Simple Summary
In humans, a dietary intake of omega-3 polyunsaturated fatty acids along with antioxidants has been shown to have anti-inflammatory and antioxidant activities. In pigs, on the other hand, there are few studies dealing with the use of omega-3 polyunsaturated fatty acids in the diet. For this reason, our study aimed to investigate the differences in gene expression of the Longissimus thoracis muscle of Italian Large White pigs fed with four different diets: a standard diet for growing-finishing pigs and three experimental diets; one supplemented with extruded linseed, a source of omega-3 polyunsaturated fatty acids, another with extruded linseed plus vitamin E and selenium as antioxidants, and another with extruded linseed plus oregano and grape skin extracts, which are natural polyphenols. From the results of the expression analysis, it was possible to deduce that, in the diets, the oxidative stability of the n-3 fatty acids increased, consistent with an increase in the fluidity of cell membranes, and increasing the anti-inflammatory potential of muscle. This can determine the high quality of the muscle tissue as regards the lipid composition; consequently, the meat will be qualitatively better for human health.
Abstract
The addition of n-3 polyunsaturated fatty acids (n-3 PUFAs) to the swine diet increases their content in muscle cells, and the additional supplementation of antioxidants promotes their oxidative stability. However, to date, the functionality of these components within muscle tissue is not well understood. Using a published RNA-seq dataset and a selective workflow, the study aimed to find the differences in gene expression and investigate how differentially expressed genes (DEGs) were implicated in the cellular composition and metabolism of muscle tissue of 48 Italian Large White pigs under different dietary conditions. A functional enrichment analysis of DEGs, using Cytoscape, revealed that the diet enriched with extruded linseed and supplemented with vitamin E and selenium promoted a more rapid and massive immune system response because the overall function of muscle tissue was improved, while those enriched with extruded linseed and supplemented with grape skin and oregano extracts promoted the presence and oxidative stability of n-3 PUFAs, increasing the anti-inflammatory potential of the muscular tissue.
1. Introduction
To date, there are multiple strategies used to improve the nutritional quality of meat and meat products [1]. These include adding sources of n-3 polyunsaturated fatty acids (n-3 PUFAs) and antioxidants, such as selenium plus vitamin E or natural polyphenols, to the diet. In humans, n-3 PUFAs co-added with antioxidants have a positive role in the metabolism by showing anti-inflammatory and antioxidant activity and have a positive effect against obesity and insulin resistance [2]. In swine, few studies in the literature examined the effects on metabolism, particularly at the molecular level, of dietary intake of n-3 PUFAs sources supplemented with antioxidants and polyphenols.
This research aims to study pig Longissimus thoracis muscle gene expression differences between pig diets through the application of a selective workflow of RNA-seq data processing. Compared with previous studies, we chose DESeq2 to identify the differential expression genes (DEGs) and we applied a strict log2 Fold Change (log2FC) to identify the DEGs. This approach, which is quite common in human research, was used in pigs to identify effects on gene expression of diets supplemented with different antioxidants. Thanks to the identification of differentially expressed genes, this paper highlights the relevance of adding antioxidants to pig diets when animals are fed with a source of polyunsaturated fatty acids, in order to increase the stability of the fat component of pork utilized both for fresh consumption and to produce high-quality pig-meat-seasoned products.
2. Materials and Methods
Forty-eight Italian Large White pigs, 24 gilts and 24 barrows, were used in the experiment. These pigs were chosen from a large group of 258 piglets, which were descended from 21 sows and 3 boars marked in the herd book of the Italian National Association of Pig Breeders (ANAS; [3]).
The 48 pigs were divided into four experimental groups of 12 animals, each balanced for weight, father, and sex. The subjects were all fed a standard diet until the start of the trial, after which each group was given its respective diet, which was a standard diet for growing-finishing pigs (D1); the same diet as D1, enriched with extruded linseed, an n-3 PUFAs source (D2); the same diet as D2, enriched with vitamin E and selenium (D3); and the same diet as D2, enriched with grape skin and oregano extracts, sources of natural polyphenols (D4). In the middle of the trial, in the experimental group D4, a pig died of natural causes. For ingredients, chemical composition, and feeding methods of the four diets administered, for the manner and timing (in relation to the weight of the animals) of pig slaughtering, and for regulations on the protection of animals at slaughter, refer to [2,4]. After slaughter, Longissimus thoracis muscle samples were taken and placed immediately into liquid nitrogen for cryopreservation. After that, they were stored at −80 °C until the time of RNA extraction. Regarding materials and methods of RNA extraction, library preparation, and sequencing, we refer to [2,4].
The forty-eight RNA-seq datasets for pigs fed with different diets were downloaded from the ArrayExpress [5] (accession: E-MTAB-7131), whose reads are 100 nucleotides paired-end sequencing reads. The quality of the raw reads was evaluated using FastQC v.0.11.5 [6] and reported in detailed files with MultiQC v.1.10.1 [7]. Then, the reads were trimmed with Trimmomatic v.0.39 [8,9] by removing Illumina adapters, deleting the final bases of the reads with quality <3, eliminating reads when their average quality was <15 in a sliding window of 4 bases, and, finally, removing reads of length <60 nucleotides to ensure the highest quality of clean reads. Following this, clean reads were mapped to the reference genome, Sus scrofa genome assembly version Sscrofa11.1 [10] using STAR v.2.6.1.d [11,12] with default parameters and uniquely mapped reads obtained after filtering were used for the quantification of gene expression. FeatureCounts was used for the evaluation of gene expression, implemented in Subread v.1.6.3 [13] using the default parameters, and based on the genomic annotation of swine (release-104) from Ensembl database [14]. The identified genes were then assessed for differential expression between experimental diets: D1 vs. D2, D1 vs. D3, D1 vs. D4, D2 vs. D3, D2 vs. D4, and D3 vs. D4, for a total of six comparisons.
DEGs were then detected using DESeq2 [15], an R package from Bioconductor v.3.14. In DESeq2, the correction method used anticipated the dietary groups as experimental factors, while father, sex, slaughter day, and hidden batch effect were fixed factors. The hidden batch effect was previously calculated with sva [16], an R package from Bioconductor v.3.14, to adjust for unknown, unmodeled, or latent sources of noise; noise that would have conditioned the effect exerted by diets [17]. Genes were assumed to be differentially expressed only in those with at least 8 samples in at least one condition, with a number of reads equal to at least 10. The same conditions were used in studies concerning humans [18]. In addition, DEGs were considered those fulfilling the criteria of log2FC ≥ |0.70| [19] and False Discovery Rate (FDR) adjusted p-value ≤ 0.1, preserving the highly expressed DEGs, and they detected and described, in particular, the most pronounced differences in gene expression between diets provided to pigs of the same breed. For the validation methodology with quantitative real-time PCR, refer to [2,4].
In order to perform a functional analysis, DEGs were considered. For the annotation of the DEGs, the pig annotation gene was used first, after which the remaining unidentified genes were named using the human homologous genes. To do this, BioMart-Ensembl [20] was employed [14]. The functional enrichment analysis of DEGs was analyzed using stringApp, an app of the Cytoscape v3.9.1 software [21], using databases Gene Ontology (GO, including Biological Process, Cellular Component, Molecular Function), KEGG Pathways, and Reactome Pathways. All the genes from Homo sapiens were used as the background for the analysis because, with this background, we obtained networks with more genes involved and more interactions than using Sus scrofa. For the realization of the network, a confidence (score) cutoff of 0.40 was used, and to favor the creation of a network that included genes relevant for functional analysis, but which were not present among the DEGs, 5 maximum additional interactor genes were added for the comparison of D1 vs. D3. No genes were summed for the comparison of D1 vs. D4, while for the comparison between D2 vs. D4, no network was built because of the small number of DEGs found therein. The functions and pathways considered in the study had a significance threshold of FDR < 0.05.
3. Results
The results of the RNA-seq data pre-processing and gene expression analysis are shown in Table 1. Not all clean reads were assigned to that feature, and this is probably because the pig genome was not completely annotated, so a part of the remaining reads was not assigned. However, through these reads, it would be possible to update the annotation of the pig genome [22].

Table 1.
Results of RNA-seq data pre-processing and gene expression analysis. The table shows, for each sample, the number of starting reads (raw reads), the number of reads remaining after the trimming step (clean reads), the percentage of reads out of the total clean reads uniquely mapped to the genome (uniquely mapped reads), and the number and percentage of reads assigned to exons out of the total clean reads for the identification of differential expression genes.
As the result of differential expression assessment, a total of 36 significant DEGs were detected, of which 34 genes were unique and non-redundant considering all comparisons between diets. Only two DEGs (transmembrane protein with EGF-like and two follistatin-like domains 2, TMEFF2, and RING1 and YY1 binding protein, RYBP) were detected for D2 vs. D4, and thus were not further considered in functional analyses. For D1 vs. D3, 22 DEGs were obtained, of which 19 were upregulated in D3. Finally, for D1 vs. D4, 12 DEGs were obtained, of which 11 were upregulated in D4. Since we did not detect significant DEGs in the comparison of D1 vs. D2, D2 vs. D3, and D3 vs. D4, these were omitted from Table 2 and Table 3. The complete list of DEGs with their average expressions and significance is reported in Table 2. The list of pathways and functions detected by functional enrichment analysis is reported in Table 3, and the description of the roles of the DEGs considered in the study is shown in Table 4.

Table 2.
Differentially expressed genes (DEGs) were obtained from D1 vs. D3, D1 vs. D4, and D2 vs. D4 comparisons. For each DEG, the ENSEMBL identity number, the mean of the normalized counts, the log2 Fold Change (log2FC), the raw p-values, the adjusted p-values (padj), and gene symbol are reported.

Table 3.
Functions and pathways generated by functional analysis with Cytoscape were used for each comparison (D1 vs. D3 and D1 vs. D4) of the respective differentially expressed genes. For each category, there are numbers and symbols of genes of the category of belonging (categories of Gene Ontology (GO) or Reactome or KEGG Pathways); description; and False Discovery Rate (FDR). The pathways marked in bold are those of interest to the study.

Table 4.
Description of the functions of the most relevant differentially expressed genes (DEGs).
From the subsequent functional analyses of the D1 vs. D3, some pathways were detected using Cytoscape (Figure 1a). Among them, the “positive regulation of the immune system process” Biological Process of the GO database (Table 3) included the DEG lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1), which is connected to the C-C motif chemokine ligand 21 (CCL21) and plasmalemma vesicle-associated protein (PLVAP) in the network (Figure 1a), and has a role in regulating immune cell migration (Table 4).
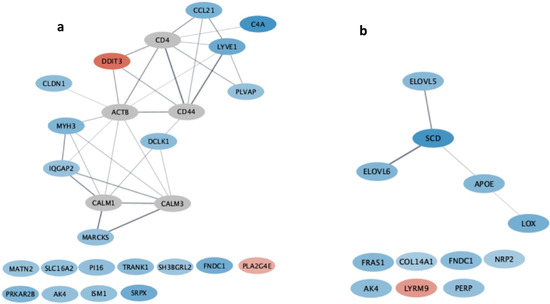
Figure 1.
Cytoscape Gene networks achieved with stringApp for the DEGs obtained comparing D1 vs. D3 diets on the left (a) and D1 vs. D4 diets on the right (b). Over-expressed genes were colored in shades of blue, from the lightest (least over-expressed) to the darkest (most over-expressed). Down-expressed genes were colored in shades of red, from the lightest, least under-expressed to the darkest, most under-expressed. The genes indicated with a gray ellipse are additional interactor genes selected by Cytoscape.
4. Discussion
The results obtained from the D1 vs. D3 comparison are consistent with the hypothesis that the cell migration process of the immune system is more activated in the diet supplemented with extruded linseed plus selenium and vitamin E (CCL21, [24]; LYVE1, [25,26]), and the filtering efficiency of lymphocytes within the blood vessels is stimulated (C4A, [23]; PLVAP, [27]). This may suggest that, in pigs, the intake of a diet enriched with n-3 PUFAs (extruded linseed) plus antioxidants (vitamin E and selenium) promotes a more rapid and massive immune system response because the overall function of muscle tissue is improved.
Considering the comparison of D1 vs. D4, the “Unsaturated fatty acid biosynthetic process” Biological Process of the GO database was detected as significant (Table 3) and included stearoyl-CoA desaturase (SCD), ELOVL fatty acid elongase 5 (ELOVL5), and ELOVL fatty acid elongase 6 (ELOVL6) genes (Figure 1b); the ELOVL5 gene codes for an enzyme acting in the metabolic path of docosahexaenoic (DHA) and eicosapentaenoic (EPA) n-3 acid formation [29] from the alpha-linoleic supplementation [28]. This is consistent with the D4 supplementation stimulated the expression of genes acting in the synthesis of eicosanoid acids. In addition, dietary D4 supplements, grape skin, and oregano, with their antioxidant effects on lipids, might preserve EPA and DHA [32,33,34,35]. This could result in a greater concentration in the phospholipid membrane of these n-3 fatty acids—which are the precursors of important anti-inflammatory metabolites released upon inflammation, such as resolvins, protectins, and marensins—in muscle cells [36]. However, the lack of phenotypes limits the full understanding of how DEGs and signaling pathways influence certain characteristic meat traits.
5. Conclusions
To summarize, we identified 34 DEGs of which 27 are new DEGs compared to the 289 DEGs in [4]. Given the source of RNA being comparable pigs under different diets, we do not expect large changes in their transcriptional landscape (reflected by the low log2FC cut-off). Hence, to retrieve a set of DEGs with a lower number of false positives we conducted the present data analysis using more conservative filters and the statistical tool DESeq2 which was shown in [37] as preferential for a moderate number of replicates to call small numbers of true positive DEGs. The current data analysis suggests that the use of antioxidants (selenium and vitamin E) or polyphenols as natural antioxidants (grape skin and oregano) in the diets enriched with n-3 PUFAs derived from extruded linseed increased both the content and oxidative stability of n-3 fatty acids. This possibly provides the cells with greater membrane fluidity and anti-inflammatory potential, important requirements for maintaining cellular physiology as reported for immune cells [28], and allows for a higher quality of muscle tissue resulting in increased meat quality for human health in relation to the lipid content and composition [38]. In general, this paper highlights the relevance of adding natural antioxidants to pig diets when animals are fed with a source of polyunsaturated fatty acids in order to increase the stability of the fat component of pork produced by heavy pigs, which can be utilized both for fresh consumption and to produce high-quality pig-meat-seasoned products.
Author Contributions
Conceptualization and methodology, J.G., R.D., S.E.S., P.Z., Y.S. and J.V.; formal analysis, Y.S. and J.V.; investigation, J.V., Y.S. and S.D.; resources, P.Z.; data curation R.D., P.Z., J.G. and J.V.; writing—original draft preparation, J.V., P.Z., S.D., M.Z. and R.D.; writing—review and editing, all authors; supervision, S.E.S., J.G. and P.Z.; funding acquisition, P.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Regione Emilia-Romagna, Italy, POR-FESR 2014-2020, grant number PG/2015/730542 and COST action CA15112.
Institutional Review Board Statement
Ethical approval was not necessary for this research because we utilized transcription profiles obtained in previous experiments (Sirri et al. 2018; Vitali et al. 2019) [2,3].
Informed Consent Statement
Not applicable.
Data Availability Statement
The forty-eight RNA-seq datasets for pigs utilized for this paper were downloaded from the ArrayExpress, https://www.ebi.ac.uk/biostudies/arrayexpress, (accessed on 29 January 2023) accession: E-MTAB-7131 [2,4].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rocchetti, G.; Vitali, M.; Zappaterra, M.; Righetti, L.; Sirri, R.; Lucini, L.; Dall’Asta, C.; Davoli, R.; Galaverna, G. A molecular insight into the lipid changes of pig Longissimus thoracis muscle following dietary supplementation with functional ingredients. PLoS ONE 2022, 17, e0264953. [Google Scholar] [CrossRef] [PubMed]
- Vitali, M.; Sirri, R.; Zappaterra, M.; Zambonelli, P.; Giannini, G.; Lo Fiego, D.P.; Davoli, R. Functional analysis finds differences in the muscle transcriptome of pigs fed an n-3 PUFA-enriched diet with or without antioxidant supplementations. PLoS ONE 2019, 14, e0212449. [Google Scholar] [CrossRef] [PubMed]
- ANAS. Available online: https://www.anas.it/ (accessed on 29 January 2023).
- Sirri, R.; Vitali, M.; Zambonelli, P.; Giannini, G.; Zappaterra, M.; Lo Fiego, D.P.; Sami, D.; Davoli, R. Effect of diets supplemented with linseed alone or combined with vitamin E and selenium or with plant extracts, on Longissimus thoracis transcriptome in growing-finishing Italian Large White pigs. J. Anim. Sci. Biotechnol. 2018, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- ArrayExpress. Available online: https://www.ebi.ac.uk/biostudies/arrayexpress (accessed on 29 January 2023).
- FastQC v.0.11.5. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 29 January 2023).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Trimmomatic v.0.39. Available online: http://www.usadellab.org/cms/?page=trimmomatic (accessed on 29 January 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Sus Scrofa Genome Assembly Version Sscrofa11.1. Available online: https://www.ensembl.org/Sus_scrofa/Info/Index (accessed on 29 January 2023).
- STAR v.2.6.1.d. Available online: https://github.com/alexdobin/STAR (accessed on 29 January 2023).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Sva. Available online: https://bioconductor.org/packages/3.14/bioc/html/sva.html (accessed on 29 January 2023).
- Parker, H.S.; Corrada Bravo, H.; Leek, J.T. Removing batch effects for prediction problems with frozen surrogate variable analysis. PeerJ 2014, 2, e561. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [PubMed]
- Markussen, L.K.; Rondini, E.A.; Johansen, O.; Madsen, J.; Sustarsic, E.G.; Marcher, A.; Hansen, J.B.; Gerhart-Hines, Z.; Granneman, J.G.; Mandrup, S. Lipolysis regulates major transcriptional programs in brown adipocytes. Nat. Commun. 2022, 13, 3956. [Google Scholar] [CrossRef] [PubMed]
- BioMart-Ensembl. Available online: https://www.ensembl.org/biomart/martview/ (accessed on 29 January 2023).
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.; Wang, L.; Wang, J.; Zhang, L.; Hou, X.; Yan, H.; Wang, L. Integrative Analysis of Nanopore and Illumina Sequencing Reveals Alternative Splicing Complexity in Pig Longissimus Dorsi Muscle. Front. Genet. 2022, 13, 877646. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, M. Complement C4, Infections, and Autoimmune Diseases. Front. Immunol. 2021, 12, 694928. [Google Scholar] [CrossRef]
- Christopherson, K.W.; Campbell, J.J.; Travers, J.B.; Hromas, R.A. Low-Molecular-Weight Heparins Inhibit CCL21-Induced T Cell Adhesion and Migration. J. Pharmacol. Exp. Ther. 2002, 302, 290–295. [Google Scholar] [CrossRef]
- Lawrance, W.; Banerji, S.; Day, A.J.; Bhattacharjee, S.; Jackson, D.G. Binding of Hyaluronan to the Native Lymphatic Vessel Endothelial Receptor LYVE-1 Is Critically Dependent on Receptor Clustering and Hyaluronan Organization. J. Biol. Chem. 2016, 291, 8014–8030. [Google Scholar] [CrossRef]
- Johnson, L.A.; Banerji, S.; Lawrance, W.; Gileadi, U.; Prota, G.; Holder, K.A.; Roshorm, Y.M.; Hanke, T.; Cerundolo, V.; Gale, N.W.; et al. Dendritic cells enter lymph vessels by hyaluronan-mediated docking to the endothelial receptor LYVE-1. Nat. Immunol. 2017, 18, 762–770. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, H.; Hou, Y.; Wei, T.; Liu, J. Plasmalemma vesicle-associated protein: A crucial component of vascular homeostasis. Exp. Ther. Med. 2016, 12, 1639–1644. [Google Scholar] [CrossRef]
- Calder, P.C. Omega-3 fatty acids and inflammatory processes: From molecules to man. Biochem. Soc. Trans. 2017, 45, 1105–1115. [Google Scholar] [CrossRef]
- Guillou, H.; Zadravec, D.; Martin, P.G.P.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, L.; Schmidt, R.E.; Su, C.; Huang, X.; Gould, K.; Cao, G. Characterization of HSCD5, a novel human stearoyl-CoA desaturase unique to primates. Biochem. Biophys. Res. Commun. 2005, 332, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Goren, M.A.; Fox, B.G. Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expr. Purif. 2008, 62, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Rosenblat, M.; Volkova, N.; Coleman, R.; Aviram, M. Pomegranate Byproduct Administration to Apolipoprotein E-Deficient Mice Attenuates Atherosclerosis Development as a Result of Decreased Macrophage Oxidative Stress and Reduced Cellular Uptake of Oxidized Low-Density Lipoprotein. J. Agric. Food Chem. 2006, 54, 1928–1935. [Google Scholar] [CrossRef] [PubMed]
- Aviram, M.; Volkova, N.; Coleman, R.; Dreher, M.; Reddy, M.K.; Ferreira, D.; Rosenblat, M. Pomegranate Phenolics from the Peels, Arils, and Flowers Are Antiatherogenic: Studies in Vivo in Atherosclerotic Apolipoprotein E-Deficient (E0) Mice and in Vitro in Cultured Macrophages and Lipoproteins. J. Agric. Food Chem. 2008, 56, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Ranucci, D.; Beghelli, D.; Trabalza-Marinucci, M.; Branciari, R.; Forte, C.; Olivieri, O.; Badillo Pazmay, G.V.; Cavallucci, C.; Acuti, G. Dietary effects of a mix derived from oregano (Origanum vulgare L.) essential oil and sweet chestnut (Castanea sativa Mill.) wood extract on pig performance, oxidative status and pork quality traits. Meat Sci. 2015, 100, 319–326. [Google Scholar] [CrossRef]
- Zou, Y.; Xiang, Q.; Wang, J.; Wei, H.; Peng, J. Effects of oregano essential oil or quercetin supplementation on body weight loss, carcass characteristics, meat quality and antioxidant status in finishing pigs under transport stress. Livest. Sci. 2016, 192, 33–38. [Google Scholar] [CrossRef]
- Ishihara, T.; Yoshida, M.; Arita, M. Omega-3 fatty acid-derived mediators that control inflammation and tissue homeostasis. Int. Immunol. 2019, 31, 559–567. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wroblel, N.; Gharbi, K.; Simpson, G.G.; Owe-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef]
- de Tonnac, A.; Guillevic, M.; Mourot, J. Fatty acid composition of several muscles and adipose tissues of pigs fed n-3 PUFA rich diets. Meat Sci. 2018, 140, 1–8. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).