Disturbances of Ruminal Microbiota and Liver Inflammation, Mediated by LPS and Histamine, in Dairy Cows Fed a High-Concentrate Diet
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Experimental Design
2.3. Sample Collection
2.4. LPS Measurement
2.5. Histamine Measurement
2.6. Histological Analysis
2.7. Radioimmunoassay
2.8. Quantitative Real-Time PCR
2.9. Western Blot Analysis
2.10. DNA Extraction, 16S rRNA Gene Amplification, Ion S5TM XL Sequencing, and Data Analysis
2.11. Statistical Analysis
3. Results
3.1. SARA Is Induced by a High-Concentrate Diet Combined with High Production of LPS and Histamine in Gut and Bloodstream
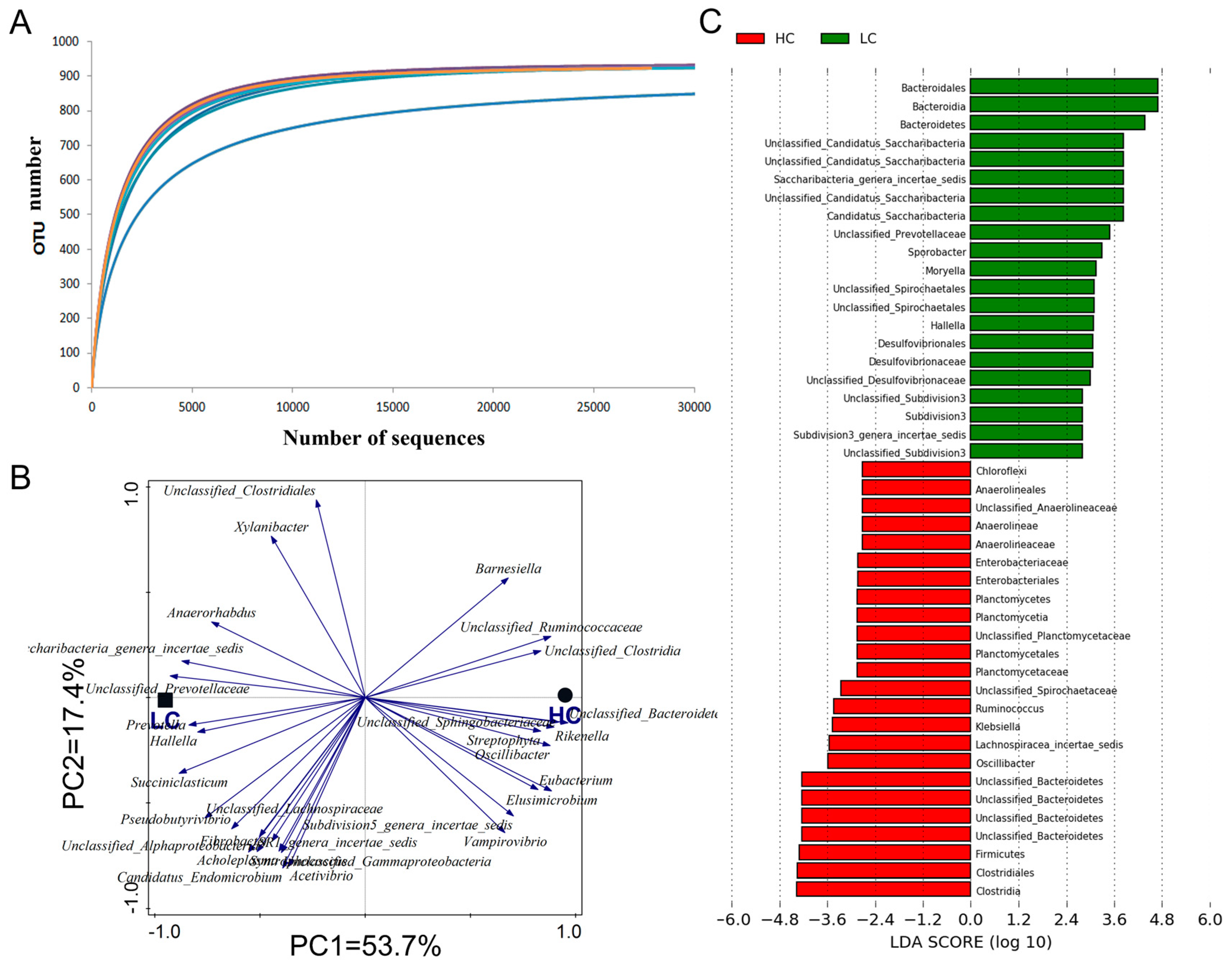
3.2. The Microbiota Composition Is Changed in the Rumen under SARA
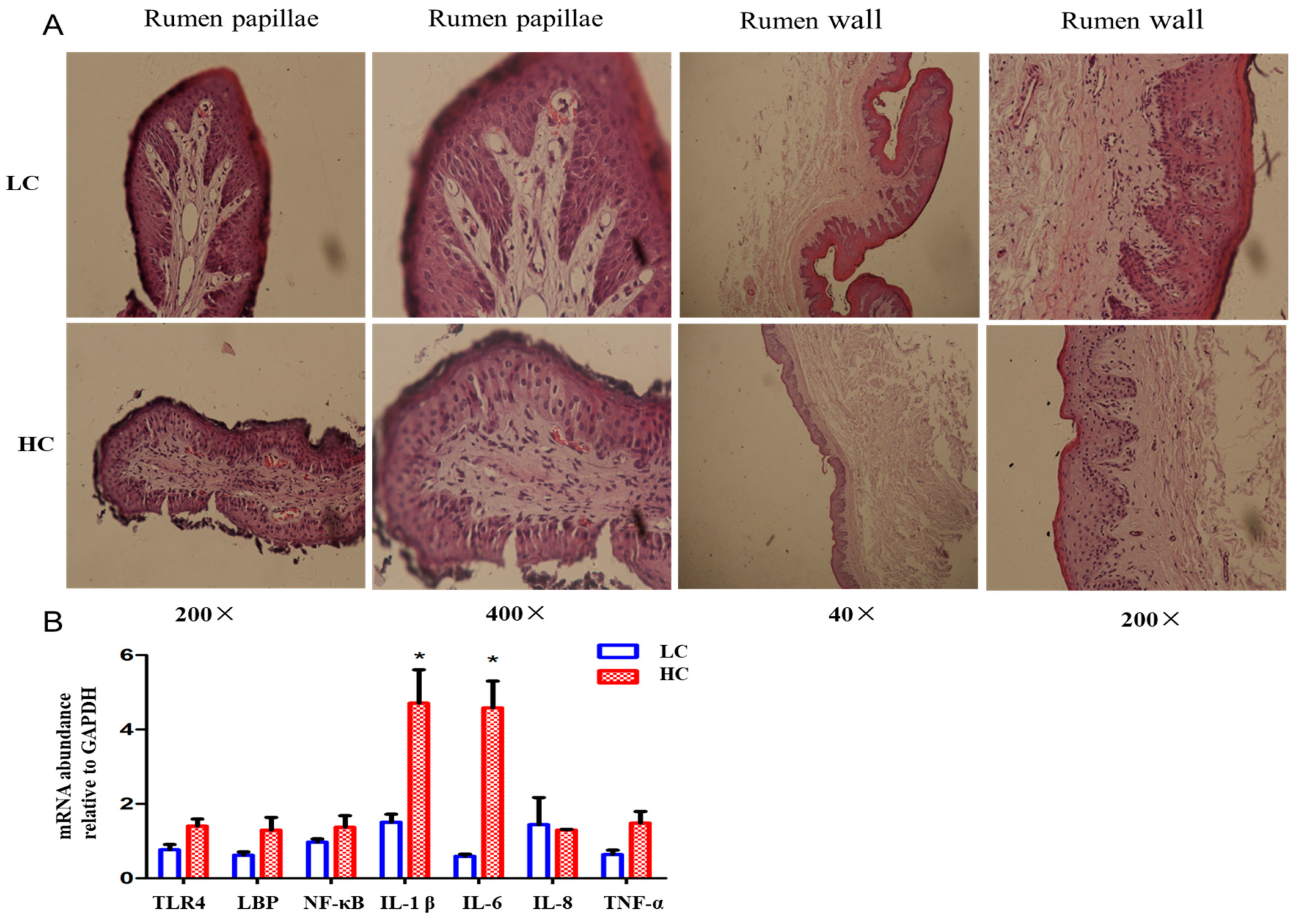
3.3. SARA Induces Histopathological Changes and Inflammatory Responses in the Rumen and Cecum
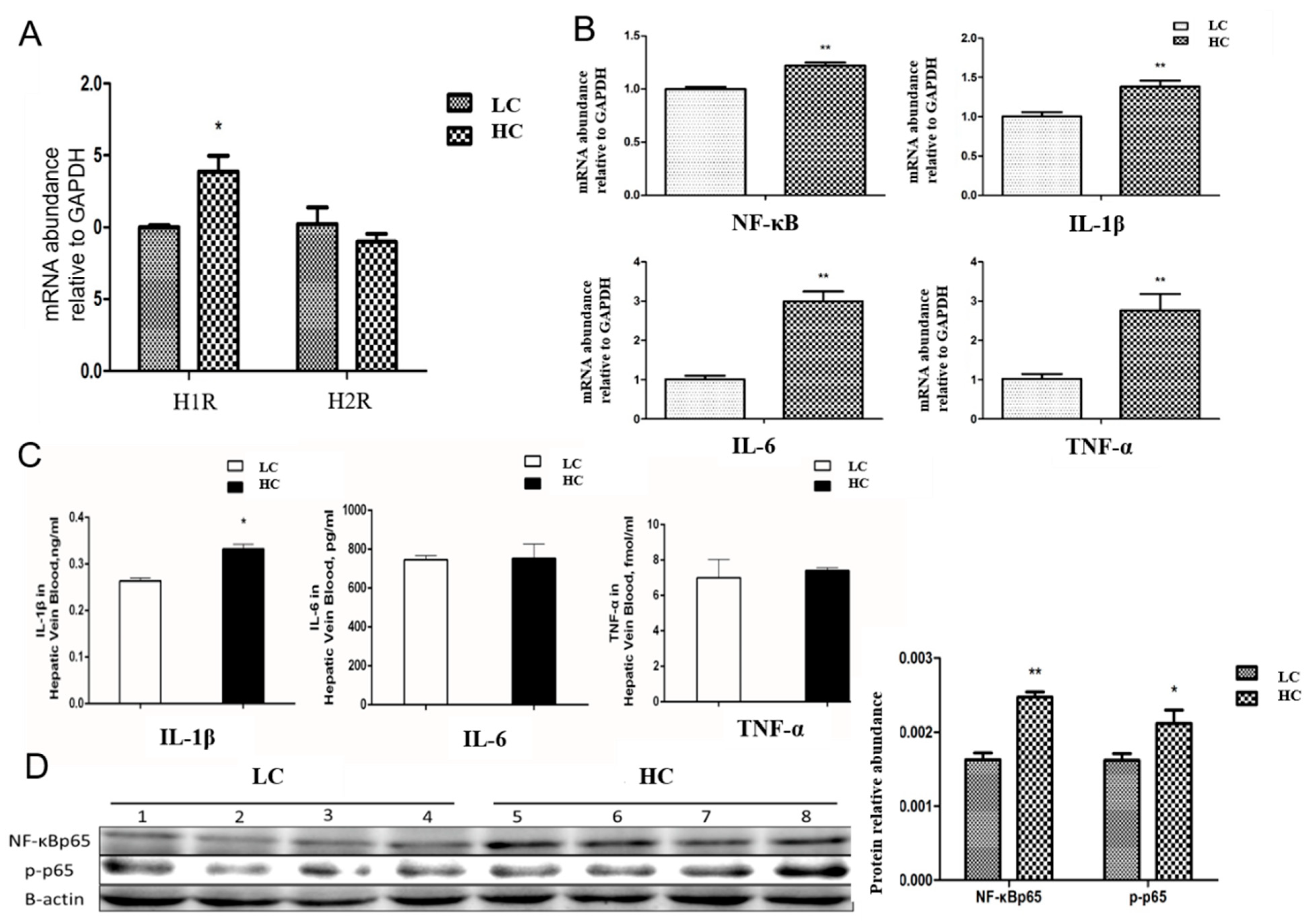
3.4. Transfer/Translocation of LPS and Histamine from the Rumen to Blood Induces an Inflammatory Response in the Liver
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Xie, F.; Sun, D.; Liu, J.; Zhu, W.; Mao, S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Liu, J.X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, E.; Neves, A.L.A.; Song, Y.; Guan, L.L. The Role of the Gut Microbiome in Cattle Production and Health: Driver or Passenger? Annu. Rev. Anim. Biosci. 2020, 8, 199–220. [Google Scholar] [CrossRef]
- Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
- Derakhshani, H.; Tun, H.M.; Cardoso, F.C.; Plaizier, J.C.; Khafipour, E.; Loor, J.J. Linking Peripartal Dynamics of Ruminal Microbiota to Dietary Changes and Production Parameters. Front. Microbiol. 2016, 7, 2143. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Abaker, J.A.; Wei, G.; Chen, H.; Shen, X.; Chang, G. A high-concentrate diet induces an inflammatory response and oxidative stress and depresses milk fat synthesis in the mammary gland of dairy cows. J. Dairy Sci. 2022, 105, 5493–5505. [Google Scholar] [CrossRef]
- Steele, M.A.; AlZahal, O.; Hook, S.E.; Croom, J.; McBride, B.W. Ruminal acidosis and the rapid onset of ruminal parakeratosis in a mature dairy cow: A case report. Acta Vet. Scand. 2009, 51, 39. [Google Scholar] [CrossRef]
- Chang, G.; Zhang, K.; Xu, T.; Jin, D.; Seyfert, H.M.; Shen, X.; Zhuang, S. Feeding a high-grain diet reduces the percentage of LPS clearance and enhances immune gene expression in goat liver. BMC Vet. Res. 2015, 11, 67. [Google Scholar] [CrossRef]
- Abaker, J.A.; Xu, T.L.; Jin, D.; Chang, G.J.; Zhang, K.; Shen, X.Z. Lipopolysaccharide derived from the digestive tract provokes oxidative stress in the liver of dairy cows fed a high-grain diet. J. Dairy Sci. 2017, 100, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Mu, Y.; Zhang, R.; Xue, Y.; Guo, C.; Qi, W.; Zhang, J.; Mao, S. Responsive changes of rumen microbiome and metabolome in dairy cows with different susceptibility to subacute ruminal acidosis. Anim. Nutr. 2022, 8, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Elmhadi, M.E.; Ali, D.K.; Khogali, M.K.; Wang, H. Subacute ruminal acidosis in dairy herds: Microbiological and nutritional causes, consequences, and prevention strategies. Anim. Nutr. 2022, 10, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Li, S.; Tun, H.M.; Khafipour, E. Nutritional Models of Experimentally-Induced Subacute Ruminal Acidosis (SARA) Differ in Their Impact on Rumen and Hindgut Bacterial Communities in Dairy Cows. Front. Microbiol. 2016, 7, 2128. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Li, S.; Danscher, A.M.; Derakshani, H.; Andersen, P.H.; Khafipour, E. Changes in Microbiota in Rumen Digesta and Feces Due to a Grain-Based Subacute Ruminal Acidosis (SARA) Challenge. Microb. Ecol. 2017, 74, 485–495. [Google Scholar] [CrossRef]
- Khafipour, E.; Li, S.; Tun, H.M.; Derakhshani, H.; Moossavi, S.; Plaizier, J.C. Effects of grain feeding on microbiota in the digestive tract of cattle. Anim. Front. 2016, 6, 13–19. [Google Scholar] [CrossRef]
- Guo, J.; Mu, R.; Li, S.; Zhang, N.; Fu, Y.; Hu, X. Characterization of the Bacterial Community of Rumen in Dairy Cows with Laminitis. Genes 2021, 12, 1996. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, S.; Mu, R.; Guo, J.; Zhao, C.; Cao, Y.; Zhang, N.; Fu, Y. The Rumen Microbiota Contributes to the Development of Mastitis in Dairy Cows. Microbiol. Spectr. 2022, 10, e0251221. [Google Scholar] [CrossRef]
- Hurley, J.C. Endotoxemia: Methods of detection and clinical correlates. Clin. Microbiol. Rev. 1995, 8, 268–292. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Khafipour, E.; Li, S.; Gozho, G.N.; Krause, D.O. Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim. Feed Sci. Technol. 2012, 172, 9–21. [Google Scholar] [CrossRef]
- Hua, C.; Tian, J.; Tian, P.; Cong, R.; Luo, Y.; Geng, Y.; Tao, S.; Ni, Y.; Zhao, R. Feeding a High Concentration Diet Induces Unhealthy Alterations in the Composition and Metabolism of Ruminal Microbiota and Host Response in a Goat Model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef]
- Huntington, G.B.; Reynolds, C.K.; Stroud, B.H. Techniques for measuring blood flow in splanchnic tissues of cattle. J. Dairy Sci. 1989, 72, 1583–1595. [Google Scholar] [CrossRef]
- Larsen, M.; Galindo, C.; Ouellet, D.R.; Maxin, G.; Kristensen, N.B.; Lapierre, H. Abomasal amino acid infusion in postpartum dairy cows: Effect on whole-body, splanchnic, and mammary amino acid metabolism. J. Dairy Sci. 2015, 98, 7944–7961. [Google Scholar] [CrossRef]
- Zhang, K.; Chang, G.; Xu, T.; Xu, L.; Guo, J.; Jin, D.; Shen, X. Lipopolysaccharide derived from the digestive tract activates inflammatory gene expression and inhibits casein synthesis in the mammary glands of lactating dairy cows. Oncotarget 2016, 7, 9652–9665. [Google Scholar] [CrossRef]
- Dai, H.; Wei, G.; Wang, Y.; Ma, N.; Chang, G.; Shen, X. Sodium butyrate promotes lipopolysaccharide-induced innate immune responses by enhancing mitogen-activated protein kinase activation and histone acetylation in bovine mammary epithelial cells. J. Dairy Sci. 2020, 103, 11636–11652. [Google Scholar] [CrossRef]
- Chang, G.; Wang, L.; Ma, N.; Zhang, W.; Zhang, H.; Dai, H.; Shen, X. Histamine activates inflammatory response and depresses casein synthesis in mammary gland of dairy cows during SARA. BMC Vet. Res. 2018, 14, 168. [Google Scholar] [CrossRef]
- Gozho, G.N.; Plaizier, J.C.; Krause, D.O.; Kennedy, A.D.; Wittenberg, K.M. Subacute ruminal acidosis induces ruminal lipopolysaccharide endotoxin release and triggers an inflammatory response. J. Dairy Sci. 2005, 88, 1399–1403. [Google Scholar] [CrossRef]
- Khafipour, E.; Li, S.; Plaizier, J.C.; Krause, D.O. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl. Environ. Microbiol. 2009, 75, 7115–7124. [Google Scholar] [CrossRef]
- Kong, Y.; Teather, R.; Forster, R. Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol. Ecol. 2010, 74, 612–622. [Google Scholar] [CrossRef]
- McCann, J.C.; Luan, S.; Cardoso, F.C.; Derakhshani, H.; Khafipour, E.; Loor, J.J. Induction of Subacute Ruminal Acidosis Affects the Ruminal Microbiome and Epithelium. Front. Microbiol. 2016, 7, 701. [Google Scholar] [CrossRef]
- McCann, J.C.; Wickersham, T.A.; Loor, J.J. High-throughput Methods Redefine the Rumen Microbiome and Its Relationship with Nutrition and Metabolism. Bioinform. Biol. Insights 2014, 8, 109–125. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Abecia, L.; Angarita, E.; Aravena, P.; Nora Arenas, G.; Ariza, C.; et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Mickdam, E.; Khiaosa-Ard, R.; Metzler-Zebeli, B.U.; Klevenhusen, F.; Chizzola, R.; Zebeli, Q. Rumen microbial abundance and fermentation profile during severe subacute ruminal acidosis and its modulation by plant derived alkaloids in vitro. Anaerobe 2016, 39, 4–13. [Google Scholar] [CrossRef]
- Fernando, S.C.; Purvis, H.T., 2nd; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; Desilva, U. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef]
- Liu, J.H.; Bian, G.R.; Zhu, W.Y.; Mao, S.Y. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front. Microbiol. 2015, 6, 167. [Google Scholar] [CrossRef]
- Russell, J.B.; Rychlik, J.L. Factors That Alter Rumen Microbial Ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef]
- Tadepalli, S.; Narayanan, S.K.; Stewart, G.C.; Chengappa, M.M.; Nagaraja, T.G. Fusobacterium necrophorum: A ruminal bacterium that invades liver to cause abscesses in cattle. Anaerobe 2009, 15, 36–43. [Google Scholar] [CrossRef]
- Abbas, W.; Keel, B.N.; Kachman, S.D.; Fernando, S.C.; Wells, J.E.; Hales, K.E.; Lindholm-Perry, A.K. Rumen epithelial transcriptome and microbiome profiles of rumen epithelium and contents of beef cattle with and without liver abscesses. J. Anim. Sci. 2020, 98, skaa359. [Google Scholar] [CrossRef]
- Maintz, L.; Novak, N. Histamine and histamine intolerance. Am. J. Clin. Nutr. 2007, 85, 1185–1196. [Google Scholar] [CrossRef]
- Ladero, V.; Calles-Enriquez, M.; Fernandez, M.; Alvarez, M.A. Toxicological Effects of Dietary Biogenic Amines. Curr. Nutr. Food Sci. 2010, 6, 145–156. [Google Scholar] [CrossRef]
- Colombo, F.M.; Cattaneo, P.; Confalonieri, E.; Bernardi, C. Histamine food poisonings: A systematic review and meta-analysis. Crit. Rev. Food Sci. Nutr. 2018, 58, 1131–1151. [Google Scholar] [CrossRef] [PubMed]
- Dain, J.A.; Neal, A.L.; Dougherty, R.W. The Occurrence of Histamine and Tyramine in Rumen Ingesta of Experimentally Over-Fed Sheep. J. Anim. Sci. 1955, 14, 930–935. [Google Scholar] [CrossRef]
- Moniente, M.; García-Gonzalo, D.; Ontañón, I.; Pagán, R.; Botello-Morte, L. Histamine accumulation in dairy products: Microbial causes, techniques for the detection of histamine-producing microbiota, and potential solutions. Compr. Rev. Food Sci. Food Saf. 2021, 20, 1481–1523. [Google Scholar] [CrossRef]
- Wang, D.S.; Zhang, R.Y.; Zhu, W.Y.; Mao, S.Y. Effects of subacute ruminal acidosis challenges on fermentation and biogenic amines in the rumen of dairy cows. Livest. Sci. 2013, 155, 262–272. [Google Scholar] [CrossRef]
- Garner, M.R.; Flint, J.F.; Russell, J.B. Allisonella histaminiformans gen. nov., sp. nov. A novel bacterium that produces histamine, utilizes histidine as its sole energy source, and could play a role in bovine and equine laminitis. Syst. Appl. Microbiol. 2002, 25, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.Y.; Huo, W.J.; Zhu, W.Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016, 18, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Aschenbach, J.R.; Gäbel, G. Effect and absorption of histamine in sheep rumen: Significance of acidotic epithelial damage. J. Anim. Sci. 2000, 78, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Silberberg, M.; Chaucheyras-Durand, F.; Commun, L.; Mialon, M.M.; Monteils, V.; Mosoni, P.; Morgavi, D.P.; Martin, C. Repeated acidosis challenges and live yeast supplementation shape rumen microbiota and fermentations and modulate inflammatory status in sheep. Animal 2013, 7, 1910–1920. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.A.; Croom, J.; Kahler, M.; AlZahal, O.; Hook, S.E.; Plaizier, K.; McBride, B.W. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1515–R1523. [Google Scholar] [CrossRef]
- Dai, H.; Ma, N.; Chang, G.; Aabdin, Z.U.; Shen, X. Long-term high-concentrate diet feeding induces apoptosis of rumen epithelial cells and inflammation of rumen epithelium in dairy cows. Anim. Biotechnol. 2020, 33, 289–296. [Google Scholar] [CrossRef]
- Gressley, T.F.; Hall, M.B.; Armentano, L.E. Ruminant nutrition symposium: Productivity, digestion, and health responses to hindgut acidosis in ruminants. J. Anim. Sci. 2011, 89, 1120–1130. [Google Scholar] [CrossRef]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef]
- Teuscher, C.; Poynter, M.E.; Offner, H.; Zamora, A.; Watanabe, T.; Fillmore, P.D.; Zachary, J.F.; Blankenhorn, E.P. Attenuation of Th1 effector cell responses and susceptibility to experimental allergic encephalomyelitis in histamine H2 receptor knockout mice is due to dysregulation of cytokine production by antigen-presenting cells. Am. J. Pathol. 2004, 164, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Sun, Z.; Li, Y.; Liu, M.; Loor, J.J.; Jiang, Q.; Liu, G.; Wang, Z.; Song, Y.; Li, X. Histamine promotes adhesion of neutrophils by inhibition of autophagy in dairy cows with subacute ruminal acidosis. J. Dairy Sci. 2022, 105, 7600–7614. [Google Scholar] [CrossRef]
- Thangam, E.B.; Jemima, E.A.; Singh, H.; Baig, M.S.; Khan, M.; Mathias, C.B.; Church, M.K.; Saluja, R. The Role of Histamine and Histamine Receptors in Mast Cell-Mediated Allergy and Inflammation: The Hunt for New Therapeutic Targets. Front. Immunol. 2018, 9, 1873. [Google Scholar] [CrossRef] [PubMed]
- Barcik, W.; Wawrzyniak, M.; Akdis, C.A.; O’Mahony, L. Immune regulation by histamine and histamine-secreting bacteria. Curr. Opin. Immunol. 2017, 48, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Meiler, F.; Zumkehr, J.; Klunker, S.; Rückert, B.; Akdis, C.A.; Akdis, M. In vivo switch to IL-10-secreting T regulatory cells in high dose allergen exposure. J. Exp. Med. 2008, 205, 2887–2898. [Google Scholar] [CrossRef]
- Frei, R.; Ferstl, R.; Konieczna, P.; Ziegler, M.; Simon, T.; Rugeles, T.M.; Mailand, S.; Watanabe, T.; Lauener, R.; Akdis, C.A.; et al. Histamine receptor 2 modifies dendritic cell responses to microbial ligands. J. Allergy Clin. Immunol. 2013, 132, 194–204. [Google Scholar] [CrossRef]
- Ferstl, R.; Frei, R.; Schiavi, E.; Konieczna, P.; Barcik, W.; Ziegler, M.; Lauener, R.P.; Chassard, C.; Lacroix, C.; Akdis, C.A.; et al. Histamine receptor 2 is a key influence in immune responses to intestinal histamine-secreting microbes. J. Allergy Clin. Immunol. 2014, 134, 744–746.e3. [Google Scholar] [CrossRef]
- Fitzsimons, C.P.; Lazar-Molnar, E.; Tomoskozi, Z.; Buzás, E.; Rivera, E.S.; Falus, A. Histamine deficiency induces tissue-specific down-regulation of histamine H2 receptor expression in histidine decarboxylase knockout mice. FEBS Lett. 2001, 508, 245–248. [Google Scholar] [CrossRef]


| Ingredient | Diets | |
|---|---|---|
| LC | HC | |
| Corn silage | 30 | 20 |
| Alfalfa hay | 30 | 20 |
| Corn | 24.3 | 32 |
| Bran | 0 | 12.4 |
| Soybean meal | 13.5 | 13 |
| Calcium phosphate dibasic | 0.85 | 0.45 |
| limestone | 0 | 0.8 |
| Salt | 0.35 | 0.35 |
| Premix a | 1 | 1 |
| F:C | 6:4 | 4:6 |
| Nutrient composition | ||
| NEI, MJ/kg CP b | 6.36 16.99 | 6.71 16.92 |
| EE b | 3.93 | 4.07 |
| NDF b | 36.54 | 31.45 |
| ADF b NFC b | 22.51 33.76 | 17.56 39.32 |
| Ca b | 0.88 | 0.89 |
| P b | 0.43 | 0.43 |
| Item | LC | HC | SEM | p-Value |
|---|---|---|---|---|
| Daily average pH | 6.02 | 5.90 | 0.03 | 0.03 |
| Duration of pH < 5.6 per day, min/d | 99.0 | 223 | 30.3 | <0.01 |
| Mean LPS in ruminal contents (EU/mL) | 4.921 × 105 | 7.855 × 105 | 0.644 × 105 | <0.05 |
| Mean LPS in cecum contents (EU/g) | 11.960 × 105 | 13.115 × 105 | 0.149 × 105 | <0.01 |
| Mean LPS in plasma of portal vein (EU/mL) | 0.106 | 0.204 | 0.019 | <0.01 |
| Mean LPS in plasma of hepatic vein (EU/mL) | 0.067 | 0.053 | 0.004 | >0.05 |
| Histamine Content (ng/mL) | Groups | SEM | p-Value | |
|---|---|---|---|---|
| LC | HC | |||
| Ruminal liquid | 1386.94 | 2575.38 | 259.74 | <0.01 |
| Plasma of Jugular vein | 1644.75 | 2380.50 | 149.08 | <0.01 |
| Liver | 133.22 | 183.03 | 4.57 | <0.01 |
| Items | LC | HC | SEM | p-Value |
|---|---|---|---|---|
| OTUs | 852.83 | 847.83 | 7.51 | 0.756 |
| ACE | 876.72 | 890.41 | 6.01 | 0.275 |
| Chao1 | 887.03 | 908.70 | 5.87 | 0.060 |
| Shannon index | 5.65 | 5.45 | 0.06 | 0.100 |
| Simpson index | 0.01 | 0.01 | 0.001 | 0.207 |
| Phylum | Family | Genus | LC | HC | SEM | p-Value |
|---|---|---|---|---|---|---|
| Bacteroidetes | Prevotellaceae | Hallella | 0.26 | 0.10 | 0.03 | 0.001 |
| Prevotella | 22.69 | 15.98 | 2.02 | 0.041 | ||
| Unclassified_Prevotellaceae | 1.99 | 1.42 | 0.13 | 0.010 | ||
| Unclassified_Bacteroidetes | Unclassified_Bacteroidetes | 2.02 | 5.21 | 1.19 | 0.027 | |
| Firmicutes | Lachnospiraceae | Unclassified_Lachnospiraceae | 0.45 | 1.19 | 0.08 | <0.001 |
| Ruminococcaceae | Oscillibacter | 1.02 | 1.75 | 0.14 | 0.007 | |
| Ruminococcus | 0.74 | 1.26 | 0.11 | 0.015 | ||
| Sporobacter | 1.10 | 0.73 | 0.08 | 0.012 | ||
| Unclassified_Clostridia | Unclassified_Clostridia | 0.26 | 0.39 | 0.04 | 0.037 | |
| Acidaminococcaceae | Succiniclasticum | 0.75 | 0.29 | 0.08 | 0.006 | |
| Planctomycetes | Planctomycetaceae | Unclassified_Planctomycetaceae | 0.11 | 0.21 | 0.02 | 0.022 |
| Spirochaetes | Spirochaetaceae | Unclassified_Spirochaetaceae | 0.19 | 0.31 | 0.01 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, N.; Guo, J.; Li, Z.; Xu, L.; Zhang, K.; Xu, T.; Chang, G.; Loor, J.J.; Shen, X. Disturbances of Ruminal Microbiota and Liver Inflammation, Mediated by LPS and Histamine, in Dairy Cows Fed a High-Concentrate Diet. Animals 2024, 14, 1495. https://doi.org/10.3390/ani14101495
Ma N, Guo J, Li Z, Xu L, Zhang K, Xu T, Chang G, Loor JJ, Shen X. Disturbances of Ruminal Microbiota and Liver Inflammation, Mediated by LPS and Histamine, in Dairy Cows Fed a High-Concentrate Diet. Animals. 2024; 14(10):1495. https://doi.org/10.3390/ani14101495
Chicago/Turabian StyleMa, Nana, Junfei Guo, Zhenfu Li, Lei Xu, Kai Zhang, Tianle Xu, Guangjun Chang, Juan J. Loor, and Xiangzhen Shen. 2024. "Disturbances of Ruminal Microbiota and Liver Inflammation, Mediated by LPS and Histamine, in Dairy Cows Fed a High-Concentrate Diet" Animals 14, no. 10: 1495. https://doi.org/10.3390/ani14101495
APA StyleMa, N., Guo, J., Li, Z., Xu, L., Zhang, K., Xu, T., Chang, G., Loor, J. J., & Shen, X. (2024). Disturbances of Ruminal Microbiota and Liver Inflammation, Mediated by LPS and Histamine, in Dairy Cows Fed a High-Concentrate Diet. Animals, 14(10), 1495. https://doi.org/10.3390/ani14101495








