14121750







