Relationship between Rumen Microbial Differences and Phenotype Traits among Hu Sheep and Crossbred Offspring Sheep
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Analysis of Body Size Indices
2.3. Analysis of Serum Indicators
2.4. Analysis of Rumen Fermentation Parameters and Enzyme Activities
2.5. DNA Extraction and Analysis of Bacterial Community in Rumen
2.6. High–Throughput Sequencing Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Effect of Crossbreeding on Body Size Indices, Serum Indices, Rumen Enzyme Activity, and Fermentation in Progeny
3.1.1. Effects of Crossbred Offspring on Body Size Indices in Sheep
3.1.2. The Effect of Crossbred Offspring on the Serum Biochemical Indices
3.1.3. The Effect of Crossbred Offspring on the Serum Immune and Antioxidant Indices
3.1.4. The Effect of Crossbred Offspring on the Rumen Digestive Enzyme Activities
3.1.5. The Effect of Crossbred Offspring on the Rumen Fermentation Parameters
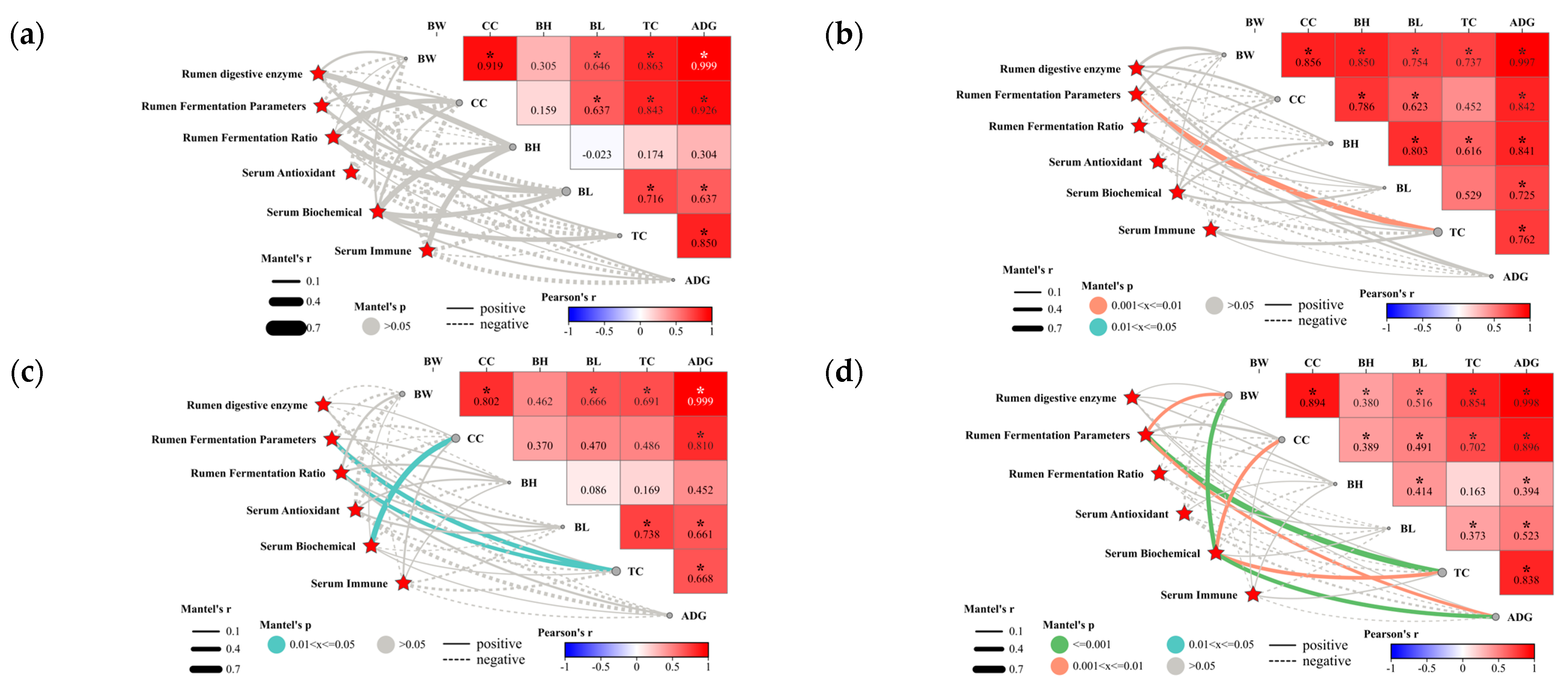
3.1.6. Relationship between Physiological Metabolic Indices and Economic Traits in Lambs
3.2. Effect of Hybridization on the Composition and Function of the Rumen Microbial Community of Progeny
3.2.1. Sequencing of Rumen Microbiota
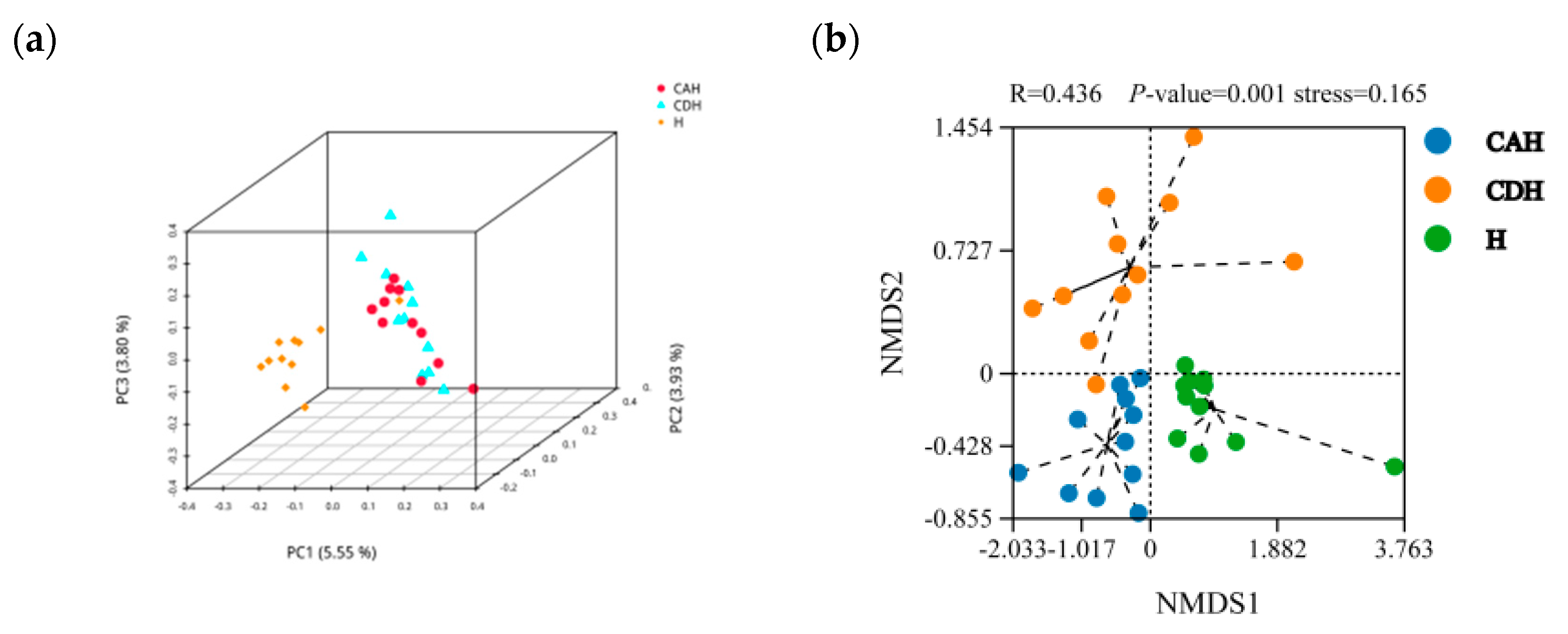
3.2.2. Diversity Analysis
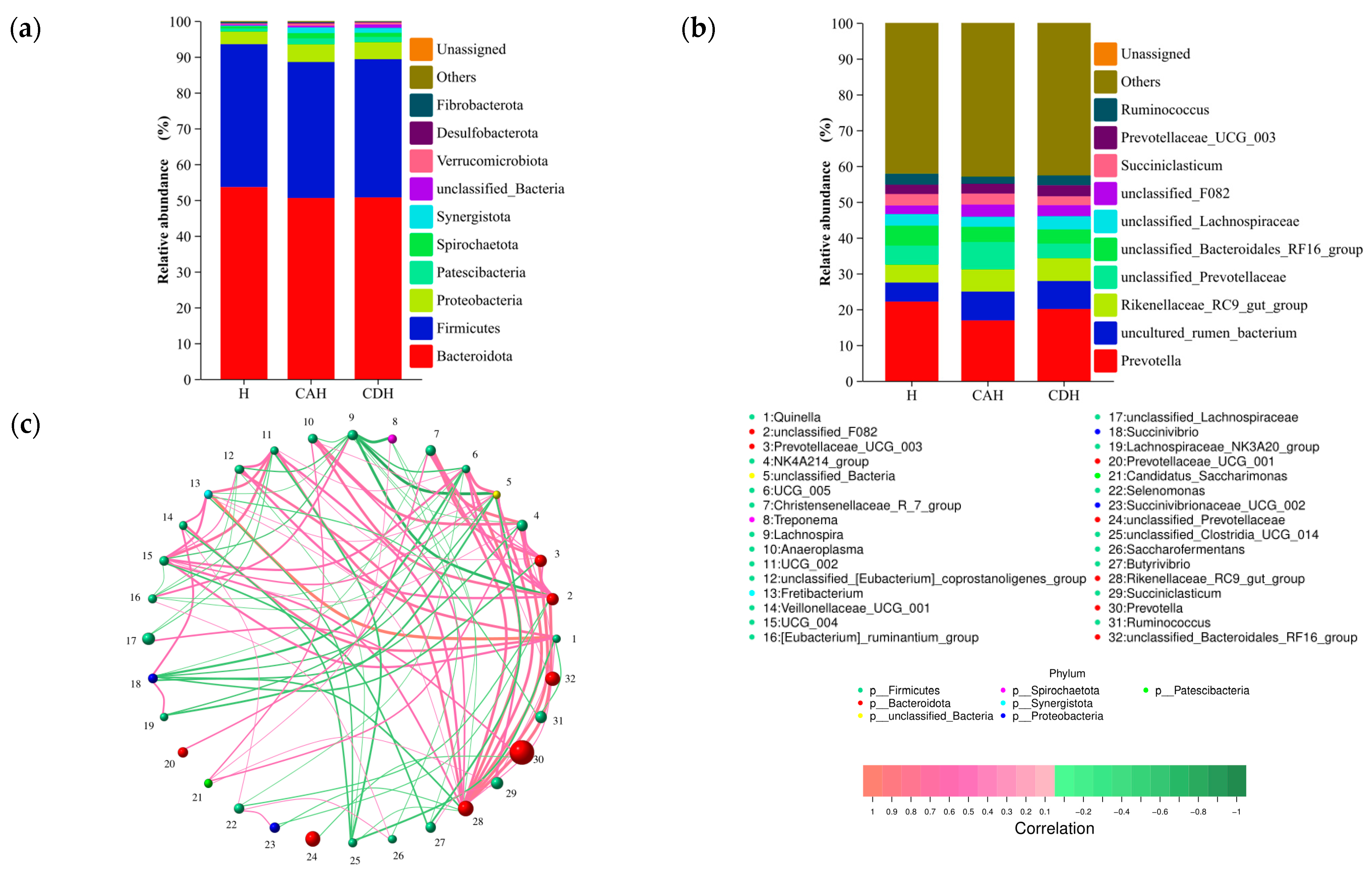
3.2.3. Composition of the Rumen Microbiota
3.2.4. Microbial Composition Drives Rumen Homeostasis
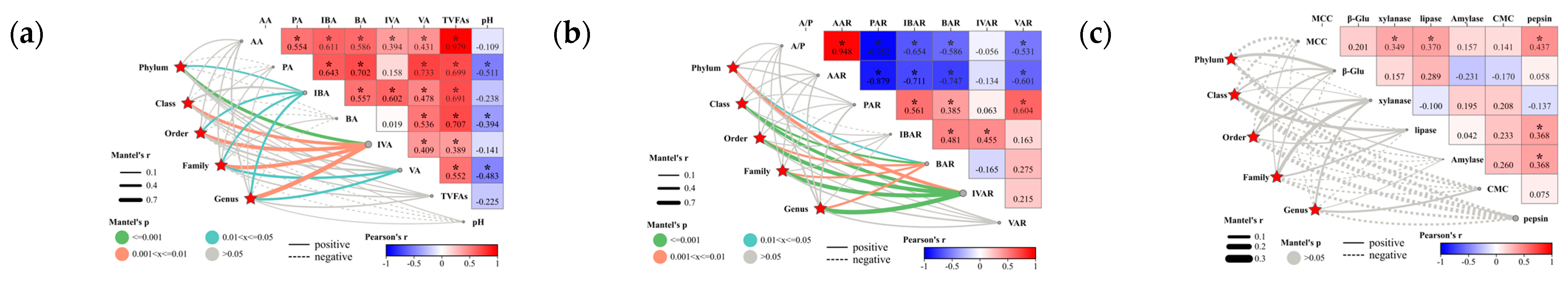
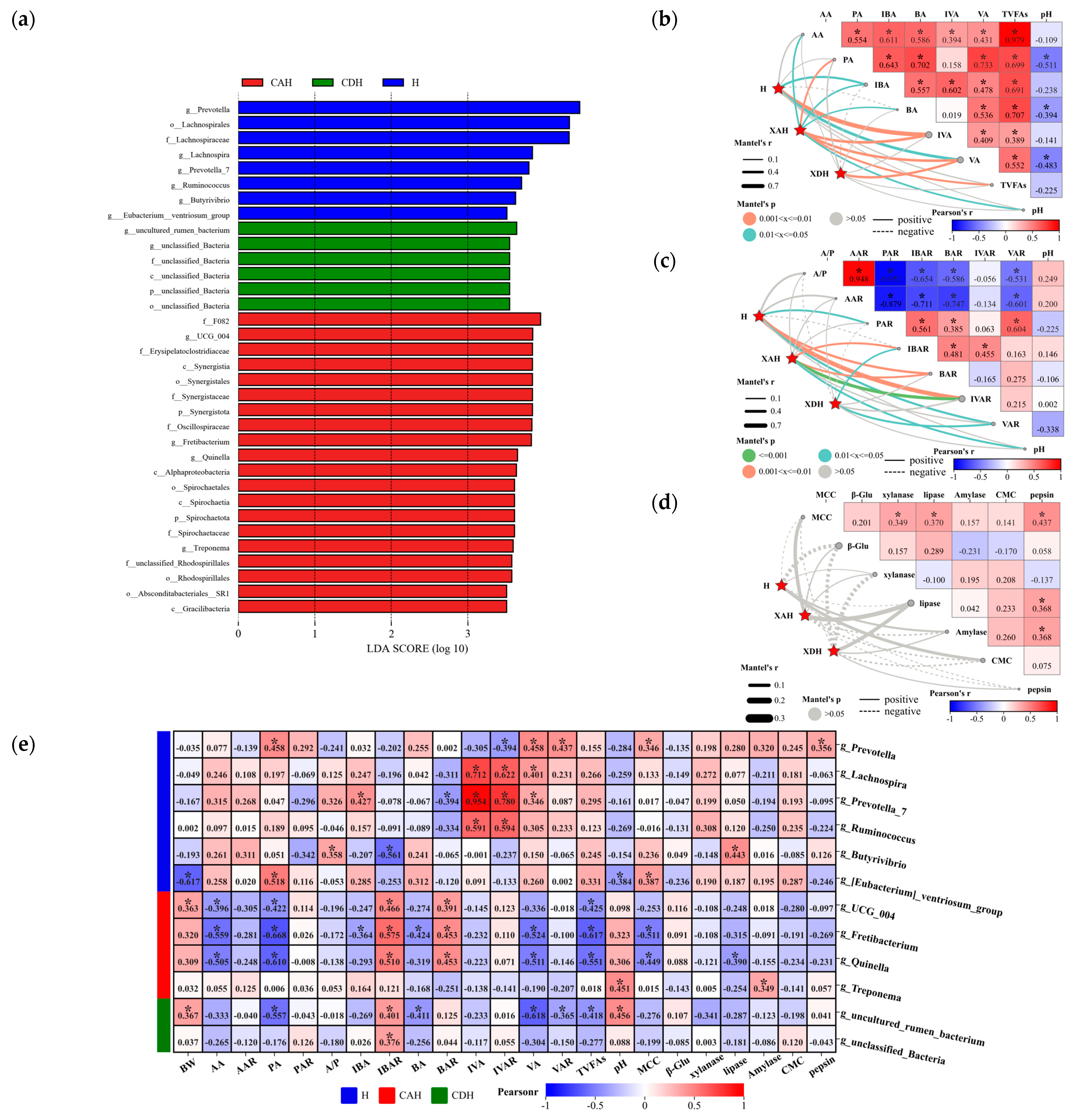
3.2.5. Relationship between Biomarkers and Sheep Rumen Parameters
3.2.6. Functional Prediction of Rumen Microbial in H, CAH and CDH
4. Discussion
4.1. Effect of Crossbreeding on Body Size Indices, Serum Indices, Rumen Enzyme Activity, and Fermentation in Progeny
4.2. Effect of Hybridization on the Composition and Function of the Rumen Microbial Community of Progeny
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Liu, G.J.; Xu, Y.X.; Lv, F.H.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole–genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef] [PubMed]
- Akinmoladun, O.F.; Muchenje, V.; Fon, F.N.; Mpendulo, C.T. Small Ruminants: Farmers’ hope in a world threatened by water scarcity. Animals 2019, 9, 456. [Google Scholar] [CrossRef]
- Kalds, P.; Zhou, S.; Cai, B.; Liu, J.; Wang, Y.; Petersen, B.; Sonstegard, T.; Wang, X.; Chen, Y. Sheep and goat genome engineering: From random transgenesis to the CRISPR Era. Front. Genet. 2019, 10, 750. [Google Scholar] [CrossRef] [PubMed]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Bickhart, D.M.; Weimer, P.J. Symposium review: Host-rumen microbe interactions may be leveraged to improve the productivity of dairy cows. J. Dairy Sci. 2018, 101, 7680–7689. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Gordon, G.L.R.; Phillips, M.W. The role of anaerobic gut fungi in ruminants. Nutr. Res. Rev. 1998, 11, 133–168. [Google Scholar] [CrossRef] [PubMed]
- van der Hee, B.; Wells, J.M. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, Y.; Zhang, X.; Li, C.; Yuan, L.; Zhang, D.; Zhao, Y.; Li, X.; Cheng, J.; Lin, C.; et al. Heritability and recursive influence of host genetics on the rumen microbiota drive body weight variance in male Hu sheep lambs. Microbiome 2023, 11, 197. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Fei, W.; Ye, Y.; Zhao, M.; Zheng, C. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit. Rev. Food Sci. Nutr. 2022, 62, 1–12. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Wang, X.; Li, F.; Zhang, D.; Li, X.; Zhao, Y.; Zhao, L.; Xu, D.; Cheng, J.; et al. Association between rumen microbiota and marbling grade in Hu sheep. Front. Microbiol. 2022, 13, 978263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.; Li, F.; Li, C.; Li, G.; Zhang, D.; Song, Q.; Li, X.; Zhao, Y.; Wang, W. Characterization of the rumen microbiota and its relationship with residual feed intake in sheep. Animal 2021, 15, 100161. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, M.A.P.; Lima, A.C.S.; Araújo, J.C.; Guimarães, C.M.C.; Joele, M.R.S.P.; Borges, I.; Daher, L.C.C.; e Silva, A.G.M. Tissue composition and allometric growth of carcass of lambs Santa Inês and crossbreed with breed Dorper. Trop. Anim. Health Prod. 2019, 51, 1903–1908. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Yue, Y.; Li, J.; Yang, B.; Chen, B.; Liu, J.; Lu, Z. Transcriptomics and metabolomics reveal improved performance of Hu sheep on hybridization with Southdown sheep. Food Res. Int. 2023, 173 Pt 1, 113240. [Google Scholar] [CrossRef] [PubMed]
- Lunesu, M.F.; Battacone, G.; Mellino, M.R.; Carta, S.; Pulina, G.; Nudda, A. The heavy suckling lamb of Sarda dairy sheep and its crossbreed with Dorper rams: Performance, meat quality and consumer perceptions. Meat Sci. 2023, 204, 109234. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, S.; Li, Z.; Huang, J.; Liu, Y.; Liu, Y.; Wang, Q.; Li, X.; Yan, Y.; Li, M. Comparison between the gut microbiota in different gastrointestinal segments of large-tailed han and small-tailed han sheep breeds with high-throughput sequencing. Indian J. Microbiol. 2020, 60, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhang, X.; Xu, D.; Zhang, D.; Zhang, Y.; Song, Q.; Li, X.; Zhao, Y.; Zhao, L.; Li, W.; et al. Relationship between rumen microbial differences and traits among Hu sheep, Tan sheep, and Dorper sheep. J. Anim. Sci. 2022, 100, skac261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.; Li, F.; Li, C.; Zhang, D.; Li, X.; Zhao, Y.; Wang, W. Exploring the ruminal microbial community associated with fat deposition in lambs. Animals 2021, 11, 3584. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Ju, L.; Cheng, Q.; Jiang, Y.; Hou, Q.; Hu, Z.; Wang, Y.; Wang, Z. Comparison of growth performance and rumen metabolic pathways in sheep and goats under the same feeding pattern. Front. Vet. Sci. 2023, 10, 1013252. [Google Scholar] [CrossRef]
- Guo, X.; Sha, Y.; Lv, W.; Pu, X.; Liu, X.; Luo, Y.; Hu, J.; Wang, J.; Li, S.; Zhao, Z. Sex differences in rumen fermentation and microbiota of Tibetan goat. Microb. Cell Factories 2022, 21, 55. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Z.; Hu, R.; Wang, X.; Li, F.; Zhang, X.; Zou, H.; Peng, Q.; Xue, B.; Wang, L. Comparative study of the bacterial communities throughout the gastrointestinal tract in two beef cattle breeds. Appl. Microbiol. Biotechnol. 2021, 105, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, H.; Hu, L.; Zhang, G.; Lu, H.; Luo, H.; Zhao, S.; Zhu, H.; Wang, Y. Characterization of the microbial communities along the gastrointestinal tract in crossbred cattle. Animals 2022, 12, 825. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Cheng, B.; Liu, Y.; Li, M.; Zhao, G. Effects of red cabbage extract rich in anthocyanins on rumen fermentation, rumen bacterial community, nutrient digestion, and plasma indices in beef bulls. Animal 2022, 16, 100510. [Google Scholar] [CrossRef]
- Mu, Y.; Qi, W.; Zhang, T.; Zhang, J.; Mao, S. Multi-omics analysis revealed coordinated responses of rumen microbiome and epithelium to high-grain-induced subacute rumen acidosis in lactating dairy cows. mSystems 2022, 7, e0149021. [Google Scholar] [CrossRef]
- Zheng, Y.; He, T.; Xie, T.; Wang, J.; Yang, Z.; Sun, X.; Wang, W.; Li, S. Hydroxy-selenomethionine supplementation promotes the in vitro rumen fermentation of dairy cows by altering the relative abundance of rumen microorganisms. J. Appl. Microbiol. 2022, 132, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Wanapat, M.; Hou, F. Rumen bacteria influence milk protein yield of yak grazing on the Qinghai–Tibet plateau. Anim. Biosci. 2021, 34, 1466–1478. [Google Scholar] [CrossRef]
- Osadchiy, V.; Martin, C.R.; Mayer, E.A. The gut-brain axis and the microbiome: Mechanisms and clinical implications. Clin. Gastroenterol. Hepatol. 2019, 17, 322–332. [Google Scholar] [CrossRef]
- Li, Z.; Wright, A.G.; Si, H.; Wang, X.; Qian, W.; Zhang, Z.; Li, G. Changes in the rumen microbiome and metabolites reveal the effect of host genetics on hybrid crosses. Environ. Microbiol. Rep. 2016, 8, 1016–1023. [Google Scholar] [CrossRef]
- Furman, O.; Shenhav, L.; Sasson, G.; Kokou, F.; Honig, H.; Jacoby, S.; Hertz, T.; Cordero, O.X.; Halperin, E.; Mizrahi, I. Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat. Commun. 2020, 11, 1904. [Google Scholar] [CrossRef]
- Li, L.-P.; Peng, K.-L.; Xue, M.-Y.; Zhu, S.-L.; Liu, J.-X.; Sun, H.-Z. An age effect of rumen microbiome in dairy buffaloes revealed by metagenomics. Microorganisms 2022, 10, 1491. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, Y.; Yu, Z.; Xu, Q.; Zheng, N.; Zhao, S.; Huang, G.; Wang, J. Ruminal microbiota–host interaction and its effect on nutrient metabolism. Anim. Nutr. 2021, 7, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef]
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Qin, X.; Chen, L.; Chen, Z.; Hao, R.; Zhang, S.; Yang, S.; Wang, L.; Cui, Y.; Li, Y.; et al. Serum biochemical parameters, rumen fermentation, and rumen bacterial communities are partly driven by the breed and sex of cattle when fed high-grain diet. Microorganisms 2022, 10, 323. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yang, P.; Li, M.; Fang, W.; Yue, X.; Nanaei, H.A.; Gan, S.; Du, D.; Cai, Y.; Dai, X.; et al. A Hu sheep genome with the first ovine Y chromosome reveal introgression history after sheep domestication. Sci. China Life Sci. 2021, 64, 1116–1130. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, X.; Li, F.; Liu, T.; Hu, Z.; Gao, N.; Yuan, L.; Li, X.; Zhao, Y.; Zhao, L.; et al. Whole-genome resequencing identified candidate genes associated with the number of ribs in Hu sheep. Genomics 2021, 113, 2077–2084. [Google Scholar] [CrossRef] [PubMed]
- Wan, Z.; Yang, H.; Cai, Y.; Ma, J.; Cheng, P.; Wang, Z.; Wang, F.; Zhang, Y. Comparative Transcriptomic Analysis of Hu Sheep Pituitary Gland Prolificacy at the Follicular and Luteal Phases. Genes 2022, 13, 440. [Google Scholar] [CrossRef]
- Wang, H.B.; Zhan, J.S.; Gu, Z.Y.; Chen, X.F.; Pan, Y.; Jia, H.B.; Zhong, X.J.; Li, K.R.; Zhao, S.G.; Huo, J.H. Comparative study on meat quality characteristics of three-way hybrid sheep Charolais×Duper×Hu and Charolais×Australian White×Hu and Hu sheep. Acta Vet. Zootech. Sin. 2024, 55, 110–119. [Google Scholar]
- Liu, X.; Sha, Y.; Dingkao, R.; Zhang, W.; Lv, W.; Wei, H.; Shi, H.; Hu, J.; Wang, J.; Li, S.; et al. Interactions between rumen microbes, VFAs, and host genes regulate nutrient absorption and epithelial barrier function during cold season nutritional stress in tibetan sheep. Front. Microbiol. 2020, 11, 593062. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PIC-RUSt2: An improved and customizable approach for metagenome inference. BioRxiv 2019, 15, 672295. [Google Scholar] [CrossRef]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M.; et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef] [PubMed]
- E Pryce, J.; Hayes, B.J.; Bolormaa, S.; E Goddard, M. Polymorphic regions affecting human height also control stature in cattle. Genetics 2011, 187, 981–984. [Google Scholar] [CrossRef]
- Talebi, R.; Ghaffari, M.R.; Fabre, S.; Mardi, M.; Alamouti, M.K. Comparison of the growth performance between pure Moghani sheep and crosses with Texel or Booroola sheep carrying major genes contributing to muscularity and prolificacy. Anim. Biotechnol. 2023, 34, 3495–3506. [Google Scholar] [CrossRef]
- Gootwine, E. Invited review: Opportunities for genetic improvement toward higher prolificacy in sheep. Small Rumin. Res. 2020, 186, 106090. [Google Scholar] [CrossRef]
- Slavin, J.L. Dietary fiber and body weight. Nutrition 2005, 21, 411–418. [Google Scholar] [CrossRef]
- Sleeth, M.L.; Thompson, E.L.; Ford, H.E.; Zac-Varghese, S.E.K.; Frost, G. Free fatty acid receptor 2 and nutrient sensing: A proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr. Res. Rev. 2010, 23, 135–145. [Google Scholar] [CrossRef]
- Hernandez-Sanabria, E.; Guan, L.L.; Goonewardene, L.A.; Li, M.; Mujibi, D.F.; Stothard, P.; Moore, S.S.; Leon-Quintero, M.C. Correlation of particular bacterial PCR-denaturing gradient gel electrophoresis patterns with bovine ruminal fermentation parameters and feed efficiency traits. Appl. Environ. Microbiol. 2010, 76, 6338–6350. [Google Scholar] [CrossRef]
- Sadan, T.; Aravindakshan, T.V.; Radhika, G.; Anand, L.F.; Ally, K. Metagenomic analysis exploring taxonomic diversity of rumen microbial communities in vechur and crossbred cattle of Kerala state, India. J. Appl. Genet. 2020, 61, 287–297. [Google Scholar] [CrossRef]
- Cholewińska, P.; Wołoszyńska, M.; Michalak, M.; Czyż, K.; Rant, W.; Janczak, M. Evaluation of changes in the levels of Firmicutes and Bacteroidetes phyla of sheep feces depending on the breed. Animals 2020, 10, 1901. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Chen, Y.; Ma, Z.; Zhang, X.; Shi, D.; Khan, J.A.; Liu, H. Gut microbiota-derived short chain fatty acids are potential mediators in gut inflammation. Anim. Nutr. 2021, 8, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Argaw-Denboba, A.; Schmidt, T.S.B.; Di Giacomo, M.; Ranjan, B.; Devendran, S.; Mastrorilli, E.; Lloyd, C.T.; Pugliese, D.; Paribeni, V.; Dabin, J.; et al. Paternal microbiome perturbations impact offspring fitness. Nature 2024, 629(8012), 652–659. [Google Scholar] [CrossRef]
- Huang, Y.; Lv, H.; Song, Y.; Sun, C.; Zhang, Z.; Chen, S. Community composition of cecal microbiota in commercial yellow broilers with high and low feed efficiencies. Poult. Sci. 2021, 100, 100996. [Google Scholar] [CrossRef]
- Szeligowska, N.; Cholewińska, P.; Czyż, K.; Wojnarowski, K.; Janczak, M. Inter and intraspecies comparison of the level of selected bacterial phyla in in cattle and sheep based on feces. BMC Vet. Res. 2021, 17, 224. [Google Scholar] [CrossRef] [PubMed]
- Konopka, A. What is microbial community ecology? ISME J. 2009, 3, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Cersosimo, L.M.; Bainbridge, M.L.; Kraft, J.; Wright, A.-D.G. Influence of periparturient and postpartum diets on rumen methanogen communities in three breeds of primiparous dairy cows. BMC Microbiol. 2016, 16, 78. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, J.; Yang, Z.; Liu, A. Dominance of the Enterocytozoon bieneusi genotype BEB6 in red deer (Cervus elaphus) and Siberian roe deer (Capreolus pygargus) in China and a brief literature review. Parasite 2017, 24, 54. [Google Scholar] [CrossRef] [PubMed]
- Gürelli, G.; Canbulat, S.; Aldayarov, N.; Dehority, B.A. Rumen ciliate protozoa of domestic sheep (Ovis aries) and goat (Capra aegagrus hircus) in Kyrgyzstan. FEMS Microbiol. Lett. 2016, 363, fnw028. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Gao, H.; Qin, W.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Wang, D.; Zhang, T. Marked seasonal variation in structure and function of gut microbiota in forest and alpine musk deer. Front. Microbiol. 2021, 12, 699797. [Google Scholar] [CrossRef] [PubMed]
- Jewell, K.A.; McCormick, C.A.; Odt, C.L.; Weimer, P.J.; Suen, G. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl. Environ. Microbiol. 2015, 81, 4697–4710. [Google Scholar] [CrossRef] [PubMed]
- Bauman, D.E.; Harvatine, K.J.; Lock, A.L. Nutrigenomics, rumen-derived bioactive fatty acids, and the regulation of milk fat synthesis. Annu. Rev. Nutr. 2011, 31, 299–319. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- Gharechahi, J.; Vahidi, M.F.; Bahram, M.; Han, J.-L.; Ding, X.-Z.; Salekdeh, G.H. Metagenomic analysis reveals a dynamic microbiome with diversified adaptive functions to utilize high lignocellulosic forages in the cattle rumen. ISME J. 2021, 15, 1108–1120. [Google Scholar] [CrossRef]
- Leong, L.E.X.; Denman, S.E.; Kang, S.; Mondot, S.; Hugenholtz, P.; McSweeney, C.S. Identification of the mechanism for dehalorespiration of monofluoroacetate in the phylum Synergistota. Anim. Biosci. 2024, 37, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.K.; Webb, R.I.; Sly, L.I.; Denman, S.E.; McSweeney, C.S. Isolation and survey of novel fluoroacetate-degrading bacteria belonging to the phylum Synergistetes. FEMS Microbiol. Ecol. 2012, 80, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Leong, L.E.X.; Denman, S.E.; Hugenholtz, P.; McSweeney, C.S. Amino acid and peptide utilization profiles of the fluoroacetate-degrading bacterium Synergistetes strain MFA1 under varying conditions. Microb. Ecol. 2016, 71, 494–504. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Sha, Y.; Qi, S.; Zhang, X.; Wang, H.; Wang, Z.; Zhao, S.; Jiao, T. Effects of isochlorogenic acid on ewes rumen fermentation, microbial diversity and ewes immunity of different physiological stages. Animals 2024, 14, 715. [Google Scholar] [CrossRef]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A key player in ruminal metabolism. Microorganisms 2022, 11, 1. [Google Scholar] [CrossRef]
- Aguilar-Marin, S.B.; Betancur-Murillo, C.L.; Isaza, G.A.; Mesa, H.; Jovel, J. Lower methane emissions were associated with higher abundance of ruminal Prevotella in a cohort of Colombian buffalos. BMC Microbiol. 2020, 20, 364. [Google Scholar] [CrossRef]
- Han, G.; Vaishnava, S. Microbial underdogs: Exploring the significance of low-abundance commensals in host-microbe interactions. Exp. Mol. Med. 2023, 55, 2498–2507. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Ocean plankton. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology: Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Amabebe, E.; Robert, F.O.; Agbalalah, T.; Orubu, E.S.F. Microbial dysbiosis-induced obesity: Role of gut microbiota in homoeostasis of energy metabolism. Br. J. Nutr. 2020, 123, 1127–1137. [Google Scholar] [CrossRef]
- Kumar, S.; Altermann, E.; Leahy, S.C.; Jauregui, R.; Jonker, A.; Henderson, G.; Kittelmann, S.; Attwood, G.T.; Kamke, J.; Waters, S.M.; et al. Genomic insights into the physiology of Quinella, an iconic uncultured rumen bacterium. Nat. Commun. 2022, 13, 6240. [Google Scholar] [CrossRef]
- Yin, Y.; Sichler, A.; Ecker, J.; Laschinger, M.; Liebisch, G.; Höring, M.; Basic, M.; Bleich, A.; Zhang, X.-J.; Kübelsbeck, L.; et al. Gut microbiota promote liver regeneration through hepatic membrane phospholipid biosynthesis. J. Hepatol. 2023, 78, 820–835. [Google Scholar] [CrossRef]
- Wanapat, M.; Anantasook, N.; Rowlinson, P.; Pilajun, R.; Gunun, P. Effect of carbohydrate sources and levels of cotton seed meal in concentrate on feed intake, nutrient digestibility, rumen fermentation and microbial protein synthesis in young dairy bulls. Asian-Australas. J. Anim. Sci. 2013, 26, 529–536. [Google Scholar] [CrossRef]
- Wang, D.; Chen, L.; Tang, G.; Yu, J.; Chen, J.; Li, Z.; Cao, Y.; Lei, X.; Deng, L.; Wu, S.; et al. Multi-omics revealed the long-term effect of ruminal keystone bacteria and the microbial metabolome on lactation performance in adult dairy goats. Microbiome 2023, 11, 215. [Google Scholar] [CrossRef]
| Items | Body Weight, kg | Chest Circumference, cm | Body Height, cm | Body Length, cm | Tube Circumference, cm | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 d | 135 d | 0 d | 135 d | 0 d | 135 d | 0 d | 135 d | 0 d | 135 d | ||
| H | 3.83 ±0.19 | 29.80 ±1.30 b | 34.31 ±0.61 | 65.83 ±1.03 | 39.54 ±0.60 | 64.45 ±0.72 | 30.46 ±0.58 | 62.74· ±1.01 | 5.62· ±0.12 | 7.79 ±0.11 | |
| CAH | 4.30 ±0.31 | 35.66 ±1.06 a | 35.28 ±1.01 | 69.64 ±1.09 | 39.11 ±0.86 | 64.08 ±1.12 | 28.31· ±0.77 | 62.28· ±1.36 | 6.15 ±0.22 | 8.70 ±0.12 | |
| CDH | 3.91 ±0.22 | 33.67 ±0.82 a | 34.51 ±0.67 | 68.35 ±0.72 | 38.15 ±0.75 | 62.72 ±0.60 | 29.18· ±1.01 | 62.42· ±1.14 | 5.86 ±0.16 | 8.42 ±0.09 | |
| Group | H | 16.81 ± 2.91 | 50.07 ± 3.49 | 51.99 ± 2.76 | 46.60 ± 3.57 | 6.70 ± 0.25 | |||||
| CAH | 19.98 ± 3.46 | 52.46 ± 3.82 | 51.60 ± 2.81 | 45.30 ± 3.78 | 7.42 ± 0.30 | ||||||
| CDH | 18.79 ± 3.27 | 51.43 ± 3.72 | 50.43 ± 2.72 | 45.80 ± 3.70 | 7.14 ± 0.29 | ||||||
| p Value | Time | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |||||
| Group | 0.003 | 0.083 | 0.242 | 0.463 | <0.001 | ||||||
| Time × Group | 0.002 | 0.106 | 0.061 | 0.685 | 0.216 | ||||||
| Items | H | CAH | CDH | p Value |
|---|---|---|---|---|
| GOT,U/L | 12.17 ± 1.84 | 8.35 ± 1.34 | 8.93 ± 1.07 | 0.150 |
| GPT, U/L | 6.08 ± 0.65 | 3.97 ± 0.6 | 5.86 ± 0.67 | 0.051 |
| LDH, U/L | 784.76 ± 40.18 b | 1107.62 ± 33.36 a | 1045.36 ± 34.95 a | <0.001 |
| UN, mmol/L | 3.32 ± 1.04 b | 10.15 ± 2.12 a | 7.08 ± 0.28 ab | 0.005 |
| Albumin, g/L | 18.66 ± 1.59 | 16.46 ± 1.27 | 15.37 ± 0.71 | 0.180 |
| BCA protein, μg/μL | 45.86 ± 2.18 | 40.56 ± 2.12 | 42.58 ± 2.05 | 0.219 |
| GLU, mmol/L | 2.06 ± 0.08 | 2.50 ± 0.26 | 2.29 ± 0.26 | 0.371 |
| HDL–C, mmol/L | 2.93 ± 0.36 | 2.48 ± 0.35 | 2.33 ± 0.31 | 0.438 |
| LDL–C, mmol/L | 2.09 ± 0.15 | 2.51 ± 0.23 | 2.19 ± 0.24 | 0.335 |
| CRE, μmol/L | 55.29 ± 2.36 b | 74.97 ± 3.18 a | 76.33 ± 2.89 a | <0.001 |
| TG, mmol/L | 0.38 ± 0.05 | 0.43 ± 0.05 | 0.35 ± 0.04 | 0.460 |
| T–CHO, mmol/L | 3.28 ± 0.23 | 3.45 ± 0.25 | 4.52 ± 0.82 | 0.198 |
| CK, U/mL | 1.97 ± 0.07 | 1.67 ± 0.10 | 1.63 ± 0.21 | 0.188 |
| PFK, U/mL | 33.56 ± 4.32 | 27.26 ± 3.41 | 21.67 ± 2.07 | 0.060 |
| Items | H | CAH | CDH | p Value |
|---|---|---|---|---|
| IgA, μg/mL | 35.08 ± 1.44 | 35.27 ± 1.49 | 35.87 ± 1.93 | 0.939 |
| IgM, μg/mL | 18.16 ± 0.63 | 18.63 ± 0.68 | 18.35 ± 1.37 | 0.939 |
| IgG, μg/mL | 1334.46 ± 69.45 | 1257.74 ± 59.32 | 1348.03 ± 68.20 | 0.584 |
| MDA, nmol/mL | 8.29 ± 0.39 | 7.84 ± 0.27 | 8.49 ± 0.31 | 0.366 |
| T–AOC, U/mL | 3.16 ± 0.07 | 3.12 ± 0.23 | 3.15 ± 0.24 | 0.987 |
| CAT, ng/L | 15.06 ± 0.38 | 15.21 ± 0.32 | 15.66 ± 0.33 | 0.452 |
| GSH–Px, ng/L | 11.64 ± 0.37 | 11.67 ± 0.22 | 11.73 ± 0.36 | 0.977 |
| T–SOD, pg/mL | 7.56 ± 0.31 | 7.57 ± 0.27 | 8.15 ± 0.31 | 0.297 |
| Items | H | CAH | CDH | p Value |
|---|---|---|---|---|
| MCC, pg/mL | 112.00 ± 2.57 a | 94.25 ± 3.32 b | 98.00 ± 3.39 b | 0.001 |
| β–glucosidase, ng/L | 960.73 ± 27.85 | 996.05 ± 40.21 | 962.21 ± 60.23 | 0.831 |
| xylanase, pg/mL | 167.96 ± 3.43 a | 150.64 ± 5.85 b | 174.26 ± 5.06 a | 0.005 |
| lipase, ng/mL | 254.31 ± 12.92 a | 221.75 ± 12.35 b | 193.00 ± 7.48 b | 0.002 |
| amylase, umol/L | 150.47 ± 4.41 | 146.49 ± 4.37 | 152.05 ± 3.84 | 0.631 |
| CMC, pg/mL | 268.32 ± 10.14 a | 223.83 ± 11.56 b | 267.32 ± 13.24 a | 0.019 |
| pepsin, ug/L | 17.85 ± 0.84 | 16.86 ± 0.34 | 16.14 ± 0.67 | 0.190 |
| Items | H | CAH | CDH | p Value |
|---|---|---|---|---|
| pH | 6.97 ± 0.05 b | 7.41 ± 0.08 a | 7.25 ± 0.05 a | <0.001 |
| molar concentration (mmol/L) | ||||
| acetic acid | 64.81 ± 5.56 a | 44.81 ± 6.08 b | 38.38 ± 2.51 b | 0.002 |
| propionic acid | 18.18 ± 0.54 a | 11.45 ± 1.03 b | 13.29 ± 0.45 b | <0.001 |
| isobutyric acid | 1.34 ± 0.05 a | 1.08 ± 0.08 b | 1.25 ± 0.05 ab | 0.021 |
| butyric acid | 13.88 ± 0.62 a | 9.80 ± 0.82 b | 10.21 ± 0.72 b | 0.001 |
| isovaleric acid | 1.77 ± 0.47 | 1.18 ± 0.11 | 1.47 ± 0.07 | 0.351 |
| valeric acid | 1.08 ± 0.11 a | 0.58 ± 0.05 b | 0.67 ± 0.03 b | <0.001 |
| TVFAs | 101.06 ± 5.46 a | 68.91 ± 7.57 b | 65.28 ± 3.39 b | <0.001 |
| acetic acid/propionic acid | 3.64 ± 0.38 | 3.93 ± 0.37 | 2.87 ± 0.13 | 0.067 |
| molar proportion | ||||
| acetic acid | 63.11 ± 2.01 | 63.66 ± 1.92 | 58.38 ± 0.95 | 0.069 |
| propionic acid | 18.61 ± 1.27 | 17.18 ± 1.06 | 20.65 ± 0.75 | 0.080 |
| isobutyric acid | 1.35 ± 0.06 c | 1.64 ± 0.10 b | 1.94 ± 0.07 a | <0.001 |
| butyric acid | 14.11 ± 0.91 | 14.87 ± 0.93 | 15.69 ± 0.81 | 0.459 |
| isovaleric acid | 1.71 ± 0.39 | 1.78 ± 0.10 | 2.30 ± 0.13 | 0.193 |
| valeric acid | 1.11 ± 0.16 | 0.87 ± 0.04 | 1.04 ± 0.05 | 0.210 |
| Items | H | CAH | CDH | p Value | ||
|---|---|---|---|---|---|---|
| H:CAH | H:CDH | CAH:CDH | ||||
| ACE index | 875.73 ± 44.20 b | 1104.71 ± 22.72 a | 1069.16 ± 50.03 a | <0.001 | 0.005 | 0.560 |
| Chao1 index | 873.60 ± 44.06 b | 1101.61 ± 22.80 a | 1066.98 ± 49.82 a | <0.001 | 0.005 | 0.610 |
| Simpson index | 9.90 × 10−1 ± 0.00 | 9.93 × 10−1 ± 0.00 | 9.91 × 10−1 ± 0.00 | 0.300 | 0.056 | 0.480 |
| Shannon index | 8.16 ± 0.17 | 8.44 ± 0.11 | 8.52 ± 0.15 | 0.920 | 0.130 | 0.120 |
| Items | H | CAH | CDH | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| Class 1 | Class 2 | Class 3 | H: CAH | H: CDH | CAH: CDH | |||
| ME | Amino acid metabolism | Cysteine and methionine metabolism | 9.92 × 10−3 | 9.74 × 10−3 | 9.81 × 10−3 | 0.035 | 0.289 | 1.101 |
| Alanine, aspartate and glutamate metabolism | 9.80 × 10−3 | 9.71 × 10−3 | 9.61 × 10−3 | 0.318 | 0.140 | 0.889 | ||
| Biosynthesis of other secondary metabolites | Biosynthesis of antibiotics | 5.80 × 10−2 | 5.79 × 10−2 | 5.79 × 10−2 | 0.448 | 0.805 | 0.946 | |
| Carbohydrate metabolism | Amino sugar and nucleotide sugar metabolism | 1.19 × 10−2 | 1.17 × 10−2 | 1.17 × 10−2 | 0.037 | 0.172 | 1.002 | |
| Glycolysis/Gluconeogenesis | 1.07 × 10−2 | 1.07 × 10−2 | 1.07 × 10−2 | 0.719 | 0.786 | 1.017 | ||
| Pyruvate metabolism | 9.59 × 10−3 | 9.79 × 10−3 | 9.73 × 10−3 | 0.022 | 0.204 | 0.947 | ||
| Energy metabolism | Oxidative phosphorylation | 1.02 × 10−2 | 1.02 × 10−2 | 1.02 × 10−2 | 0.751 | 0.827 | 0.990 | |
| Carbon fixation pathways in prokaryotes | 9.55 × 10−3 | 9.71 × 10−3 | 9.68 × 10−3 | 0.318 | 0.578 | 0.992 | ||
| Global and overview maps | Metabolic pathways | 1.75 × 10−1 | 1.74 × 10−1 | 1.74 × 10−1 | 0.299 | 0.446 | 0.967 | |
| Biosynthesis of secondary metabolites | 7.98 × 10−2 | 7.94 × 10−2 | 7.95 × 10−2 | 0.032 | 0.302 | 0.968 | ||
| Biosynthesis of amino acids | 4.06 × 10−2 | 4.03 × 10−2 | 4.02 × 10−2 | 0.461 | 0.444 | 0.979 | ||
| Microbial metabolism in diverse environments | 3.71 × 10−2 | 3.76 × 10−2 | 3.75 × 10−2 | 0.030 | 0.176 | 0.952 | ||
| Carbon metabolism | 2.63 × 10−2 | 2.65 × 10−2 | 2.64 × 10−2 | 0.033 | 0.550 | 0.982 | ||
| Nucleotide metabolism | Purine metabolism | 2.16 × 10−2 | 2.15 × 10−2 | 2.16 × 10−2 | 0.171 | 0.581 | 1.032 | |
| Pyrimidine metabolism | 1.87 × 10−2 | 1.86 × 10−2 | 1.85 × 10−2 | 0.476 | 0.460 | 1.014 | ||
| GIP | Translation | Ribosome | 2.59 × 10−2 | 2.57 × 10−2 | 2.59 × 10−2 | 0.332 | 1.000 | 0.957 |
| Aminoacyl–tRNA biosynthesis | 1.09 × 10−2 | 1.09 × 10−2 | 1.10 × 10−2 | 0.709 | 0.461 | 0.951 | ||
| EIP | Membrane transport | ABC transporters | 2.16 × 10−2 | 2.21 × 10−2 | 2.18 × 10−2 | 0.366 | 0.711 | 0.967 |
| Signal transduction | Two–component system | 1.71 × 10−2 | 1.72 × 10−2 | 1.72 × 10−2 | 0.642 | 0.697 | 0.997 | |
| CPr | Cellular community–prokaryotes | Quorum sensing | 1.13 × 10−2 | 1.17 × 10−2 | 1.15 × 10−2 | 0.040 | 0.176 | 0.945 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Zhan, J.; Jia, H.; Jiang, H.; Pan, Y.; Zhong, X.; Zhao, S.; Huo, J. Relationship between Rumen Microbial Differences and Phenotype Traits among Hu Sheep and Crossbred Offspring Sheep. Animals 2024, 14, 1509. https://doi.org/10.3390/ani14101509
Wang H, Zhan J, Jia H, Jiang H, Pan Y, Zhong X, Zhao S, Huo J. Relationship between Rumen Microbial Differences and Phenotype Traits among Hu Sheep and Crossbred Offspring Sheep. Animals. 2024; 14(10):1509. https://doi.org/10.3390/ani14101509
Chicago/Turabian StyleWang, Haibo, Jinshun Zhan, Haobin Jia, Haoyun Jiang, Yue Pan, Xiaojun Zhong, Shengguo Zhao, and Junhong Huo. 2024. "Relationship between Rumen Microbial Differences and Phenotype Traits among Hu Sheep and Crossbred Offspring Sheep" Animals 14, no. 10: 1509. https://doi.org/10.3390/ani14101509
APA StyleWang, H., Zhan, J., Jia, H., Jiang, H., Pan, Y., Zhong, X., Zhao, S., & Huo, J. (2024). Relationship between Rumen Microbial Differences and Phenotype Traits among Hu Sheep and Crossbred Offspring Sheep. Animals, 14(10), 1509. https://doi.org/10.3390/ani14101509







