Simple Summary
Morphological conservation has always been a problem in the identification of sibling species. In this study, molecular, karyotype and morphological methods are integrated to revise the species classification and distribution of the Crocidura attenuata species complex in mainland China. The results show that there are five species in the C. attenuata species complex. In addition to four known species, namely, C. attenuata, C. tanakae, C. anhuiensis and C. dongyangjiangensis, there was an undescribed species distributed in Guangxi. Among them, C. tanakae is widely distributed in Southern China, C. attenuata is situated only around the Sichuan Basin, and C. anhuiensis and C. dongyangjiangensis are present in Southeast China. The effectiveness of molecular, karyotypic and morphological methods in the taxonomy of the C. attenuata species complex is also discussed in the present study, which will provide a reference for the taxonomic study of other morphologically conserved species.
Abstract
The conservation of morphology has resulted in considerable issues in the taxonomy of small mammals, especially for the identification of sibling species. Moreover, it is often difficult to completely solve such taxonomic problems by relying only on a single research method. The genus Crocidura is one of the genera with a conservative morphology and high species diversity. Among them, Crocidura attenuata has been considered in the field as the most widely distributed and common species. In fact, it is a species complex containing multiple species, and the classification and distribution of this species is controversial. In this study, the species and distribution of the Crocidura attenuata species complex experienced an integrated revision using three different levels of research methods: molecular, karyotype and morphology. The results show that (1) the C. attenuata species complex contains four known species (C. attenuata, C. tanakae, C. anhuiensis and C. dongyangjiangensis) and a cryptic species distributed in Guangxi, which may be the same undescribed species as the “C. attenuata” distributed in Vietnam. (2) C. attenuata is only distributed around the Sichuan Basin, C. tanakae is the most widely distributed throughout Southern China, and C. anhuiensis and C. dongyangjiangensis are almost sympatric in Southeast China. Furthermore, (3) although the molecular method lacks a unified threshold for species classification, it can rapidly and effectively identify the species of the C. attenuata species complex. Although karyotype and morphology methods cannot completely solve the species classification issues in respect of the C. attenuata species complex, they can provide supplemental information for taxonomic purposes. Therefore, the integrated taxonomic method can present the advantages of different methodological levels, and will provide further evidence for the taxonomy of sibling species with a conservative morphology.
1. Introduction
One of the difficulties of taxonomy is the conservation of morphology among sibling species. However, the use of morphological methods alone results in the underestimation of the species diversity of morphologically conservative groups, especially in small mammals, which leads to species complexes, including cryptic and sibling species [1,2,3,4,5,6]. Therefore, integrated taxonomy based on molecular, cellular and morphological methods has been increasingly applied in the literature to perform the taxonomy of species complexes [7,8,9,10].
The genus of white-toothed shrews, Crocidura, is the most speciose genus of mammals, with 197 recognized species to date [11]. According to the statistics of the mammal diversity database of the American Society of Mammalogists (URL: https://www.mammaldiversity.org/, accessed on 31 December 2022), the genus Crocidura increased to 203 species in 2022. The taxonomy and distribution of Crocidura are complex and inconclusive because of their high diversity level and generally conservative morphology [12,13,14]. In China, the latest catalogue of mammals includes 14 species of Crocidura, comprising of C. anhuiensis, C. attenuata, C. dongyangjiangensis, C. tanakae, C. lasiura, C. shantungensis, C. wuchihensis, C. vorax, C. suaveolens, C. rapax, C. dracula, C. sibirica, C. indochinensis and C. tadae [15]. Although this catalogue contains most species of Crocidura, according to the multilocus molecular result, the species diversity of Crocidura in China remains underestimated in the literature [16].
C. attenuata has always been regarded as the most common species of Crocidura in China and is widely distributed in Southern China and surrounding countries and regions [17]. Due to the high conservation of its morphology, C. attenuata has become a species complex that includes both cryptic and sibling species. One of the most important cryptic species is C. tanakae, which has long been regarded as a Taiwanese subspecies or synonym of C. attenuata due to their extremely similar morphology [18,19,20,21,22,23]. In 2001, C. tanakae was again promoted to the status of an effective species and was only confirmed as distributed in Taiwan based on the chromosome data [24]. Subsequently, the status of C. tanakae as a distinct species was also supported by the molecular data [25]. In recent years, with the extensive application of molecular technology, it has been observed that C. tanakae is widely distributed in Vietnam, Laos and Philippines, as well as Hunan and Guizhou in Southern China [26,27,28,29]. Li et al. [30] revised the distribution of C. attenuata and C. tanakae in mainland China based on molecular methods, and observed that the distribution of C. attenuata is apparently limited to around the Sichuan Basin and Southeast China. The natural range of this species is much smaller than that of C. tanakae, which is distributed almost all over Southern China. Therefore, many specimens of C. attenuata that have been reported to be widely distributed in Southern China and Southeast Asia are in fact C. tanakae. Recently, two new species of Crocidura were reported in the Anhui and Zhejiang Provinces of China, namely C. anhuiensis [31] and C. dongyangjiangensis [32]. They are distributed in areas that have experienced relatively sufficient investigations and are sympatrically distributed with C. attenuata. However, the main reason they have not been described until recently may be that they have been regarded in the literature as the cryptic species of C. attenuata. C. anhuiensis and C. attenuata are sibling species at the molecular level and are very similar in morphology [31]. Although the body size of C. dongyangjiangensis is smaller than that of C. attenuata, it is also similar to C. attenuata in other morphological characteristics, and the relatively low numbers of C. dongyangjiangensis may have been regarded in the literature as the juveniles of C. attenuata. Therefore, we believe that C. attenuata populations widely distributed in Southern China are actually a species complex containing at least C. attenuata, C. tanakae, C. anhuiensis and C. dongyangjiangensis. It is also necessary to revise the ambivalence regarding the species classification and distribution of the C. attenuata species complex.
In this study, molecular, karyotype and morphological methods were integrated to conduct taxonomic research on the C. attenuata species complex to revise its species classification and distribution. Moreover, the effects of different research methods on the taxonomy of related species with a conservative morphology were discussed.
2. Materials and Methods
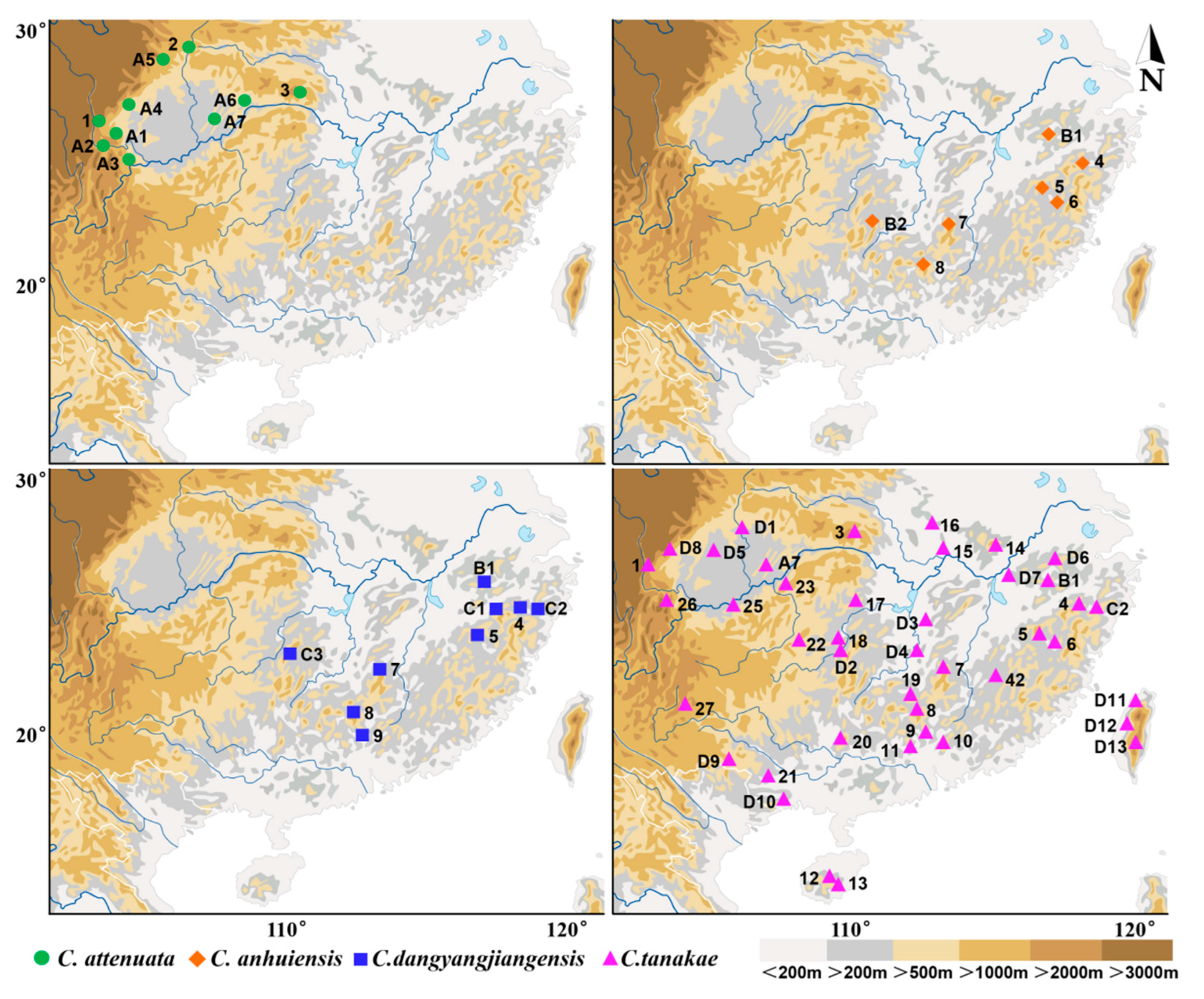
The specimens of the Crocidura attenuata species complex used in this study were obtained using a combination of Sherman live cages and pitfall traps during small-mammal surveys conducted in mainland China from the years 2000 to 2020 (Figure 1). A total of 242 specimens were used in this study, including 53 from C. attenuata, 164 from C. tanakae, 12 from C. anhuiensis and 13 from C. dongyangjiangensis. All specimens were deposited with the Zoological Research Team at Marine College, Shandong University (Weihai), China, and most of the specimens consisted of a carcass (stored in 95% alcohol), dried skin and a cleaned skull. Detailed information on the specimens is listed in Table S1. The field methods followed the relevant Chinese laws, and all animals were handled in a manner consistent with the guidelines approved by the American Society of Mammalogists [33].
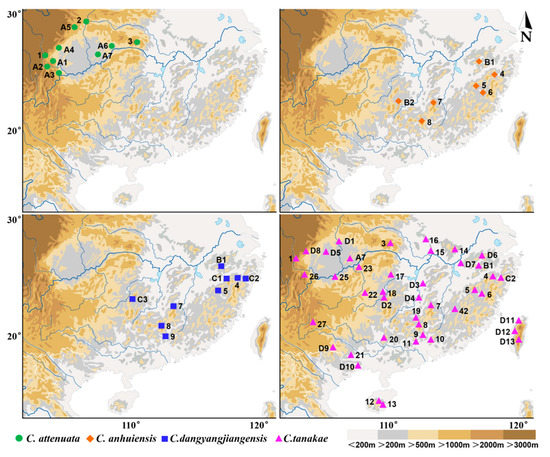
Figure 1.
Specimen locality of Crocidura attenuata species complex in China.
2.1. Molecular Analysis
We amplified and sequenced the complete mitochondrial protein-coding gene cytochrome b (Cytb, 1140 bp) obtained from 65 individuals (19 from C. attenuata, 28 from C. tanakae, 5 from C. anhuiensis and 13 from C. dongyangjiangensis) following the protocol described by Li et al. [30] and using a mammalian universal primer modified by our laboratory, including M13: GTAAAACGACGGCCAGTCCAATGACATGAAAAATCATCGTT and M14: CAGGAAACAGCTATGACTCTCCATTTCTGGTTTACAAGAC. Simultaneously, we used 177 Cytb sequences that had been previously sequenced in our laboratories [30,34]. To revise the species classification and distribution of the C. attenuata species complex, we downloaded 101 available Cytb sequences of the C. attenuata species complex (20 for C. attenuata, 57 for C. tanakae, 6 for C. anhuiensis and 18 for C. dongyangjiangensis) distributed in China for our phylogenetic analyses (Table S1).
The sequences were manually edited in BioEdit v.7.2.5 [35] and aligned and examined using MEGA X software [36]. The best model of sequence evolution estimated by the Bayesian information criterion (BIC) in jModelTest v.2.1.4. [37] was GTR + G + I. We created the phylogenetic trees using the maximum likelihood (ML) in MEGA X software [36] and Bayesian inference (BI) in MrBayes 3.2 [38] based on the sequences of all Cytb haplotypes, with C. Shantungensis and Suncus murinus as the outgroups. The bootstraps were obtained using a rapid bootstrapping algorithm with 1000 replicates. For the Bayesian analysis, we performed four Markov chain Monte Carlo (MCMC) runs with 4 chains for 10,000,000 generations, sampling every 1000 trees and discarding the first 25% as burn-in. The interspecific and intraspecific genetic distances were calculated using the Kimura 2-parameter model implemented in MEGA X software [36].
2.2. Karyotype Analysis
According to the latest taxonomic catalogue of the Crocidura genus in China, the karyotypes of C. attenuata, C. tanakae and C. anhuiensis were reported in our previous studies [34]. Therefore, we only needed to analyze the karyotypes of C. dongyangjiangensis. Preparation of conventional karyotypes was conducted using femoral bone marrow in the field [39,40,41]. CytoVision System (Applied Imaging, Newcastle upon Tyne, UK) Software was used to shoot and analyze chromosomes with good dispersion outcomes in the mitotic phase. Each specimen was observed and counted for 30–80 metaphase cells, and the mode was used to determine the diploid chromosome number (2 n) and fundamental chromosome number (FN). The karyotypes of 5 C. dongyangjiangensis specimens were analyzed, including 3 obtained from Guangdong Province (G09247, G09248 and G12189) and 2 from Zhejiang Province (S1413 and S1431). (Table S1).
2.3. Morphological Analysis
External measurements were taken directly in the field. Four measurements with a small coefficient of variation (CV < 10%) were selected: head and body length (HB), tail length (Tail), ear length (Ear), and hind-foot length with claw (HF). We identified 65 adults (16 from C. attenuata, 30 from C. tanakae, 8 from C. anhuiensis and 11 from C. dongyangjiangensis) for the morphological analysis and excluded individuals with severe skull damage and juveniles. Juveniles were excluded according to the degree of tooth wear and the presence of fused basioccipital and basisphenoid bones [42,43,44,45].
All 20 cranial characters were measured using an electronic digital caliper graduated to 0.01 mm. Among them, the following 13 measurements were identical to those presented by Hutterer et al. [45]: condylo-incisive length (CIL), height of cranial capsule (HCC), rostrum width (RW), maxillary breadth (MB), least interorbital width (IO), greatest width of skull (GW), upper toothrow length (UTR), length of anterior tip of P4 to posterior border of M3 (P4–M3), breadth of palate between the buccal margins of second molars (PW1), postglenoid width (PGL), length of lower molar series (m1–m3), length of mandible from tip of incisor to posterior edge of condyle (ML) and height of coronoid process (COR). Five measurements were identical to those from Meegaskumbura et al. [46]: length of maxillary tooth row (MTR), palatilar length (PAL), postpalatal length (PPL), length of dentary teeth excluding incisors (LDT1), and length of dentary teeth including incisors (LDT2). The remaining two measurements were the same as those presented in the study conducted by Jiang and Hoffmann [12], those being palato-incisor length (PIL) and breadth of coronoid process (BCP).
We calculated the mean and standard deviation of all external and skull morphological measurements, tested a univariate analysis of variance (ANOVA) and multiple comparisons, and conducted principal component analyses (PCAs) using SPSS Statistics 24.0 (SPSS, Chicago, IL, USA). Due to the likely existence of considerable interobserver variations in the external measurements, 3 PCA analyses were conducted based on 4 external, 20 cranial and all 24 morphological measurements.
3. Results
3.1. Sequence, Phylogeny and Genetic Distances
In this study, we successfully amplified the complete 1140-bp mitochondrial Cytb sequence for 65 individuals, with GenBank numbers OP594738-OP594802. Based on the Cytb data previously reported, we obtained 343 complete Cytb sequences for C. attenuata species complexes, which were divided into 132 haplotypes (Table S1).
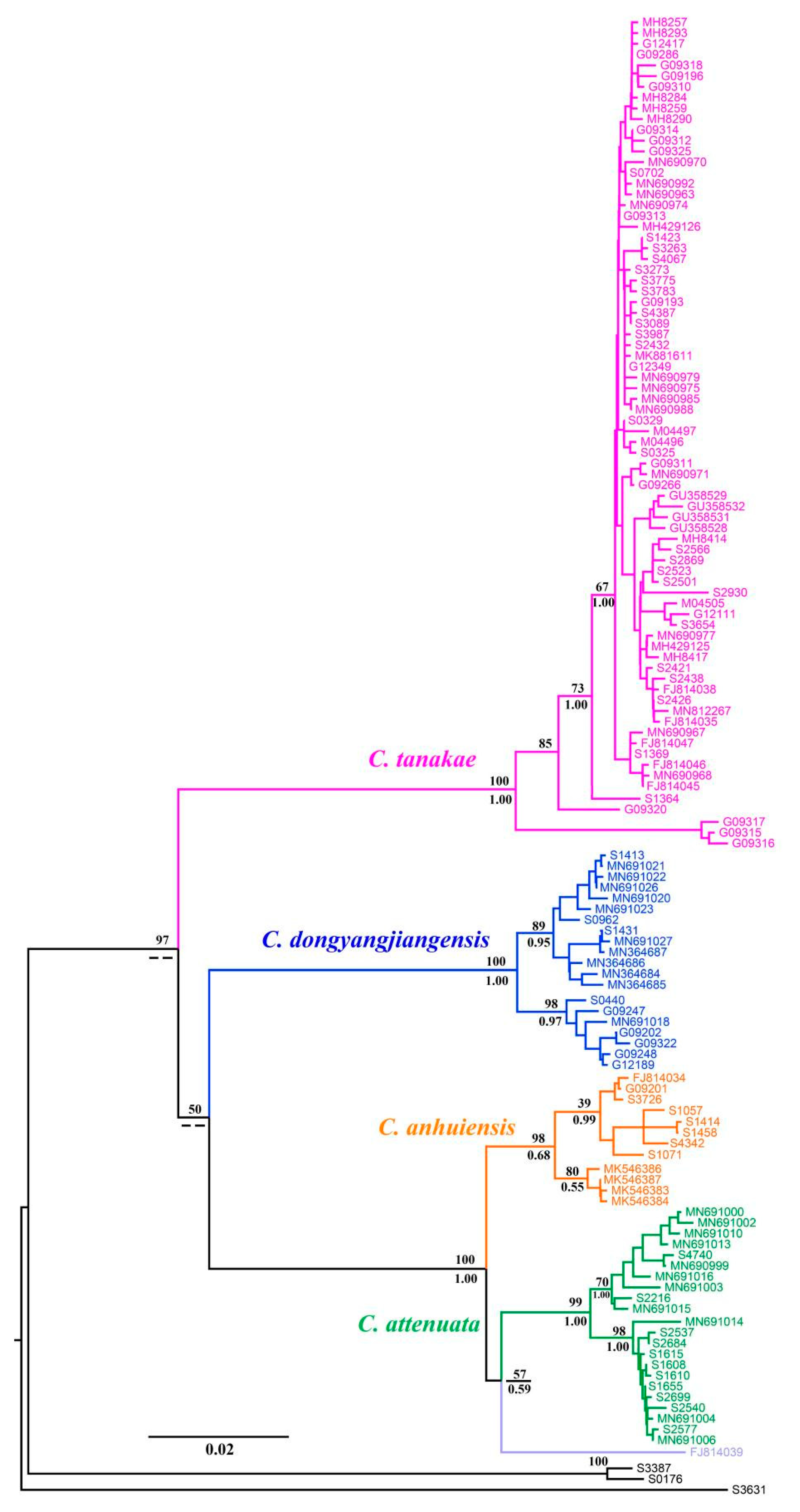
The BI and ML molecular phylogenetic trees presented nearly identical results. Although the phylogenetic tree did not describe the interspecies relationships of C. attenuata species complexes well, it had considerable support at almost the species level (BS = 100, PP = 1.00), with the exception of C. anhuiensis, which ensured the accuracy of the species identification of these specimens (Figure 2). According to the phylogenetic position of the topotype for C. attenuata, and the holotype of C. anhuiensis and C. dongyangjiangensis, we correctly classified the species for the specimens of the C. attenuata species complex and observed that more than half of the specimens belonged to C. tanakae. The four reported species all formed a monophyletic lineage, among which the closest relative to C. attenuata was C. anhuiensis, followed by C. dongyangjiangensis. Significantly, we observed that the samples of C. attenuata distributed in Southeast China in some previous reports were clustered in the same lineage as C. anhuiensis in the phylogenetic tree (Table S1), which also verified that C. anhuiensis has always been a cryptic species of C. attenuata. In addition, the sequence FJ814039 of “C. attenuata” distributed in Guangxi gathered into a single lineage, forming a sister relationship with C. attenuata.
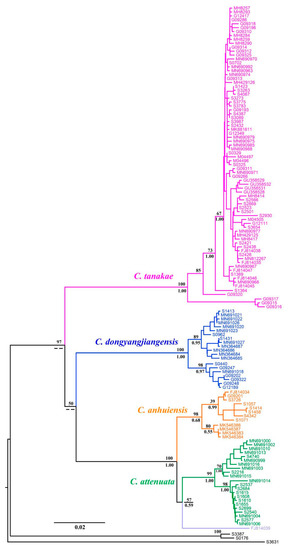
Figure 2.
Phylogenetic tree of C. attenuata species complex based on Cytb gene. Numbers above and below branches represent bootstrap support (BS) of ML and posterior probabilities (PP) of BI, respectively.
The interspecific genetic distance of the four species was greater than 10%, except for C. attenuata and C. anhuiensis, which were smaller (4.1%), while the intraspecific genetic distance of the four species was approximately 1% (Table 1). Therefore, the molecular method can rapidly identify the four species of the C. attenuata species complex.

Table 1.
The interspecific and intraspecific genetic distances (p-distances) of Crocidura attenuata species complex based on mitochondrial cytochrome-b (Cytb) sequence data.
3.2. Karyotype and Karyogram
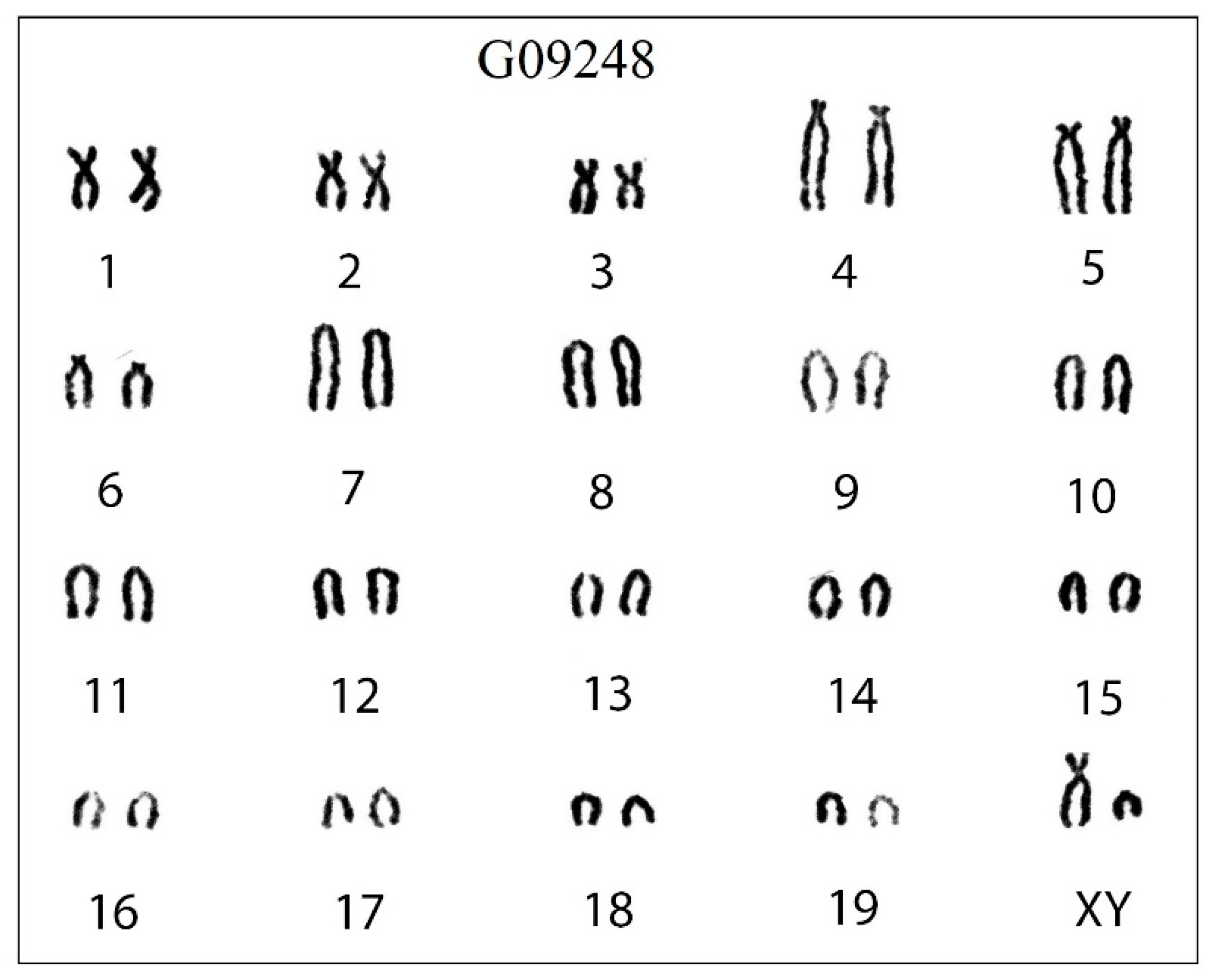
The conventional karyotype of 5 individuals of C. dongyangjiangensis had a uniform arrangement of diploid chromosome number (2 n) = 40 and fundamental chromosome number (FN) = 54, 6(m + sm) + 6 st + 26 t + X(sm) + Y(t), with 3 pairs of metacentric or submetacentric, 3 pairs of subtelocentric, and 13 pairs of acrocentric autosomes, a submetacentric X chromosome, and an acrocentric Y chromosome (Figure 3).
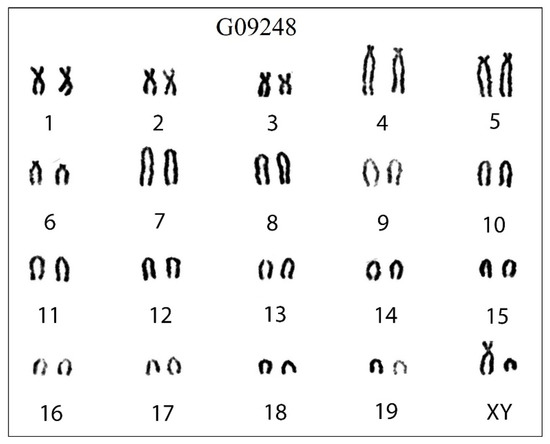
Figure 3.
The karyotype of C. dongyangjiangensis in mainland China. The diploid chromosome number (2 n) is 40 and fundamental arm number (FN) is 54, with three pairs of metacentric or submetacentric (m/sm), three pairs of subtelocentric (st), and 13 pairs of acrocentric autosomes (t), a submetacentric X chromosome (X), and an acrocentric Y chromosome (Y). The order of the chromosomes is m/sm, st, t, X, Y and then from large to small chromosomes. The numbers 1–19 represent the number of chromosome pairs, and G09248 is the label of the specimen.
Li et al. [34] reported that C. tanakae has karyotype polymorphisms and observed 32 karyotypes (2 n = 24−40, FN = 45−56) in 82 individuals, and the karyotypes of 11 C. attenuata were consistent, which means 2 n = 40, FN = 54. According to our molecular results, 5 of the 11 C. attenuata should belong to C. anhuiensis. Therefore, the karyotype of C. dongyangjiangensis is the same as that of C. attenuata and C. anhuiensis, which is consistent with the common karyotype of the genus Crocidura in China (Table S2). Therefore, it is almost impossible to accurately classify the species of the C. attenuata species complex based on the karyotype alone, but some specimens of C. tanakae can be identified by some of its unique karyotypes.
3.3. Metrological Morphology and Morphological Characteristics
The external and skull measurement indices of 65 adult C. attenuata species complex specimens were obtained (Table 2), and almost all measurement indices conformed to a normal distribution (p > 0.05). Therefore, we conducted a univariate analysis of variance (ANOVA) and multiple comparisons on four species of the C. attenuata species complex using parameter statistical analysis. The ANOVA results show that almost all indices, except ear length, present significant morphological differences among the species (Table S3). The results of multiple comparisons show that the measurement indices of C. dongyangjiangensis are significantly smaller than those of the other three species in C. attenuata species complex, belonging to the smaller shrew (Table S4). C. attenuata, C. tanakae and C. anhuiensis are similar in most indices belonging to the medium-sized shrew (Table 2 and Table S4). Overall, among the three species, C. anhuiensis is slightly larger, followed by C. attenuata, and C. tanakae is the smallest. The main performance is that C. anhuiensis is significantly larger than C. tanakae in all indices, except head and body lengths, and is significantly larger than C. attenuata in 15 measurement indices. C. attenuata is more similar to C. anhuiensis in its external morphology, which shows that none of their external indices are significantly different. However, it is closer to C. tanakae in terms of the skull indices because there are 9 skull indices that have no significant differences from C. tanakae and only 4 skull indices that have no differences from C. anhuiensis (Table S4).

Table 2.
External and skull measurement indices of C. attenuata species complex.
Although the statistical analysis determined that there were significant interspecific differences in most of the measurement indices of the C. attenuata species complex, there were many overlaps between species in the range of all the measurement indices. Therefore, according to the numerical value of the measurement indices, it is difficult to identify the species of the C. attenuata species complex, especially C. attenuata, C. tanakae and C. anhuiensis.
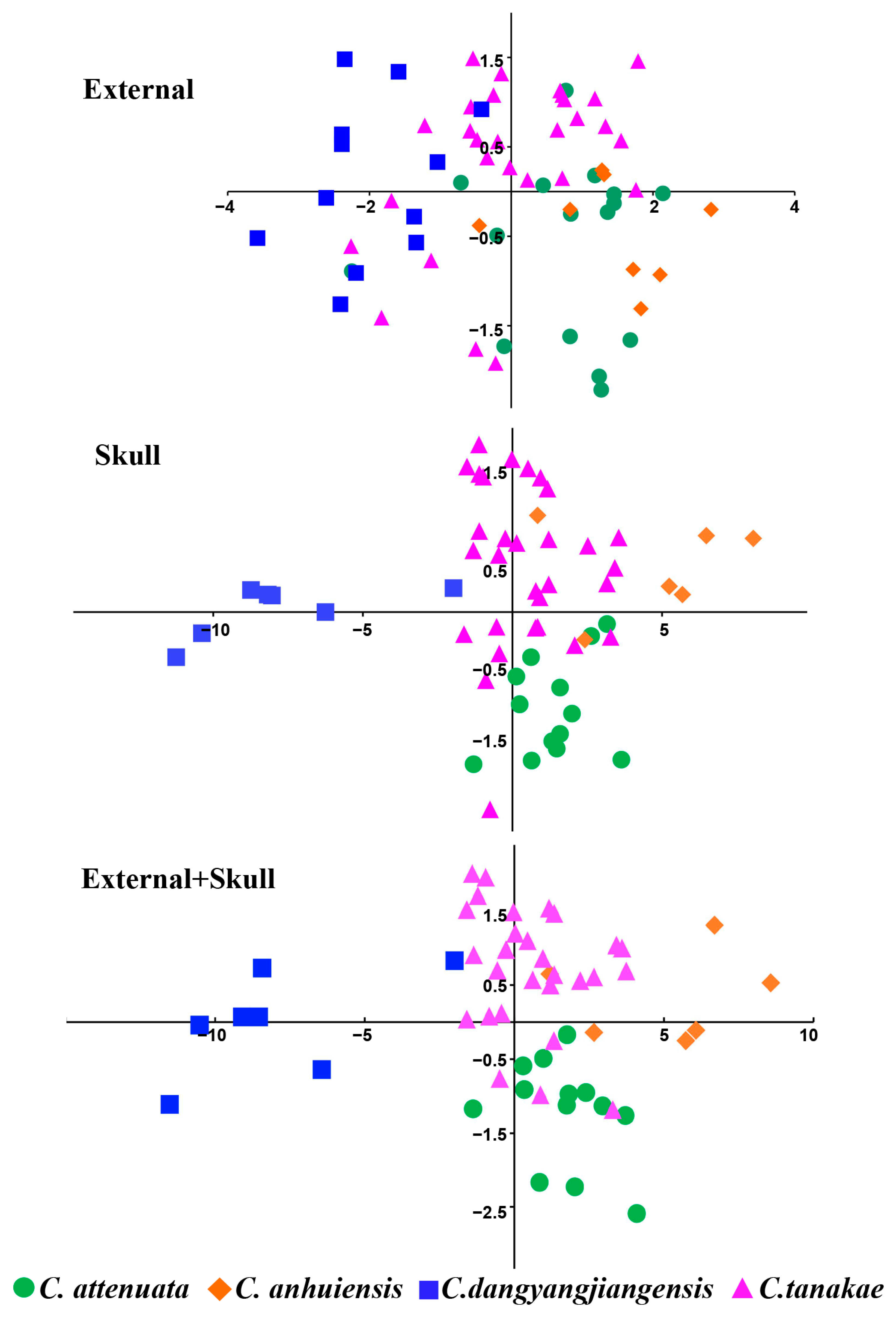
In the three PCA analyses, the first two principal components accounted for 76.37%, 88.48% and 80.91% of the variation based on 4 external, 20 cranial and all 24 morphological measurements, respectively (Table S5). Almost all the variables were strongly correlated with the first principal component, which was mainly related to the size factor, and the skull indices presented a stronger correlation than the external indices. From the main scatter plots of the PCA, it can be observed that the skull indices are better at distinguishing the four species of the C. attenuata species complex, while the external indices are the worst (Figure 4). The external main scatter plots can only distinguish C. dongyangjiangensis from C. attenuata and C. anhuiensis. At the same time, it was observed that the external indices of C. tanakae were highly variable and overlapped with those of the other three species. The results of the skull main scatter plots can completely distinguish C. dongyangjiangensis and can also distinguish most samples of the other three species. The distinguishable effect of the combined indices of skull and external measurement is similar to that of the skull, but the effect is slightly worse. In conclusion, the PCA of the C. attenuata species complex can distinguish the smaller C. dongyangjiangensis, but cannot completely distinguish the other three species.
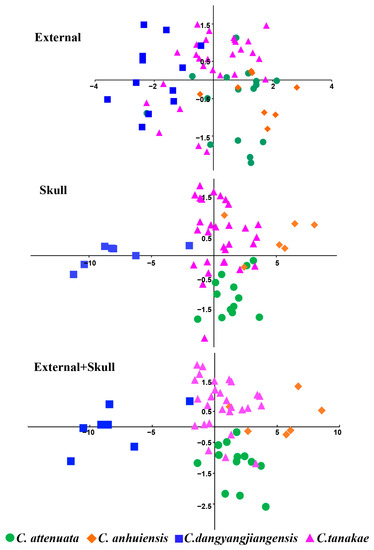
Figure 4.
Principal component plots of C. attenuata species complex.
In terms of the external morphological features, the four species of the C. attenuata species complex are extremely similar, except for C. dongyangjiangensis, and it is difficult to observe significant differences between species (Figure S1). Compared with the other three species, C. dongyangjiangensis is significantly smaller, darker in pelage color and thinner in the tail with shorter hair. The most obvious feature of C. dongyangjiangensis is that the palms and soles of the feet usually feature many small, black granules, while the fore and hind feet of C. attenuata, C. tanakae and C. anhuiensis have little pigmentation, and the palms and soles of the feet are relatively smooth without obvious granular protrusions (Figure S2). Although the somatotype of C. tanakae is similar to that of C. attenuata and C. anhuiensis, there are only differences in its tail and hind-foot lengths (Table 2): the tail (tail/head–body length = 69 ± 8%, n = 28) and hind-foot (12.97 ± 0.65 mm, n = 30) lengths are relatively short, while the tail and hind-foot lengths of C. attenuata and C. anhuiensis are 76 ± 8% (n = 16) and 77 ± 8% (n = 8), 14.19 ± 1.09 mm (n = 16) and 14.76 ± 0.70 mm (n = 8), respectively. However, no obvious morphological characteristics were observed in the external characteristics of C. attenuata and C. anhuiensis.
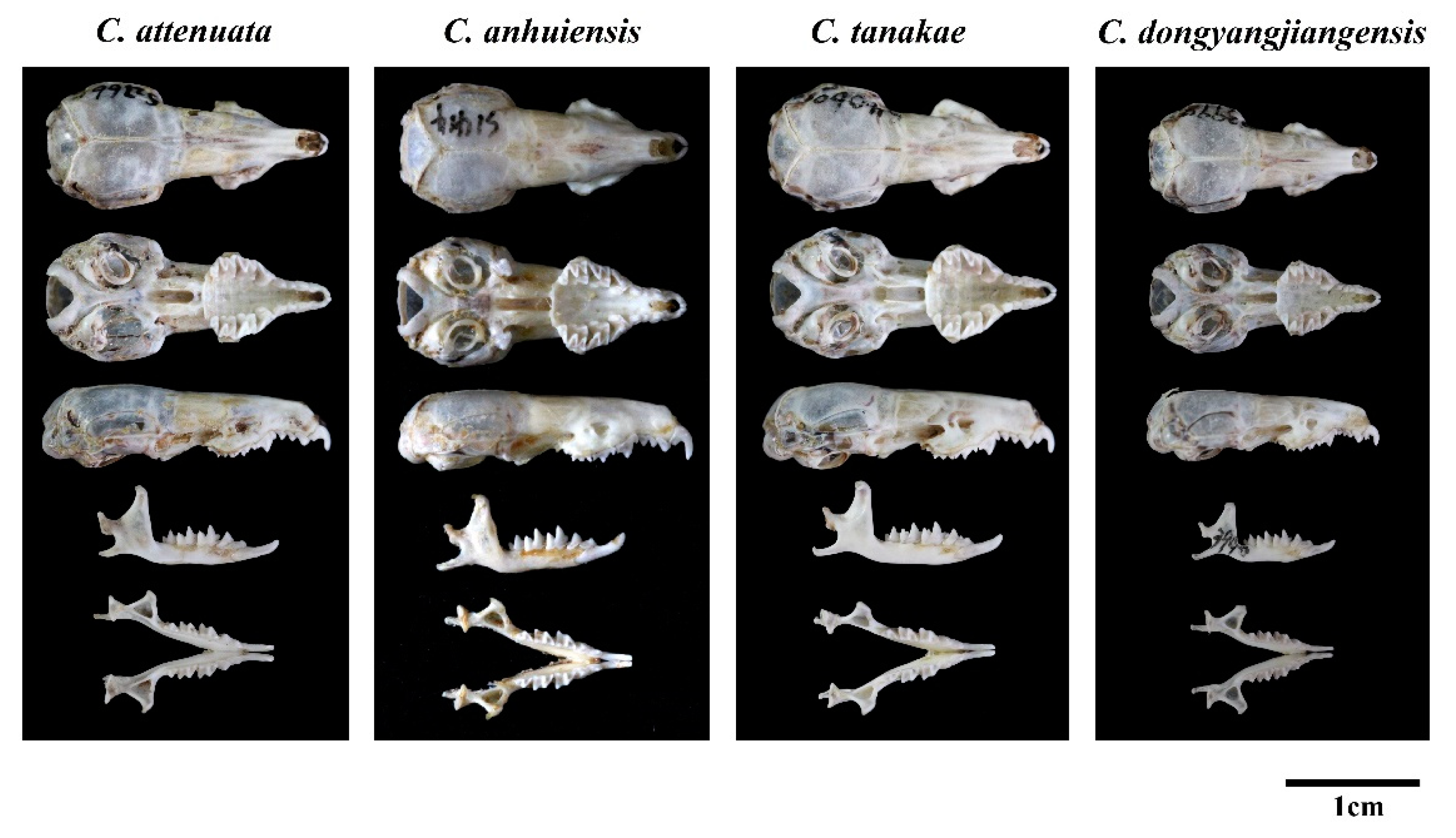
Regarding the characteristics of the skull of C. dongyangjiangensis, it is also significantly smaller, while the sizes of C. attenuata, C. tanakae and C. anhuiensis are similar (Figure 5). In addition, the most upper dentitions of C. dongyangjiangensis is closely spaced (7/9 = 77.8%), especially P4 and U3, so that the anterior margin of P4 covers almost the entire posterior margin of U3, which is closer than C. attenuata, C. tanakae and C. anhuiensis. However, in most specimens of C. attenuata (15/16 = 93.8%), C. tanakae (26/30 = 86.7%) and C. anhuiensis (7/8 = 87.5%), there is a small gap between P4 and U3, or the two just touch. Moreover, the anterolingual margin of P4 in C. dongyangjiangensis is closest to the midline of the skull, and the protocone is almost parallel to the inside of the paracone, while in C. attenuata, C. tanakae and C. anhuiensis, the closest position to the central axis of the skull is the middle lingual margin of P4, and the protocone is in the posterior position of the paracone. The basioccipital region of most C. tanakae specimens is relatively flat and broad, while most of the C. attenuata, C. anhuiensis and C. dongyangjiangensis specimens are similar, and are narrow and ridged. The skulls of C. anhuiensis are stronger than those of the other three species in both the maxilla and mandible, and are significantly larger than those of C. attenuata and C. tanakae in many skull measurement indices (Table S4). Moreover, the suborbital foramina of C. anhuiensis are slightly larger than those of the other three species. However, most of the interspecific morphological differences are based on the majority of the samples, rather than the absolute differences between species.
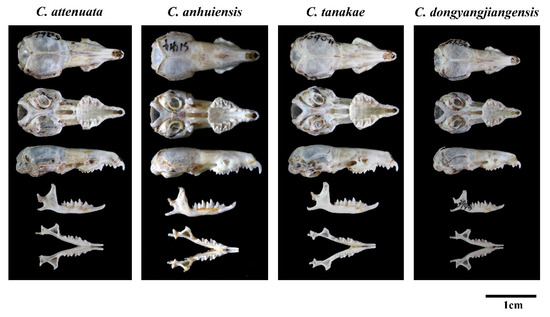
Figure 5.
Skull photos of four species of C. attenuata species complex.
In conclusion, morphological methods can intuitively and effectively identify C. dongyangjiangensis in the C. attenuata species complex, but it is difficult to accurately identify the other three species.
4. Discussion
4.1. Species Classification and Distribution Revision of the C. attenuata Species Complex
In this study, we mainly integrative revised the four species discovered in the C. attenuata species complex using molecular, karyotype and morphological methods. There is no obvious difference in morphology between C. attenuata and C. anhuiensis, the karyotype is also consistent, and the genetic distance between them is less than that between other species (Table S2 and Table 1). However, they are clustered into separate clades, and their interspecific genetic distance (4.1%) is also close to the empirical threshold mammalian Cytb gene interspecific gap of 5% [47,48]. Compared with the smaller intraspecific genetic distance (Table 1), this indicates that there are considerable genetic differences between them, thus supporting C. anhuiensis as a distinct species.
It was also determined in the molecular phylogenetic tree that “C. attenuata” (FJ814039) distributed in Guangxi gathered into a separate clade and formed a sister lineage with C. attenuata (Figure 2). The BLAST results show that the similarity between the sequence and C. attenuata is less than 96%, while the sequence is the most similar (98%) to “C. attenuata” distributed in Vietnam (GU358515; GU358516; AB175082; AB175083; JX181935). According to the latest molecular research results, “C. attenuata” distributed in Vietnam should be an undescribed species [16]. Therefore, the C. attenuata species complex distributed in mainland China includes four reported species, namely, C. attenuata, C. tanakae, C. anhuiensis and C. dongyangjiangensis, as well as a cryptic species distributed in Guangxi.
In mainland China, C. tanakae has been confirmed to be more widely distributed than C. attenuata, which is only found in Western and Southeastern China [30]. However, the sample of C. attenuata in the abovementioned study included some samples of C. anhuiensis, because C. anhuiensis was considered a distinct species in subsequent studies [32]. Therefore, the distribution of C. attenuata needs to be re-evaluated in the literature. Only one report exists, to date, on the distribution of C. anhuiensis, and it is found only in Huangshan in the Anhui Province [32]. C. dongyangjiangensis is reported to be distributed in Chun’an County and Dongyang City in the Zhejiang Province [31]. In addition, a study on the synonyms of C. dongyangjiangensis exists, which also involves its distribution in Huangshan, Anhui Province [49].
In this study, we revised the distribution of four species in mainland China based on the most abundant sample of the C. attenuata species complex (Figure 1). C. attenuata is only distributed around the Sichuan Basin in China, especially in the west and northeast of the Sichuan Basin. C. tanakae is widely distributed in Southern mainland China. C. anhuiensis is distributed in Southeastern China, including Anhui, Zhejiang, Fujian, Jiangxi, Hunan and Guangdong Provinces. The distribution of C. dongyangjiangensis is basically sympatric with that of C. anhuiensis, including Anhui, Zhejiang, Jiangxi, Hunan and Guangdong Provinces.
4.2. The Confusion in the Taxonomy of the Crocidura attenuata Species Complex
In the present study, the revised distribution area of C. attenuata is limited to only around the Sichuan Basin. Therefore, the widely distributed “C. attenuata” mentioned in previous reports may be C. anhuiensis, C. tanakae or other cryptic species, and the results of the C. attenuata species complex in these reports also require further modifications. For example, (1) in our previous study conducted on the distribution and karyotype of C. attenuata [30,34], some samples of “C. attenuata” should have been C. anhuiensis, which have been corrected in this study (Table S1). In addition, (2) regarding the morphological identification characteristics of C. attenuata [50] and the description of the morphological differences between C. attenuata and C. tanakae shrews [30,51], the samples of C. attenuata used were mixed with C. anhuiensis and the cryptic species in Vietnam, so these results also need to be verified further. Furthermore, (3) Zhang et al. [31] described a new species of C. anhuiensis obtained from China based on molecular and morphological data. In their report, the molecular specimen of “C. attenuata” was a cryptic species collected from Vietnam, and the molecular specimen of “C. attenuata” should have been C. tanakae, which is sympatrically distributed with C. anhuiensis. Although the taxonomic results of C. anhuiensis in this study are questionable, our results support the status of C. anhuiensis as a valid species. In addition, other studies have been conducted on the misuse of specimens involving the C. attenuata species complex [24,26,28,46,49], which have not been presented.
The two main reasons leading to the taxonomic confusion are as follows: first, most of the previous research only relied on morphology, which led to the misclassification of some species that were very morphologically similar. The second reason is that some small mammals lack extensive attention, leading to insufficient field investigations. Therefore, we appeal increasing the investigation and protection of these under-researched animals, and minimizing the harm to these animals during research [52].
4.3. The Taxonomic Effectiveness of Molecular, Karyotype and Morphological Methods
For the integrative taxonomic case of the C. attenuata species complex, only the molecular method can rapidly and effectively identify the species because all species can gather into an independent clade in the phylogenetic tree and obtain considerable support. The interspecific genetic distance also supports the valid species status of each species.
Although the karyotypic method is considered to be the most consistent research method for the concept of species and plays an important role in many taxonomic studies [7,8,53], it does not play a significant role in the taxonomy of the C. attenuata species complex and can only classify some specimens of C. tanakae according to its unique karyotype. There are two opposite extremes in the karyotype of the genus Crocidura in China. One is that the karyotype of the genus is conservative. This is to say that most species in the genus have the same karyotype (2 n = 40, FN = 54), and C. attenuata, C. anhuiensis and C. dongyangjiangensis all belong to this karyotype (Table S2). However, the karyotypes in C. tanakae showed a high degree of polymorphism [34]. Therefore, it is inevitable to erroneously perform the species identification of the C. attenuata species complex based only on the karyotype method. For example, C. tanakae was promoted to a distinct species based on the karyotype difference between C. attenuata and C. tanakae [24], and this karyotype difference has been proven to be the result of a karyotype polymorphism occurring in C. tanakae [34]. In small mammals, karyotype polymorphisms may be a relatively common situation [54], which also poses a challenge to taxonomy based on karyotypes.
Morphological methods have always been considered the most intuitive and effective classical taxonomic methods, but some difficulties remain in the taxonomy of the C. attenuata species complex with a conservative morphology. In this study, we observed that with the increase in the sample size and distribution range of species, some interspecific morphological differences reported in previous studies have become intraspecific variations, especially the morphological differences reported among C. attenuata, C. tanakae and C. anhuiensis [30,31,52]. There are obvious molecular differences among species of the C. attenuata species complex, but there are no obvious interspecific morphological differences. We speculated that the reason for this was that Crocidura has experienced rapid evolution [16], which led to species differentiation without synchronous morphological variations [55].
5. Conclusions
In conclusion, there may be five species of the C. attenuata species complex distributed in mainland China, comprising of C. attenuata, C. tanakae, C. anhuiensis, C. dongyangjiangensis and a cryptic species distributed in Guangxi. The four reported species were revised and we observed that C. attenuata was distributed in the northeast and west around the Sichuan Basin in Western China, while C. tanakae was distributed throughout the Southern mainland of China. The distribution ranges of C. anhuiensis and C. dongyangjiangensis almost overlapped and were distributed in Southeast China. In the taxonomy of the sibling species with a conservative morphology, such as the C. attenuata species complex, molecular methods can often play an effective role and rapidly produce desirable results. Although the molecular classification threshold is not uniform, the integrated taxonomy combined with morphology, karyotype or other research methods will achieve more comprehensive and accurate results.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani13040643/s1, Figure S1: External morphological characters of the four species of C. attenuata species complex; Figure S2: Hindfoot characteristics of the four species of C. attenuata species complex; Table S1: Sample information of Chinese Crocidura attenuata species complex used in Current study; Table S2: Karyotype of the C. attenuata species complex and other reported Crocidura species in China; Table S3: One-way ANOVA among four species of C. attenuata species complex; Table S4: The interspecific comparison of morphological measurement indexes among four species of C. attenuata species complex; Table S5: Factor loading matrix of measurement indicators of C. attenuata species complex.
Author Contributions
Conceptualization, Y.L. (Yuchun Li) and H.L.; methodology, H.L., M.M. and M.H.; software, H.L.; investigation, Y.L. (Yaoyao Li) and Y.W.; resources, Y.L. (Yuchun Li), Y.L. (Yaoyao Li), Y.W., M.M. and M.H.; writing—original draft preparation, H.L.; writing—review and editing, H.L. and Y.L. (Yuchun Li); funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32270480, 31672254).
Institutional Review Board Statement
The sampling and experiment procedures complied with internationally recognized standards and were approved by the Special Committee on Scientific Research Ethic of Liaocheng University (Licence No. 2023020401).
Informed Consent Statement
Not applicable.
Data Availability Statement
All the sequences used in this study were accessed through the GenBank database and the accession numbers are listed in Table S1. Morphological specimens and chromosome materials were deposited in the Zoological Research Team at Marine College, Shandong University (Weihai).
Acknowledgments
We want to acknowledge all researchers who participated in the sample collection, and thank the National Center for Biotechnology Information (NCBI) Database for providing the molecular data.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- Wan, T.; He, K.; Jiang, X.L. Multilocus Phylogeny and Cryptic Diversity in Asian Shrew-like Moles (Uropsilus, Talpidae): Implications for Taxonomy and Conservation. BMC Evol. Biol. 2013, 13, 232. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Shinohara, A.; Helgen, K.M.; Springer, M.S.; Jiang, X.-L.; Campbell, K.L. Talpid Mole Phylogeny Unites Shrew Moles and Illuminates Overlooked Cryptic Species Diversity. Mol. Biol. Evol. 2017, 34, 78–87. [Google Scholar] [CrossRef]
- Koju, N.P.; He, K.; Chalise, M.K.; Ray, C.; Chen, Z.; Zhang, B.; Wan, T.; Chen, S.; Jiang, X. Multilocus Approaches Reveal Underestimated Species Diversity and Inter-Specific Gene Flow in Pikas (Ochotona) from Southwestern China. Mol. Phylogenet. Evol. 2017, 107, 239–245. [Google Scholar] [CrossRef]
- Li, H.; Kong, L.; Wang, K.; Zhang, S.; Motokawa, M.; Wu, Y.; Wang, W.; Li, Y. Molecular Phylogeographic Analyses and Species Delimitations Reveal That Leopoldamys Edwardsi (Rodentia: Muridae) Is a Species Complex. Integr. Zool. 2019, 14, 494–505. [Google Scholar] [CrossRef]
- Bugarski-Stanojević, V.; Stamenković, G.; Jojić, V.; Ćosić, N.; Ćirović, D.; Stojković, O.; Veličković, J.; Savić, I. Cryptic Diversity of the European Blind Mole Rat Nannospalax Leucodon Species Complex: Implications for Conservation. Animals 2022, 12, 1097. [Google Scholar] [CrossRef]
- Yu, W.; Lin, C.; Huang, Z.; Liu, S.; Wang, Q.; Quan, R.; Li, S.; Wu, Y. Discovery of Kerivoula Kachinensis and a Validity of K. Titania (Chiroptera: Vespertilionidae) in China. Mammalia 2022, 86, 303–308. [Google Scholar] [CrossRef]
- Huang, C.; Yu, W.; Xu, Z.; Qiu, Y.; Chen, M.; Qiu, B.; Motokawa, M.; Harada, M.; Li, Y.; Wu, Y. A Cryptic Species of the Tylonycteris Pachypus Complex (Chiroptera: Vespertilionidae) and Its Population Genetic Structure in Southern China and Nearby Regions. Int. J. Biol. Sci. 2014, 10, 200–211. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Li, H.; Wang, J.; Rong, X.; Li, Y. Niviventer Confucianus Sacer (Rodentia, Muridae) Is a Distinct Species Based on Molecular, Karyotyping, and Morphological Evidence. Zookeys 2020, 959, 137–159. [Google Scholar] [CrossRef]
- Lim, K.C.; White, W.T.; Then, A.Y.H.; Naylor, G.J.P.; Arunrugstichai, S.; Loh, K.-H. Integrated Taxonomy Revealed Genetic Differences in Morphologically Similar and Non-Sympatric Scoliodon Macrorhynchos and S. Laticaudus. Animals 2022, 12, 681. [Google Scholar] [CrossRef]
- Liu, S.; Zhou, C.; Meng, G.; Wan, T.; Tang, M.; Yang, C.; Murphy, R.W.; Fan, Z.; Liu, Y.; Zeng, T.; et al. Evolution and Diversification of Mountain Voles (Rodentia: Cricetidae). Commun. Biol. 2022, 5, 1417. [Google Scholar] [CrossRef] [PubMed]
- Burgin, C.J.; He, K. Family Soricidae (shrew). In Hand Book of the Mammals of the World: Insectivores, Sloths and Colugos; Wilson, D.E., Russell, A.M., Eds.; Barcelona: Lynx Edicions, Spain, 2018; pp. 474–530. [Google Scholar]
- Jiang, X.; Hoffmann, R.S. A revision of the white-toothed shrews (Crocidura) of Southern China. J. Mammal. 2001, 82, 1059–1079. [Google Scholar] [CrossRef]
- Stanley, W.T.; Hutterer, R.; Giarla, T.C.; Esselstyn, J.A. Phylogeny, Phylogeography and Geographical Variation in the Crocidura Monax (Soricidae) Species Complex from the Montane Islands of Tanzania, with Descriptions of Three New Species. Zool. J. Linn. Soc. 2015, 174, 185–215. [Google Scholar] [CrossRef]
- Nicolas, V.; Jacquet, F.; Hutterer, R.; Konečný, A.; Kouassi, S.K.; Durnez, L.; Lalis, A.; Colyn, M.; Denys, C. Multilocus Phylogeny of the Crocidura Poensis Species Complex (Mammalia, Eulipotyphla): Influences of the Palaeoclimate on Its Diversification and Evolution. J. Biogeogr. 2019, 46, 871–883. [Google Scholar] [CrossRef]
- Wei, F.; Yang, Q.; Wu, Y.; Jiang, X.; Liu, S.; Li, B.; Yang, G.; Li, M.; Zhou, J.; Li, S.; et al. Catalogue of mammals in China (2021). Acta Theriol. Sin. 2021, 41, 487–501. [Google Scholar]
- Chen, S.; Qing, J.; Liu, Z.; Liu, Y.; Tang, M.; Murphy, R.W.; Pu, Y.; Wang, X.; Tang, K.; Guo, K.; et al. Multilocus Phylogeny and Cryptic Diversity of White-Toothed Shrews (Mammalia, Eulipotyphla, Crocidura) in China. BMC Evol. Biol. 2020, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Hutterer, R. Order Soricomorpha. In Mammal Species of the World a Taxonomic and Geographic Reference, 3rd ed.; Wilson, D.E., Reeder, D.M., Eds.; The Johns Hopkins University Press: Baltimore, MD, USA, 2005. [Google Scholar]
- Ellerman, J.; Morrison-Scott, T. Checklist of Palaearctic and Indian Mammals, 1758 to 1946; British Museum, Natural History: London, UK, 1951. [Google Scholar]
- Jameson, E.; Jones, G. The Soricidae of Taiwan. Proc. Biol. Soc. Wash. 1977, 90, 459–482. [Google Scholar]
- Corbet, G.B.; Hill, J.E. The Mammals of the Indomalayan Region: A Systematic Review; Natural History Museum publications; Oxford University Press: Oxford, UK, 1992. [Google Scholar]
- Hutterer, R. Order Insectivora. In Mammal Species of the World: A Taxonomic and Geographic Reference, 2nd ed.; Wilson, D.E., Reeder, D.M., Eds.; Smithsonian Institution Press: Washington, DC, USA, 1993; pp. 69–130. [Google Scholar]
- Fang, Y.; Lee, L.; Yew, F.; Yu, H. Systematics of White-toothed Shrews (Crocidura) (Mammalia: Insectivora: Soricidae) of Taiwan: Karyological and Morphological Studies. J. Zool. 1997, 242, 151–166. [Google Scholar] [CrossRef]
- Motokawa, M.; Harada, M.; Lin, L.K.; Koyasu, K.; Hattori, S. Karyological Study of the Gray Shrew Crocidura attenuata (Mammalia: Insectivora) from Taiwan. Zool. Stud. 1997, 36, 70–73. [Google Scholar]
- Motokawa, M.; Harada, M.; Wu, Y.; Lin, L.-K.; Suzuki, H. Chromosomal Polymorphism in the Gray Shrew Crocidura Attenuata (Mammalia: Insectivora). Zool. Sci. 2001, 18, 1153–1160. [Google Scholar] [CrossRef]
- Ohdachi, S.D.; Hasegawa, M.; Iwasa, M.A.; Vogel, P.; Oshida, T.; Lin, L.K.; Abe, H. Molecular Phylogenetics of Soricid Shrews (Mammalia) Based on Mitochondrial Cytochrome b Gene Sequences: With Special Reference to the Soricinae. J. Zool. 2006, 270, 177–191. [Google Scholar] [CrossRef]
- Esselstyn, J.A.; Timm, R.M.; Brown, R.M. Do Geological or Climatic Processes Drive Speciation in Dynamic Archipelagos? The Tempo and Mode of Diversification in Southeast Asian Shrews. Evolution 2009, 63, 2595–2610. [Google Scholar] [CrossRef]
- Esselstyn, J.A.; Oliveros, C.H. Colonization of the Philippines from Taiwan: A Multi-Locus Test of the Biogeographic and Phylogenetic Relationships of Isolated Populations of Shrews. J. Biogeogr. 2010, 37, 1504–1514. [Google Scholar] [CrossRef]
- Bannikova, A.A.; Abramov, A.V.; Borisenko, A.V.; Lebedev, V.S.; Rozhnov, V.V. Mitochondrial Diversity of the White-Toothed Shrews (Mammalia, Eulipotyphla, Crocidura) in Vietnam. Zootaxa 2011, 2812, 1–20. [Google Scholar] [CrossRef]
- Abramov, A.V.; Bannikova, A.A.; Rozhnov, V.V. White-Toothed Shrews (Mammalia, Soricomorpha, Crocidura) of Coastal Islands of Vietnam. Zookeys 2012, 207, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, H.; Motokawa, M.; Wu, Y.; Harada, M.; Sun, H.; Mo, X.; Wang, J.; Li, Y. A Revision of the Geographical Distributions of the Shrews Crocidura tanakae and C. attenuata Based on Genetic Species Identification in the Mainland of China. Zookeys 2019, 869, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, G.Y.; Wu, Y.Q.; Yao, J.F.; You, S.; Wang, C.C.; Cheng, F.; Chen, J.J.; Tang, M.X.; Li, C.L.; et al. A new species of the genus Crocidura from China based on molecular and morphological data (Eulipotyphla: Soricidae). Zool. Syst. 2019, 4, 279–293. [Google Scholar]
- Liu, Y.; Chen, S.; Liu, B.; Liao, R.; Liu, Y.; Liu, S. A New Species of the Genus Crocidura (Eulipotyphla: Soricidae) from Zhejiang Pvovince, Eastern China. Acta Theriol. Sin. 2020, 40, 1–12. [Google Scholar]
- Sikes, R.S. Animal Care and Use Committee of the American Society of Mammalogists 2016 Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research and Education. J. Mammal. 2016, 97, 663–688. [Google Scholar] [CrossRef]
- Li, H.; Mo, X.; Sun, H.; Wang, J.; Motokawa, M.; Harada, M.; Wu, Y.; Li, Y. Karyotypic Polymorphism of Crocidura tanakae (Eulipotyphla: Soricidae) and Revision of the Karyotype of C. attenuata in Mainland China. J. Mammal. 2020, 101, 1548–1560. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Patton, J.L. Chromosome Studies of Certain Pocket Mice, Genus Perognathus (Rodentia: Heteromyidae). J. Mammal. 1967, 48, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Harada, M.; Yosida, T.H. Karyological Study of Four Japanese Myotis Bats (Chiroptera, Mammalia). Chromosoma 1978, 65, 283–291. [Google Scholar] [CrossRef]
- Searle, J.B. Factors Responsible for a Karyotypic Polymorphism in the Common Shrew, Sorex araneus. Proc. R. Soc. Lond. B Biol. Sci. 1986, 229, 277–298. [Google Scholar]
- Kitchener, D.J.; Schmitt, L.H. Maharadatunkamsi Morphological and Genetic Variation in Suncus murinus (Soricidae: Crocidurinae) from Java, Lesser Sunda Islands, Maluku and Sulawesi, Indonesia. Mammalia 1994, 58, 433–451. [Google Scholar] [CrossRef]
- Motokawa, M.; Hattori, S.; Ota, H.; Hikida, T. Geographic Variation in the Watase’s Shrew Crocidura watasei (Insectivora, Soricidae) from the Ryukyu Archipelago, Japan. Mammalia 1996, 60, 243–254. [Google Scholar] [CrossRef]
- Motokawa, M. Geographic Variation in the Japanese White-Toothed Shrew Crocidura dsinezumi. Acta Theriol. 2003, 48, 145–156. [Google Scholar] [CrossRef]
- Hutterer, R.; Balete, D.S.; Giarla, T.C.; Heaney, L.R.; Esselstyn, J.A. A New Genus and Species of Shrew (Mammalia: Soricidae) from Palawan Island, Philippines. J. Mammal. 2018, 99, 518–536. [Google Scholar] [CrossRef]
- Meegaskumbura, S.; Meegaskumbura, M.; Pethiyagoda, R.; Manamendra-Arachchi, K.; Schneider, C.J. Crocidura hikmiya, a New Shrew (Mammalia: Soricomorpha: Soricidae) from Sri Lanka. Zootaxa 2007, 1665, 19–30. [Google Scholar]
- Bradley, R.D.; Baker, R.J. A test of the Genetic Species Concept: Cytochrome-b sequences and mammals. J. Mammal. 2001, 82, 960–973. [Google Scholar] [CrossRef]
- Baker, R.J.; Bradley, R.D. Speciation in mammals and the genetic species concept. J. Mammal. 2006, 87, 643–662. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, H.; Zhang, C.L.; Wu, J.; Wang, Z.C.; Li, C.L.; Zhang, B.W. A new species of the genus Crocidura (Mammalia: Eulipotyphla: Soricidae) from Mount Huang, China. Zool. Syst. 2020, 45, 1–14. [Google Scholar]
- Jenkins, P.D.; Lunde, D.P.; Moncrieff, C.B. Chapter 10. Descriptions of New Species of Crocidura (Soricomorpha: Soricidae) from Mainland Southeast Asia, with Synopses of Previously Described Species and Remarks on Biogeography. Bull. Am. Mus. Nat. Hist. 2009, 331, 356–405. [Google Scholar] [CrossRef]
- Jenkins, P.D.; Abramov, A.V.; Bannikova, A.A.; Rozhnov, V.V. Bones and Genes: Resolution Problems in Three Vietnamese Species of Crocidura (Mammalia, Soricomorpha, Soricidae) and the Description of an Additional New Species. Zookeys 2013, 313, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Russo, D.; Ancillotto, L.; Hughes, A.C.; Galimberti, A.; Mori, E. Collection of Voucher Specimens for Bat Research: Conservation, Ethical Implications, Reduction, and Alternatives. Mammal. Review. 2017, 47, 237–246. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Harada, M.; Lin, L.-K.; Motokawa, M. Karyotypes of Three Rat Species (Mammalia: Rodentia: Muridae) from Hainan Island, China, and the Valid Specific Status of Niviventer Lotipes. Zoolog Sci. 2008, 25, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Zima, J. Chromosomal Evolution in Small Mammals (Insectivora, Chiroptera, Rodentia). Hystrix Ital. J. Mammal. 2000, 11, 5–15. [Google Scholar]
- Adams, D.C.; Berns, C.M.; Kozak, K.H.; Wiens, J.J. Are Rates of Species Diversification Correlated with Rates of Morphological Evolution? Proc. Biol. Sci. 2009, 276, 2729–2738. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Z.; Tang, H.Y.; Tang, X.F.; Liu, M.W.; Man, X.M.; Zhao, H.T.; Wu, X.B.; Wu, H.L. Discovery of Crocidura tanakae (Mammalia: Soricidae) in Huangshan and Xuancheng, Anhui Province. Chin. J. Zool. 2019, 54, 815–819. [Google Scholar]
- Lei, B.; Yue, Y.; Cui, J.; Ji, S.; Yu, W.; Han, W.; Zhou, Y. A New Record of the Taiwanese Gray Shrew (Crocidura Tanakae Kuroda, 1938) in Hubei Province. Acta Theriol. Sin. 2019, 39, 218–223. [Google Scholar]
- Tu, Y.F. Taiwanese Gray Shrew (Crocidura tanakae) Found in Taohongling National Nature Reserve, Jiangxi. Chin. J. Zool. 2020, 55, 681–682. [Google Scholar]
- Chen, S.; Zhang, Q.; Li, F.; Wang, X.; Wang, Q.; Liu, S. A New Record of Crocidura tanakae kuroda, 1938 in Sichuan and Guizhou Provinces. Acta Theriol. Sin. 2018, 38, 211–216. [Google Scholar]
- Iwasa, M.A.; Ohdachi, S.; Han, S.H.; Oh, H.S.; Abe, H.; Suzuki, H. Karyotype and RFLP of the Nuclear RDNA of Crocidura Sp. on Cheju Island, South Korea (Mammalia, Insectivora). Mammalia 2001, 65, 451–459. [Google Scholar] [CrossRef]
- Zima, J.; Lukácová, L.; Macholán, M. Chromosomal evolution in shrews. In Evolution of Shrews; Wójcik, J.M., Wolsan, M., Eds.; Mammal Research Institute, Polish Academy of Sciences: Bialystok, Poland, 1998; pp. 175–218. [Google Scholar]
- Ruedi, M.; Vogel, P. Chromosomal Evolution and Zoogeographic Origin of Southeast Asian Shrews (Genus Crocidura). Experientia 1995, 51, 174–178. [Google Scholar] [CrossRef]
- Grafodatskiĭ, A.S.; Radzhabli, S.I.; Sharshov, A.V.; Zaĭtsev, M.V. Karyotypes of 5 Species of Soricidae-Crocidura in the Fauna of the USSR. Tsitologiia 1988, 30, 1247–1250. [Google Scholar]
- Maddalena, T.; Ruedi, M. Chromosomal evolution in the genus Crocidura (Insectivora: Soricidae). Adv. Biol. Shrews. Spec. Publ. Carnegie Mus. Nat. Hist. 1994, 18, 335–344. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).