Assessing the Effects of Dietary Cadmium Exposure on the Gastrointestinal Tract of Beef Cattle via Microbiota and Transcriptome Profile
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Feeds, and Experimental Design
2.2. Growth Performance, Morphology, Serum Parameters, and Cadmium Accumulation
2.3. DNA Extraction and Real-Time PCR
2.4. Amplicon Sequencing
2.5. Metataxonomic Data Processing and Analysis
2.6. RNA Isolation and Transcriptome Data Processing and Analysis
2.7. Statistical Analysis
3. Results
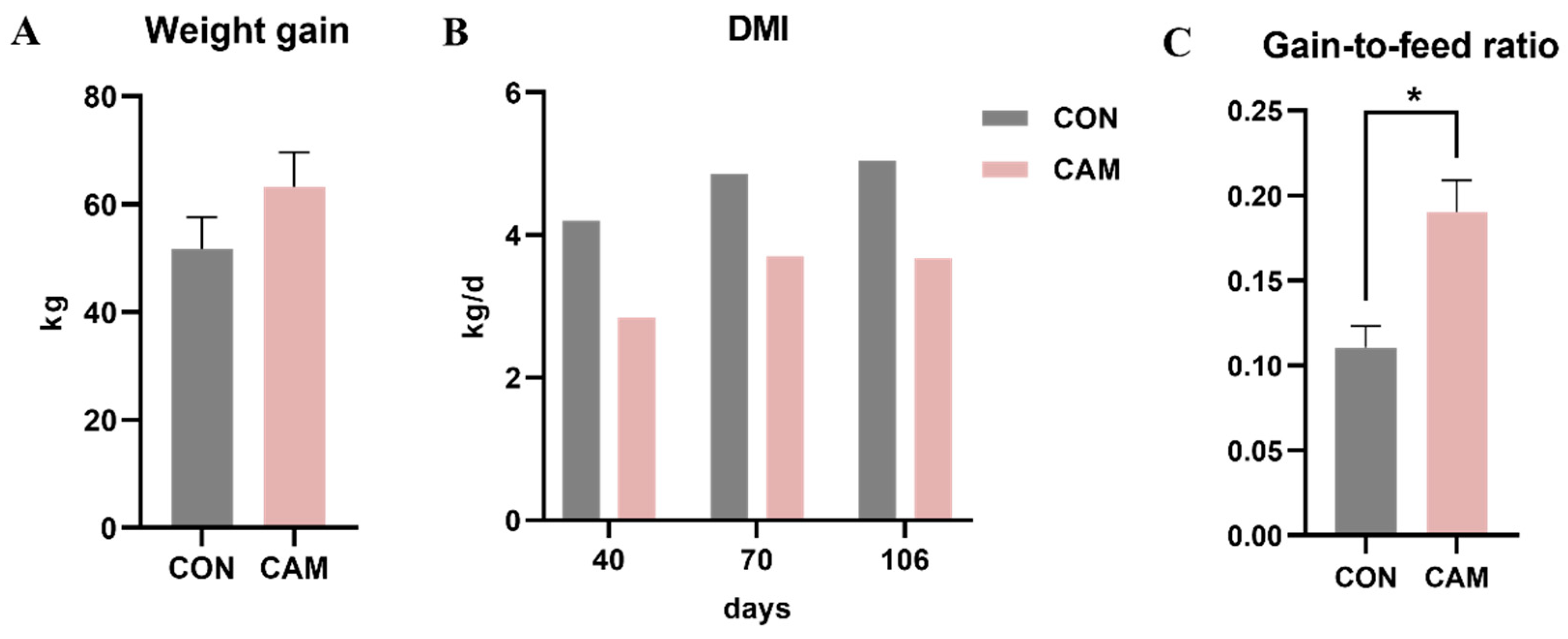
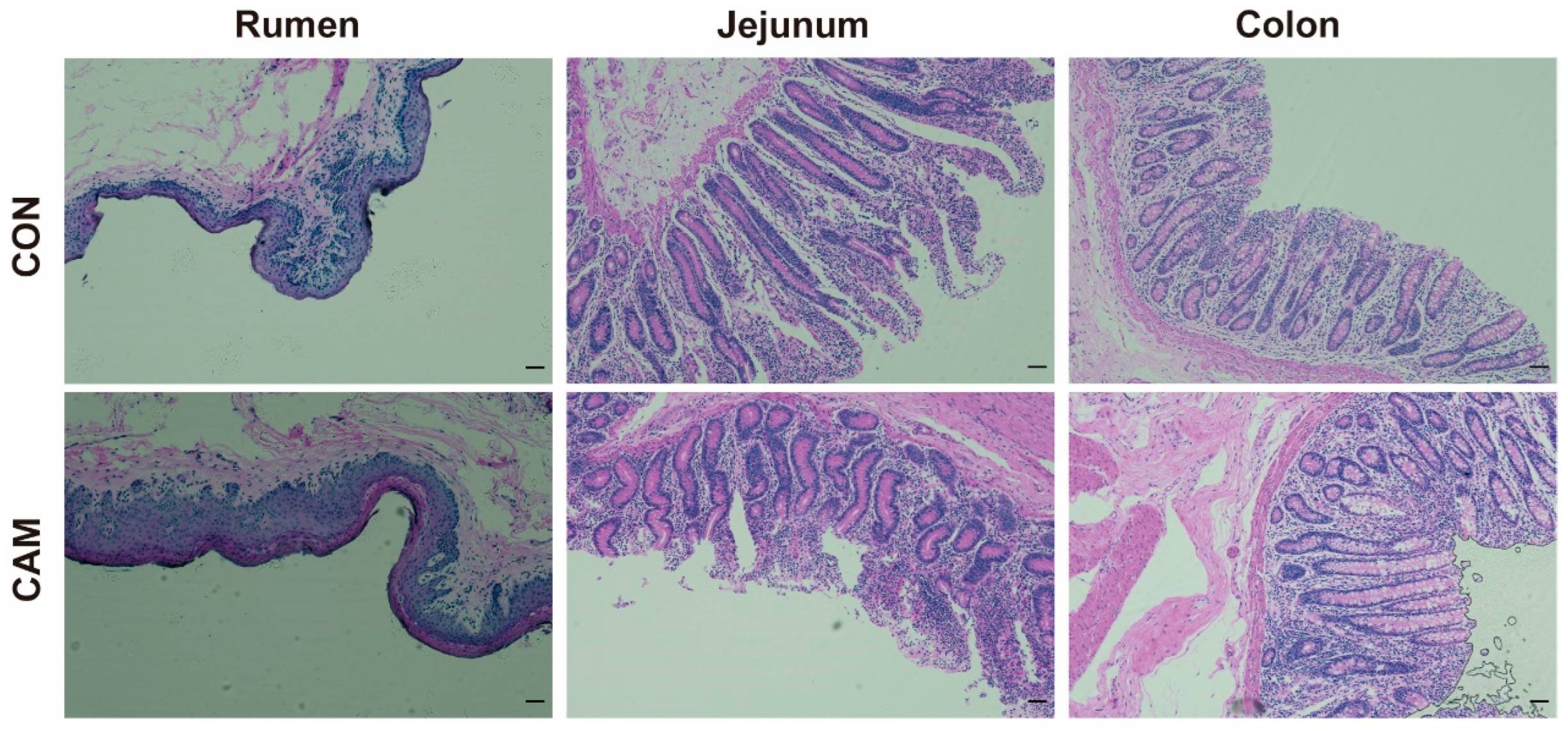
3.1. Growth Performance, Morphology, and Serum Parameters
3.2. Cadmium Concentration in Blood and in Gastrointestinal Luminal Contents and Tissues
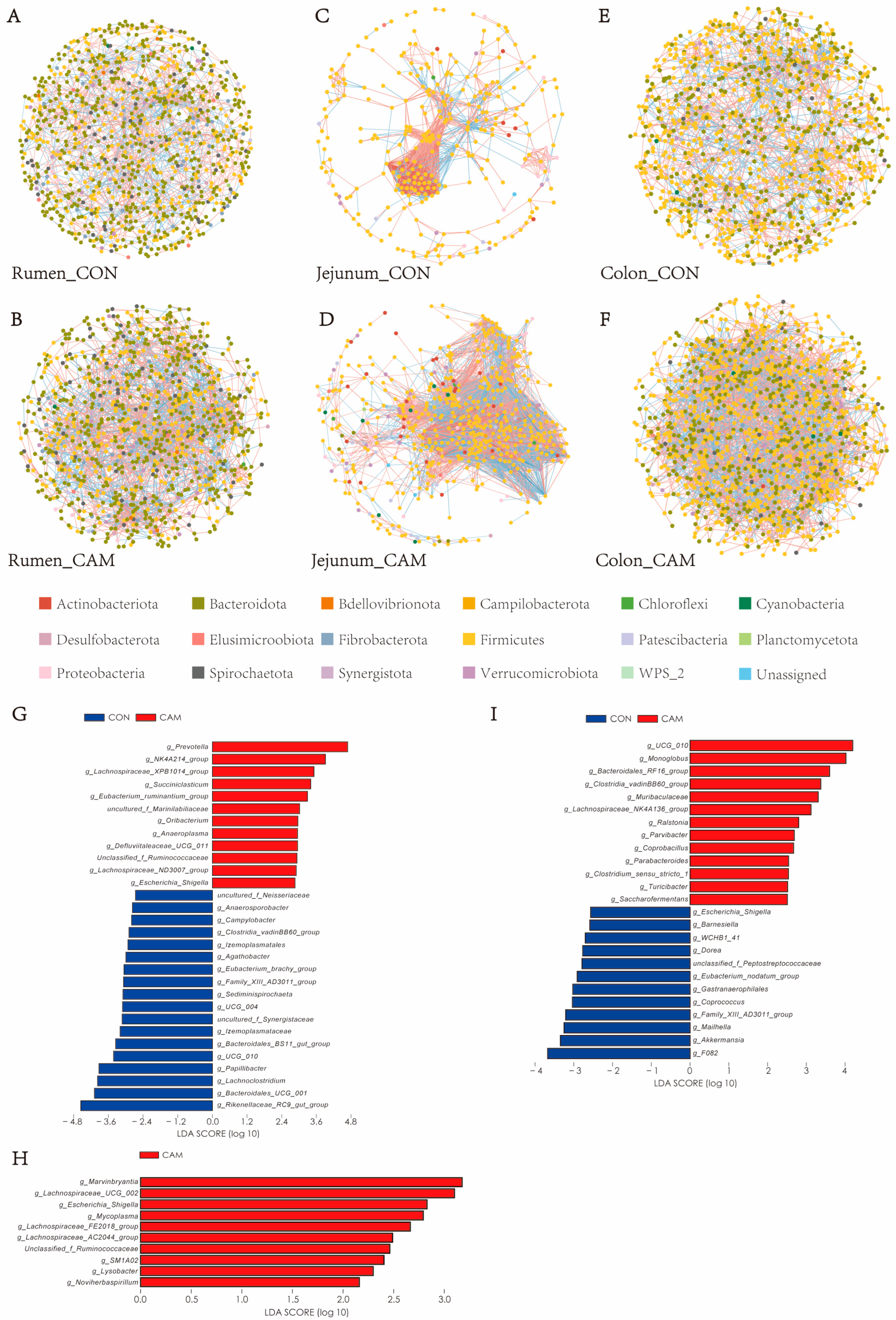
3.3. Profiles of Gastrointestinal Bacteria
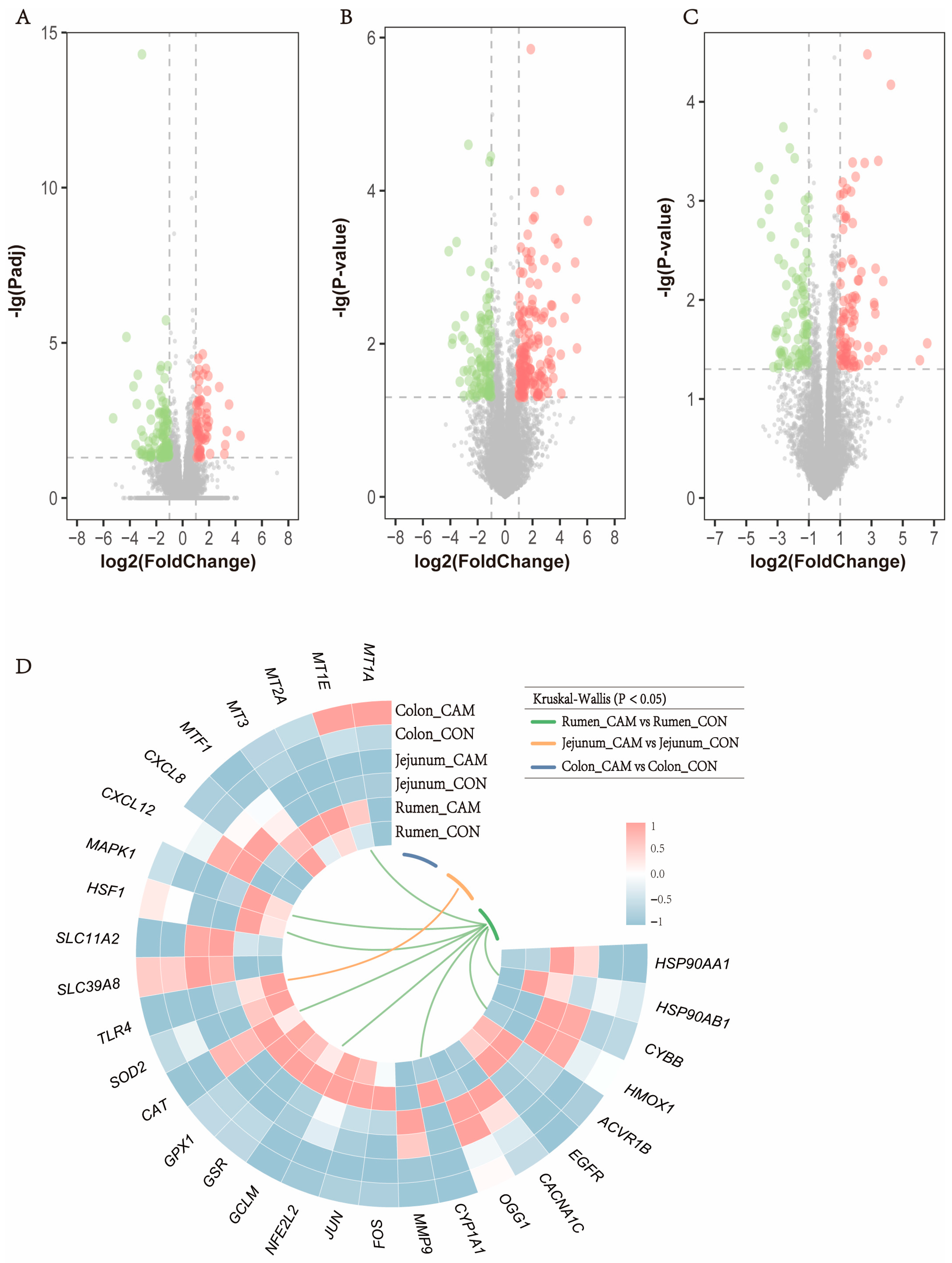
3.4. Profiles of Gastrointestinal Transcriptome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- International Agency for Research on Cancer. Arsenic, metal, fibres, and dusts. IARC Monogr. Identif. Carcinog. Hazards Hum. 2012, 100 Pt C, 121–145. [Google Scholar]
- Ba, Q.; Li, M.; Chen, P.; Huang, C.; Duan, X.; Lu, L.; Li, J.; Chu, R.; Xie, D.; Song, H.; et al. Sex-dependent effects of cadmium exposure in early life on gut microbiota and fat accumulation in mice. Environ. Health Perspect. 2017, 125, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Kumari, V.; Ram, C.; Thakur, K.; Tomar, S.K. Bio-prospectus of cadmium bioadsorption by lactic acid bacteria to mitigate health and environmental impacts. Appl. Microbiol. Biotechnol. 2018, 102, 1599–1615. [Google Scholar] [CrossRef]
- Yang, S.; Xiong, Z.; Xu, T.; Peng, C.; Hu, A.; Jiang, W.; Xiong, Z.; Wu, Y.; Yang, F.; Cao, H. Compound probiotics alleviate cadmium-induced intestinal dysfunction and microbiota disorders in broilers. Ecotoxicol. Environ. Saf. 2022, 234, 113374. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Jiang, L.; Wang, M.; Sun, W.; Yu, S.; Turner, J.R.; Yu, Q. Cadmium ingestion exacerbates Salmonella infection, with a loss of goblet cells through activation of Notch signaling pathways by ROS in the intestine. J. Hazard. Mater. 2020, 391, 122262. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bi, M.; Yang, J.; Cai, J.; Zhang, H.; Zhu, Y.; Zheng, Y.; Liu, Q.; Shi, G.; Zhang, Z. Cadmium exposure triggers oxidative stress, necroptosis, Th1/Th2 imbalance and promotes inflammation through the TNF-α/NF-κB pathway in swine small intestine. J. Hazard. Mater. 2022, 421, 126704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Jin, Y.; Zeng, Z.; Liu, Z.; Fu, Z. Subchronic exposure of mice to cadmium perturbs their hepatic energy metabolism and gut microbiome. Chem. Res. Toxicol 2015, 28, 2000–2009. [Google Scholar] [CrossRef]
- Smith, G.S. Toxification and detoxification of plant compounds by ruminants: An overview. J. Range Manag. 1992, 45, 25–30. [Google Scholar] [CrossRef]
- Yang, N.; Wang, H.; Wang, H. Screening maize (Zea mays L.) varieties with low accumulation of cadmium, arsenic, and lead in edible parts but high accumulation in other parts: A field plot experiment. Environ. Sci. Pollut. Res. 2021, 28, 33583–33598. [Google Scholar] [CrossRef]
- Ferraretto, L.F.; Shaver, R.D.; Luck, B.D. Silage review: Recent advances and future technologies for whole-plant and fractionated corn silage harvesting. J. Dairy Sci. 2018, 101, 3937–3951. [Google Scholar] [CrossRef]
- Xie, S.; Zhang, R.; Li, Z.; Liu, C.; Xiang, W.; Lu, Q.; Chen, Y.; Yu, Q. Indispensable role of melatonin, a scavenger of reactive oxygen species (ROS), in the protective effect of Akkermansia muciniphila in cadmium-induced intestinal mucosal damage. Free Radic. Biol. Med. 2022, 193 Pt 1, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.F.; Fan, G.Y.; Liu, D.Y.; Zhang, H.; Xu, Z.; Ge, Y.; Wang, Z. Effect of cadmium exposure on the histopathology of cerebral cortex in juvenile mice. Biol. Trace Elem. Res. 2015, 165, 167–172. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, B.; Yi, K.; Chen, T.; Xu, X.; Sun, A.; Li, H.; Li, J.; He, F.; Huan, C.; et al. Feasibility of feeding cadmium accumulated maize (Zea mays L.) to beef cattle: Discovering a strategy for eliminating phytoremediation residues. Anim. Nutr. 2023, 15, 1–9. [Google Scholar] [CrossRef]
- Gagen, E.J.; Denman, S.E.; Padmanabha, J.; Zadbuke, S.; Al Jassim, R.; Morrison, M.; McSweeney, C.S. Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl. Environ. Microbiol. 2010, 76, 7785–7795. [Google Scholar] [CrossRef] [PubMed]
- Nadkarni, M.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148 Pt 1, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Hübschmann, D. Simplify enrichment: A bioconductor package for clustering and visualizing functional enrichment results. Genom. Proteom. Bioinform. 2022, 21, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. vegan: Community Ecology Package. 2019. Available online: https://cran.r-project.org/package=vegan (accessed on 29 November 2022).
- Cai, Y.; Yin, Y.; Li, Y.; Guan, L.; Zhang, P.; Qin, Y.; Wang, Y.; Li, Y. Cadmium exposure affects growth performance, energy metabolism, and neuropeptide expression in Carassius auratus gibelio. Fish Physiol. Biochem. 2020, 46, 187–197. [Google Scholar] [CrossRef]
- Yu, Y.Y.; Chen, S.J.; Chen, M.; Tian, L.X.; Niu, J.; Liu, Y.J.; Xu, D.H. Effect of cadmium-polluted diet on growth, salinity stress, hepatotoxicity of juvenile Pacific white shrimp (Litopenaeus vannamei): Protective effect of Zn(II)-curcumin. Ecotoxicol. Environ. Saf. 2016, 125, 176–183. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Liu, K.; Shen, J. Exposing to cadmium stress cause profound toxic effect on microbiota of the mice intestinal tract. PLoS ONE 2014, 9, e85323. [Google Scholar] [CrossRef]
- Xue, Y.; Huang, F.; Tang, R.; Fan, Q.; Zhang, B.; Xu, Z.; Sun, X.; Ruan, Z. Chlorogenic acid attenuates cadmium-induced intestinal injury in Sprague-Dawley rats. Food Chem. Toxicol. 2019, 133, 110751. [Google Scholar] [CrossRef]
- Candelli, M.; Franza, L.; Pignataro, G.; Ojetti, V.; Covino, M.; Piccioni, A.; Gasbarrini, A.; Franceschi, F. Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int. J. Mol. Sci. 2021, 22, 6242. [Google Scholar] [CrossRef]
- Zalups, R.K.; Ahmad, S. Molecular handling of cadmium in transporting epithelia. Toxicol. Appl. Pharmacol. 2003, 186, 163–188. [Google Scholar] [CrossRef]
- Mirkov, I.; Aleksandrov, A.P.; Ninkov, M.; Tucovic, D.; Kulas, J.; Zeljkovic, M.; Popovic, D.; Kataranovski, M. Immunotoxicology of cadmium: Cells of the immune system as targets and effectors of cadmium toxicity. Food Chem. Toxicol. 2021, 149, 112026. [Google Scholar] [CrossRef] [PubMed]
- Kwong, R.W.; Andrés, J.A.; Niyogi, S. Effects of dietary cadmium exposure on tissue-specific cadmium accumulation, iron status and expression of iron-handling and stress-inducible genes in rainbow trout: Influence of elevated dietary iron. Aquat. Toxicol. 2011, 102, 1–9. [Google Scholar] [CrossRef]
- Duan, H.; Yu, L.; Tian, F.; Zhai, Q.; Fan, L.; Chen, W. Gut microbiota: A target for heavy metal toxicity and a probiotic protective strategy. Sci. Total Environ. 2020, 742, 140429. [Google Scholar] [CrossRef] [PubMed]
- Trevors, J.T.; Stratton, G.W.; Gadd, G.M. Cadmium transport, resistance, and toxicity in bacteria, algae, and fungi. Can. J. Microbiol. 1986, 32, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Deng, H.; Zhong, X.; Li, C.; Luo, Y.; Chen, L.; Zhang, B.; Wang, D.; Huang, Y.; et al. In situ analysis of variations of arsenicals, microbiome and transcriptome profiles along murine intestinal tract. J. Hazard. Mater. 2022, 427, 127899. [Google Scholar] [CrossRef]
- Bernad-Roche, M.; Bellés, A.; Grasa, L.; Casanova-Higes, A.; Mainar-Jaime, R.C. Effects of dietary supplementation with protected sodium butyrate on gut microbiota in growing-finishing pigs. Animals 2021, 11, 2137. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.R.; He, P.; Lei, S.F. Novel microbiota-related gene set enrichment analysis identified osteoporosis associated gut microbiota from autoimmune diseases. J. Bone Miner. Metab. 2021, 39, 984–996. [Google Scholar] [CrossRef]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A key player in ruminal metabolism. Microorganisms 2022, 11, 1. [Google Scholar] [CrossRef]
- Shi, W.; Hu, Y.; Ning, Z.; Xia, F.; Wu, M.; Hu, Y.O.; Chen, C.; Prast-Nielsen, S.; Xu, B. Alterations of gut microbiota in patients with active pulmonary tuberculosis in China: A pilot study. Int. J. Infect. Dis. 2021, 111, 313–321. [Google Scholar] [CrossRef]
- Stadlbauer, V.; Engertsberger, L.; Komarova, I.; Feldbacher, N.; Leber, B.; Pichler, G.; Fink, N.; Scarpatetti, M.; Schippinger, W.; Schmidt, R.; et al. Dysbiosis, gut barrier dysfunction and inflammation in dementia: A pilot study. BMC Geriatr. 2020, 20, 248. [Google Scholar] [CrossRef]
- Zhang, J.; Jin, W.; Jiang, Y.; Xie, F.; Mao, S. Response of milk performance, rumen and hindgut microbiome to dietary supplementation with Aspergillus oryzae fermentation extracts in dairy cows. Curr. Microbiol. 2022, 79, 113. [Google Scholar] [CrossRef] [PubMed]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.-J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, L.; Fu, P. Gut microbiota-derived short-chain fatty acids and kidney diseases. Drug Des. Dev. Ther. 2017, 11, 3531–3542. [Google Scholar] [CrossRef]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Zhao, Y.; Bai, L.; Hao, X.; Li, X.; et al. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int. J. Mol. Sci. 2020, 21, 6356. [Google Scholar] [CrossRef]
- Xia, T.; Duan, W.; Zhang, Z.; Li, S.; Zhao, Y.; Geng, B.; Zheng, Y.; Yu, J.; Wang, M. Polyphenol-rich vinegar extract regulates intestinal microbiota and immunity and prevents alcohol-induced inflammation in mice. Food Res. Int. 2021, 140, 110064. [Google Scholar] [CrossRef] [PubMed]
- Companys, J.; Gosalbes, M.J.; Pla-Pagà, L.; Calderón-Pérez, L.; Llauradó, E.; Pedret, A.; Valls, R.M.; Jiménez-Hernández, N.; Sandoval-Ramirez, B.A.; del Bas, J.M.; et al. Gut microbiota profile and its association with clinical variables and dietary intake in overweight/obese and lean subjects: A cross-sectional study. Nutrients 2021, 13, 2032. [Google Scholar] [CrossRef]
- He, X.; Qi, Z.; Hou, H.; Gao, J.; Zhang, X.-X. Effects of chronic cadmium exposure at food limitation-relevant levels on energy metabolism in mice. J. Hazard. Mater. 2020, 388, 121791. [Google Scholar] [CrossRef]
- Jiang, Z.; Mu, W.; Yang, Y.; Sun, M.; Liu, Y.; Gao, Z.; Li, J.; Gu, P.; Wang, H.; Lu, Y.; et al. Cadmium exacerbates dextran sulfate sodium-induced chronic colitis and impairs intestinal barrier. Sci. Total Environ. 2020, 744, 140844. [Google Scholar] [CrossRef]
- Benvenga, S.; Marini, H.R.; Micali, A.; Freni, J.; Pallio, G.; Irrera, N.; Squadrito, F.; Altavilla, D.; Antonelli, A.; Ferrari, S.M.; et al. Protective effects of myo-inositol and selenium on cadmium-induced thyroid toxicity in mice. Nutrients 2020, 12, 1222. [Google Scholar] [CrossRef]
- Xie, D.; Li, Y.; Liu, Z.; Chen, Q. Inhibitory effect of cadmium exposure on digestive activity, antioxidant capacity and immune defense in the intestine of yellow catfish (Pelteobagrus fulvidraco). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 222, 65–73. [Google Scholar] [CrossRef]
- Berentsen, S.; Hill, A.; Hill, Q.A.; Tvedt, T.H.A.; Michel, M. Novel insights into the treatment of complement-mediated hemolytic anemias. Ther. Adv. Hematol. 2019, 10, 2040620719873321. [Google Scholar] [CrossRef]
- Eskandari, S.K.; Seelen, M.A.J.; Lin, G.; Azzi, J.R. The immunoproteasome: An old player with a novel and emerging role in alloimmunity. Am. J. Transplant. 2017, 17, 3033–3039. [Google Scholar] [CrossRef]
- Butt, H.Z.; Sylvius, N.; Salem, M.K.; Wild, J.; Dattani, N.; Sayers, R.; Bown, M. Microarray-based gene expression profiling of abdominal aortic aneurysm. Eur. J. Vasc. Endovasc. Surg. 2016, 52, 47–55. [Google Scholar] [CrossRef]
- Chen, D.; Jin, C.; Dong, X.; Wen, J.; Xia, E.; Wang, Q.; Wang, O. Pan-cancer analysis of the prognostic and immunological role of PSMB8. Sci. Rep. 2021, 11, 20492. [Google Scholar] [CrossRef]
- Sun, L.; Gang, X.; Li, Z.; Zhao, X.; Zhou, T.; Zhang, S.; Wang, G. Advances in understanding the roles of CD244 (SLAMF4) in immune regulation and associated diseases. Front. Immunol. 2021, 12, 648182. [Google Scholar] [CrossRef]
- Feng, H.; Zhang, Y.B.; Gui, J.F.; Lemon, S.M.; Yamane, D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 2021, 17, e1009220. [Google Scholar] [CrossRef]
- Tokunaga, R.; Zhang, W.; Naseem, M.; Puccini, A.; Berger, M.D.; Soni, S.; McSkane, M.; Baba, H.; Lenz, H.-J. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—A target for novel cancer therapy. Cancer Treat. Rev. 2018, 63, 40–47. [Google Scholar] [CrossRef]
- Gonçalves, C.M.; Henriques, S.N.; Santos, R.F.; Carmo, A.M. CD6, a rheostat-type signalosome that tunes T cell activation. Front. Immunol. 2018, 9, 2994. [Google Scholar] [CrossRef]
- Chen, D.; Lei, X.; Hu, Y.; Li, J. Advances in CD247. Scand. J. Immunol. 2022, 96, e13170. [Google Scholar] [CrossRef]
- Yang, Y.; Zang, Y.; Zheng, C.; Li, Z.; Gu, X.; Zhou, M.; Wang, Z.; Xiang, J.; Chen, Z.; Zhou, Y. CD3D is associated with immune checkpoints and predicts favorable clinical outcome in colon cancer. Immunotherapy 2020, 12, 25–35. [Google Scholar] [CrossRef]
- Petibon, C.; Ghulam, M.M.; Catala, M.; Elela, S.A. Regulation of ribosomal protein genes: An ordered anarchy. Wiley Interdiscip. Rev. RNA 2021, 12, e1632. [Google Scholar] [CrossRef]



| Item | CON | CAM | SEM | p-Value |
|---|---|---|---|---|
| Ingredients (fresh basis, %) | ||||
| Straw | 2.84 | 2.84 | - | - |
| Corn silage | 85.82 | 85.82 | - | - |
| Pelleted concentrate 1 | 11.34 | 11.34 | - | - |
| Total | 100 | 100 | - | - |
| Chemical composition 2 (DM basis, %) | ||||
| GE, MJ/kg | 12.00 | 13.36 | 0.406 | 0.09 |
| CP | 10.79 | 11.45 | 0.292 | 0.30 |
| EE | 11.00 | 13.00 | 0.730 | 0.20 |
| NDF | 49.33 | 50.67 | 1.732 | 0.74 |
| ADF | 25.00 | 25.33 | 1.195 | 0.91 |
| Ca | 0.62 | 0.62 | 0.028 | 0.95 |
| P | 0.42 | 0.37 | 0.022 | 0.32 |
| Cd, mg/kg | 0.72 | 6.74 | 1.516 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Xu, Z.; Yang, B.; Yi, K.; He, F.; Sun, A.; Li, J.; Luo, Y.; Wang, J. Assessing the Effects of Dietary Cadmium Exposure on the Gastrointestinal Tract of Beef Cattle via Microbiota and Transcriptome Profile. Animals 2023, 13, 3104. https://doi.org/10.3390/ani13193104
Xu X, Xu Z, Yang B, Yi K, He F, Sun A, Li J, Luo Y, Wang J. Assessing the Effects of Dietary Cadmium Exposure on the Gastrointestinal Tract of Beef Cattle via Microbiota and Transcriptome Profile. Animals. 2023; 13(19):3104. https://doi.org/10.3390/ani13193104
Chicago/Turabian StyleXu, Xinxin, Zebang Xu, Bin Yang, Kangle Yi, Fang He, Ao Sun, Jianbo Li, Yang Luo, and Jiakun Wang. 2023. "Assessing the Effects of Dietary Cadmium Exposure on the Gastrointestinal Tract of Beef Cattle via Microbiota and Transcriptome Profile" Animals 13, no. 19: 3104. https://doi.org/10.3390/ani13193104
APA StyleXu, X., Xu, Z., Yang, B., Yi, K., He, F., Sun, A., Li, J., Luo, Y., & Wang, J. (2023). Assessing the Effects of Dietary Cadmium Exposure on the Gastrointestinal Tract of Beef Cattle via Microbiota and Transcriptome Profile. Animals, 13(19), 3104. https://doi.org/10.3390/ani13193104






