Molecular Epidemiological Investigation of Cyclospora spp. in Holstein Cattle in Partial Areas of the Yunnan Province, China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. DNA Extraction and PCR
2.3. Restriction Fragment Length Polymorphism (RFLP) Analysis
2.4. Sequence Analysis and Phylogenetic Analysis
2.5. Statistical Analysis
3. Results
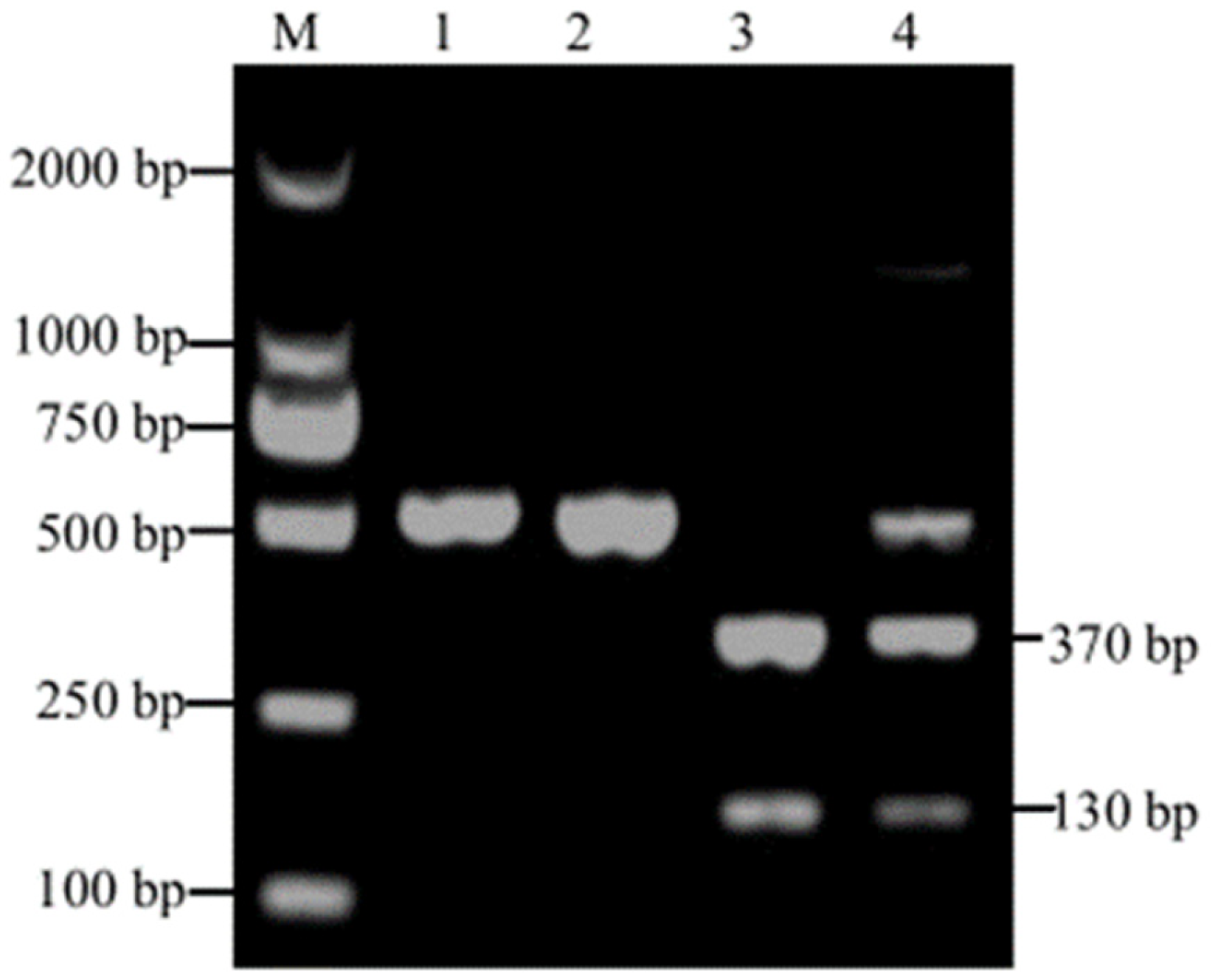
3.1. PCR Amplification and RFLP Analysis
3.2. Risk Factors of Cyclospora spp. Infection
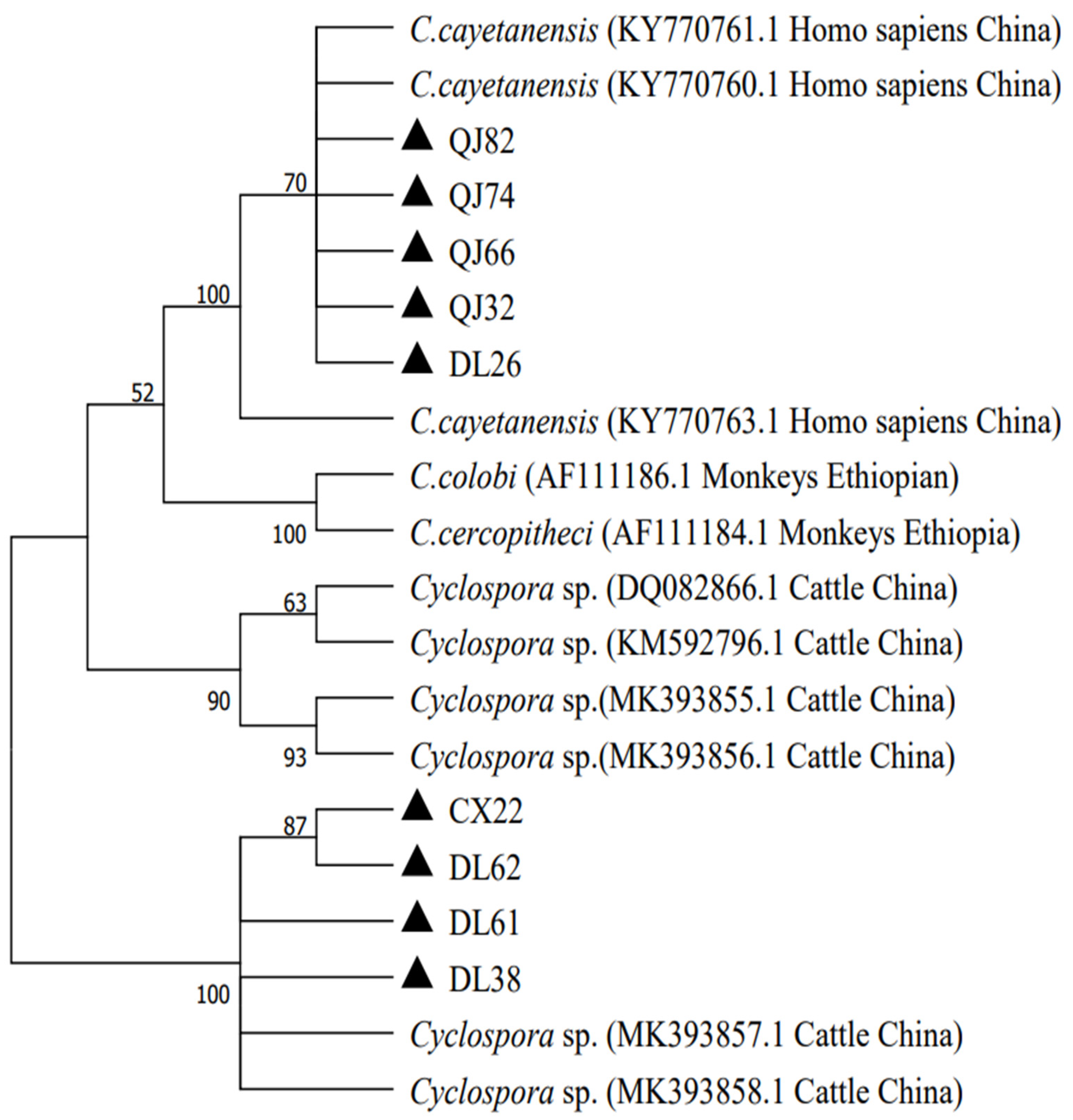
3.3. Phylogenetic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shields, J.M.; Olson, B.H. Cyclosporacayetanensis: A review of an emerging parasitic coccidian. Int. J. Parasitol. 2003, 33, 371–391. [Google Scholar] [CrossRef]
- Lainson, R. The genus Cyclospora (Apicomplexa: Eimeriidae), with a description of Cyclospora schneideri n.sp. in the snake Anilius scytale scytale (Aniliidae) from Amazonian Brazil—A review. Mem. Inst. Oswaldo Cruz 2005, 100, 103–110. [Google Scholar] [CrossRef]
- Insulander, M.; Svenungsson, B.; Lebbad, M.; Karlsson, L.; de Jong, B. A foodborne outbreak of Cyclospora infection in Stockholm, Sweden. Foodborne Pathog. Dis. 2010, 7, 1585–1587. [Google Scholar] [CrossRef]
- Ortega, Y.R.; Sanchez, R. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin. Microbiol. Rev. 2010, 23, 218–234. [Google Scholar] [CrossRef]
- Ortega, Y.R.; Nagle, R.; Gilman, R.H.; Watanabe, J.; Miyagui, J.; Quispe, H.; Kanagusuku, P.; Roxas, C.; Sterling, C.R. Pathologic and clinical findings in patients with cyclosporiasis and a description of intracellular parasite life-cycle stages. J. Infect. Dis. 1997, 176, 1584–1589. [Google Scholar] [CrossRef]
- Hart, A.S.; Ridinger, M.T.; Soundarajan, R.; Peters, C.S.; Swiatlo, A.L.; Kocka, F.E. Novel organism associated with chronic diarrhoea in AIDS. Lancet 1990, 335, 169–170. [Google Scholar] [CrossRef]
- Chacin-Bonilla, L. Transmission of Cyclospora cayetanensis infection: A review focusing on soil-borne cyclosporiasis. Trans. R Soc. Trop. Med. Hyg. 2008, 102, 215–216. [Google Scholar] [CrossRef]
- Totton, S.C.; O’Connor, A.M.; Naganathan, T.; Martinez, B.A.F.; Sargeant, J.M. A review of Cyclospora cayetanensis in animals. Zoonoses Public Health 2021, 68, 861–867. [Google Scholar] [CrossRef]
- Long, E.G.; White, E.H.; Carmichael, W.W.; Quinlisk, P.M.; Raja, R.; Swisher, B.L.; Daugharty, H.; Cohen, M.T. Morphologic and staining characteristics of a cyanobacterium-like organism associated with diarrhea. J. Infect. Dis. 1991, 164, 199–202. [Google Scholar] [CrossRef]
- Onstad, N.H.; Miller, M.R.; Green, M.L.; Witola, W.H.; Davidson, P.C. A Review of Cyclospora cayetanensis Transport in the Environment. Trans. ASABE 2019, 62, 795–802. [Google Scholar] [CrossRef]
- Li, N.; Ye, J.; Arrowood, M.J.; Ma, J.; Wang, L.; Xu, H.; Feng, Y.; Xiao, L. Identification and morphologic and molecular characterization of Cyclospora macacae n. sp. from rhesus monkeys in China. Parasitol. Res. 2015, 114, 1811–1816. [Google Scholar] [CrossRef]
- Dubey, J.P.; Khan, A.; Rosenthal, B.M. Life Cycle and Transmission of Cyclospora cayetanensis: Knowns and Unknowns. Microorganisms 2022, 10, 118. [Google Scholar] [CrossRef]
- Onstad, N.H.; Beever, J.E.; Miller, M.R.; Green, M.L.; Witola, W.H.; Davidson, P.C. CyclosporaCayetanensis Presence in the Environment—A Case Study in the Chicago Metropolitan Area. Environments 2019, 6, 80. [Google Scholar] [CrossRef]
- Chu, D.M.; Sherchand, J.B.; Cross, J.H.; Orlandi, P.A. Detection of Cyclospora cayetanensis in animal fecal isolates from Nepal using an FTA filter-base polymerase chain reaction method. Am. J. Trop. Med. Hyg. 2004, 71, 373–379. [Google Scholar] [CrossRef]
- Wang, K.X.; Li, C.P.; Wang, J.; Tian, Y. Cyclospore cayetanensis in Anhui, China. World J. Gastroenterol. 2002, 8, 1144–1148. [Google Scholar] [CrossRef]
- Zhou, Y.; Lv, B.; Wang, Q.; Wang, R.; Jian, F.; Zhang, L.; Ning, C.; Fu, K.; Wang, Y.; Qi, M.; et al. Prevalence and molecular characterization of Cyclospora cayetanensis, Henan, China. Emerg. Infect. Dis. 2011, 17, 1887–1890. [Google Scholar] [CrossRef]
- Eberhard, M.L.; da Silva, A.J.; Lilley, B.G.; Pieniazek, N.J. Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp.n., C. colobi sp.n., and C. papionis sp.n. Emerg. Infect. Dis. 1999, 5, 651–658. [Google Scholar] [CrossRef]
- Lopez, F.A.; Manglicmot, J.; Schmidt, T.M.; Yeh, C.; Smith, H.V.; Relman, D.A. Molecular characterization of Cyclospora-like organisms from baboons. J. Infect. Dis. 1999, 179, 670–676. [Google Scholar] [CrossRef]
- Zhao, G.H.; Cong, M.M.; Bian, Q.Q.; Cheng, W.Y.; Wang, R.J.; Qi, M.; Zhang, L.X.; Lin, Q.; Zhu, X.Q. Molecular characterization of Cyclospora-like organisms from golden snub-nosed monkeys in Qinling Mountain in Shaanxi province, northwestern China. PLoS ONE 2013, 8, e58216. [Google Scholar] [CrossRef]
- Li, G.; Xiao, S.; Zhou, R.; Li, W.; Wadeh, H. Molecular characterization of Cyclospora-like organism from dairy cattle. Parasitol. Res. 2007, 100, 955–961. [Google Scholar] [CrossRef]
- Eberhard, M.L.; Pieniazek, N.J.; Arrowood, M.J. Laboratory diagnosis of Cyclospora infections. Arch. Pathol. Lab. Med. 1997, 121, 792–797. [Google Scholar] [PubMed]
- Khanna, V.; Tilak, K.; Ghosh, A.; Mukhopadhyay, C. Modified negative staining of heine for fast and inexpensive screening of Cryptosporidium, Cyclospora, and Cystoisospora spp. Int. Sch. Res. Not. 2014, 2014, 165424. [Google Scholar] [CrossRef]
- Relman, D.A.; Schmidt, T.M.; Gajadhar, A.; Sogin, M.; Cross, J.; Yoder, K.; Sethabutr, O.; Echeverria, P. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J. Infect. Dis. 1996, 173, 440–445. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Ortega, Y.; Simpson, S.; Kerdahi, K. Genetic characterization of human-pathogenic Cyclospora cayetanensis parasites from three endemic regions at the 18S ribosomal RNA locus. Infect. Genet. Evol. 2014, 22, 229–234. [Google Scholar] [CrossRef]
- Pieniazek, N.J.; Slemenda, S.B.; da Silva, A.J.; Alfano, E.M.; Arrowood, M.J. PCR confirmation of infection with Cyclospora cayetanensis. Emerg. Infect. Dis. 1996, 2, 357–359. [Google Scholar] [CrossRef]
- Jinneman, K.C.; Wetherington, J.H.; Hill, W.E.; Adams, A.M.; Johnson, J.M.; Tenge, B.J.; Dang, N.L.; Manger, R.L.; Wekell, M.M. Template preparation for PCR and RFLP of amplification products for the detection and identification of Cyclospora sp. and Eimeria spp. Oocysts directly from raspberries. J. Food Prot. 1998, 61, 1497–1503. [Google Scholar] [CrossRef]
- Zhang, B.X.; Yu, H.; Zhang, L.L.; Tao, H.; Li, Y.Z.; Li, Y.; Cao, Z.K.; Bai, Z.M.; He, Y.Q. Prevalence survey on Cyclospora cayetanensis and Cryptosporidium ssp. in diarrhea cases in Yunnan Province. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 2002, 20, 106–108. [Google Scholar]
- Ye, J.; Xiao, L.; Li, J.; Huang, W.; Amer, S.E.; Guo, Y.; Roellig, D.; Feng, Y. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol. Int. 2014, 63, 132–137. [Google Scholar] [CrossRef]
- Li, J.; Shi, K.; Sun, F.; Li, T.; Wang, R.; Zhang, S.; Jian, F.; Ning, C.; Zhang, L. Identification of human pathogenic Enterocytozoon bieneusi, Cyclospora cayetanensis, and Cryptosporidium parvum on the surfaces of vegetables and fruits in Henan, China. Int. J. Food Microbiol. 2019, 307, 108292. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, X.; Yang, R.; Zhao, W.; Li, N.; Guo, Y.; Xiao, L.; Feng, Y. Molecular characterization of the waterborne pathogens Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. in wastewater and sewage in Guangzhou, China. Parasit. Vectors 2021, 14, 66. [Google Scholar] [CrossRef]
- Basnett, K.; Nagarajan, K.; Soundararajan, C.; Vairamuthu, S.; Rao, G.V.S. Morphological and molecular identification of Cyclospora species in sheep and goat at Tamil Nadu, India. J. Parasit. Dis. 2018, 42, 604–607. [Google Scholar] [CrossRef]
- Helmy, M.M. Cyclospora cayetanensis: A review, focusing on some of the remaining questions about cyclosporiasis. Infect. Disord. Drug. Targets 2010, 10, 368–375. [Google Scholar] [CrossRef]
- Li, J.; Wang, R.; Chen, Y.; Xiao, L.; Zhang, L. Cyclospora cayetanensis infection in humans: Biological characteristics, clinical features, epidemiology, detection method and treatment. Parasitology 2020, 147, 160–170. [Google Scholar] [CrossRef]
- Frickmann, H.; Alker, J.; Hansen, J.; Dib, J.C.; Aristizabal, A.; Concha, G.; Schotte, U.; Kann, S. Seasonal Differences in Cyclospora cayetanensis Prevalence in Colombian Indigenous People. Microorganisms 2021, 9, 627. [Google Scholar] [CrossRef]
- Ortega, Y.R.; Sterling, C.R.; Gilman, R.H. Cyclospora cayetanensis. Adv. Parasitol. 1998, 40, 399–418. [Google Scholar] [CrossRef]
- Almeria, S.; Cinar, H.N.; Dubey, J.P. Cyclospora cayetanensis and Cyclosporiasis: An Update. Microorganisms 2019, 7, 317. [Google Scholar] [CrossRef]
- Connor, B.A. Cyclospora infection: A review. Ann. Acad. Med. Singap. 1997, 26, 632–636. [Google Scholar]
- Hall, R.L.; Jones, J.L.; Hurd, S.; Smith, G.; Mahon, B.E.; Herwaldt, B.L. Population-based active surveillance for Cyclospora infection--United States, Foodborne Diseases Active Surveillance Network (FoodNet), 1997–2009. Clin. Infect. Dis. 2012, 54 (Suppl. S5), S411–S417. [Google Scholar] [CrossRef]
- Kaminsky, R.G.; Lagos, J.; Santos, G.R.; Urrutia, S. Marked seasonality of Cyclospora cayetanensis infections: Ten-year observation of hospital cases, Honduras. BMC Infect. Dis. 2016, 16, 66. [Google Scholar] [CrossRef]
- Jiang, Y.; Yuan, Z.; Zang, G.; Li, D.; Wang, Y.; Zhang, Y.; Liu, H.; Cao, J.; Shen, Y. Cyclospora cayetanensis infections among diarrheal outpatients in Shanghai: A retrospective case study. Front. Med. 2018, 12, 98–103. [Google Scholar] [CrossRef]
| Factors | Category | No. Tested | No. Positive | Infection Rate (%) (95%CI) | OR (95%, CI) | p-Value |
|---|---|---|---|---|---|---|
| Region | Dali | 274 | 6 | 2.19 [0.45–3.93] | Reference | 0.339 |
| Kunmin | 57 | 0 | 0 [0.00–0.00] | - | ||
| Qujing | 158 | 5 | 3.16 [0.41–5.92] | 1.46 (0.44–4.86) | ||
| Chuxiong | 35 | 2 | 5.71 [0.00–13.80] | 2.71 (0.53–13.96) | ||
| Age (M) | 0–3 | 344 | 6 | 1.74 [0.35–3.13] | Reference | 0.204 |
| 3–6 | 57 | 3 | 5.26 [0.00–11.24] | 3.13 (0.76–12.89) | ||
| 6–12 | 11 | 1 | 9.09 [0.00–29.35] | 5.63 (0.62–51.27) | ||
| >12 | 112 | 3 | 2.68 [0.00–5.72] | 1.55 (0.38–6.30) | ||
| Sex | Female | 422 | 10 | 2.37 [0.91–3.82] | Reference | 0.739 |
| Male | 102 | 3 | 2.94 [0.00–6.28] | 1.25 (0.34–4.62) | ||
| Total | 524 | 13 | 2.48 | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.-F.; Heng, Z.-J.; Shu, F.-F.; Mao, H.-M.; Su, Y.-S.; He, J.-J.; Zou, F.-C. Molecular Epidemiological Investigation of Cyclospora spp. in Holstein Cattle in Partial Areas of the Yunnan Province, China. Animals 2023, 13, 1527. https://doi.org/10.3390/ani13091527
Yang J-F, Heng Z-J, Shu F-F, Mao H-M, Su Y-S, He J-J, Zou F-C. Molecular Epidemiological Investigation of Cyclospora spp. in Holstein Cattle in Partial Areas of the Yunnan Province, China. Animals. 2023; 13(9):1527. https://doi.org/10.3390/ani13091527
Chicago/Turabian StyleYang, Jian-Fa, Zhao-Jun Heng, Fan-Fan Shu, Hua-Ming Mao, Yong-Sheng Su, Jun-Jun He, and Feng-Cai Zou. 2023. "Molecular Epidemiological Investigation of Cyclospora spp. in Holstein Cattle in Partial Areas of the Yunnan Province, China" Animals 13, no. 9: 1527. https://doi.org/10.3390/ani13091527
APA StyleYang, J.-F., Heng, Z.-J., Shu, F.-F., Mao, H.-M., Su, Y.-S., He, J.-J., & Zou, F.-C. (2023). Molecular Epidemiological Investigation of Cyclospora spp. in Holstein Cattle in Partial Areas of the Yunnan Province, China. Animals, 13(9), 1527. https://doi.org/10.3390/ani13091527








