Exploring the Impact of Insertion/Deletion in FTO and PLIN1 Genes on Morphometric Traits in Sheep
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Treatment
2.2. Experimental Animals and Morphometric Trait Data Collection
2.3. Primer Designing, PCR Amplification, and Indel Genotyping
2.4. Population Genetic Analyses
2.5. Statistical Analysis
3. Results
3.1. Identification of InDel Variants in the FTO and PLIN1 Genes
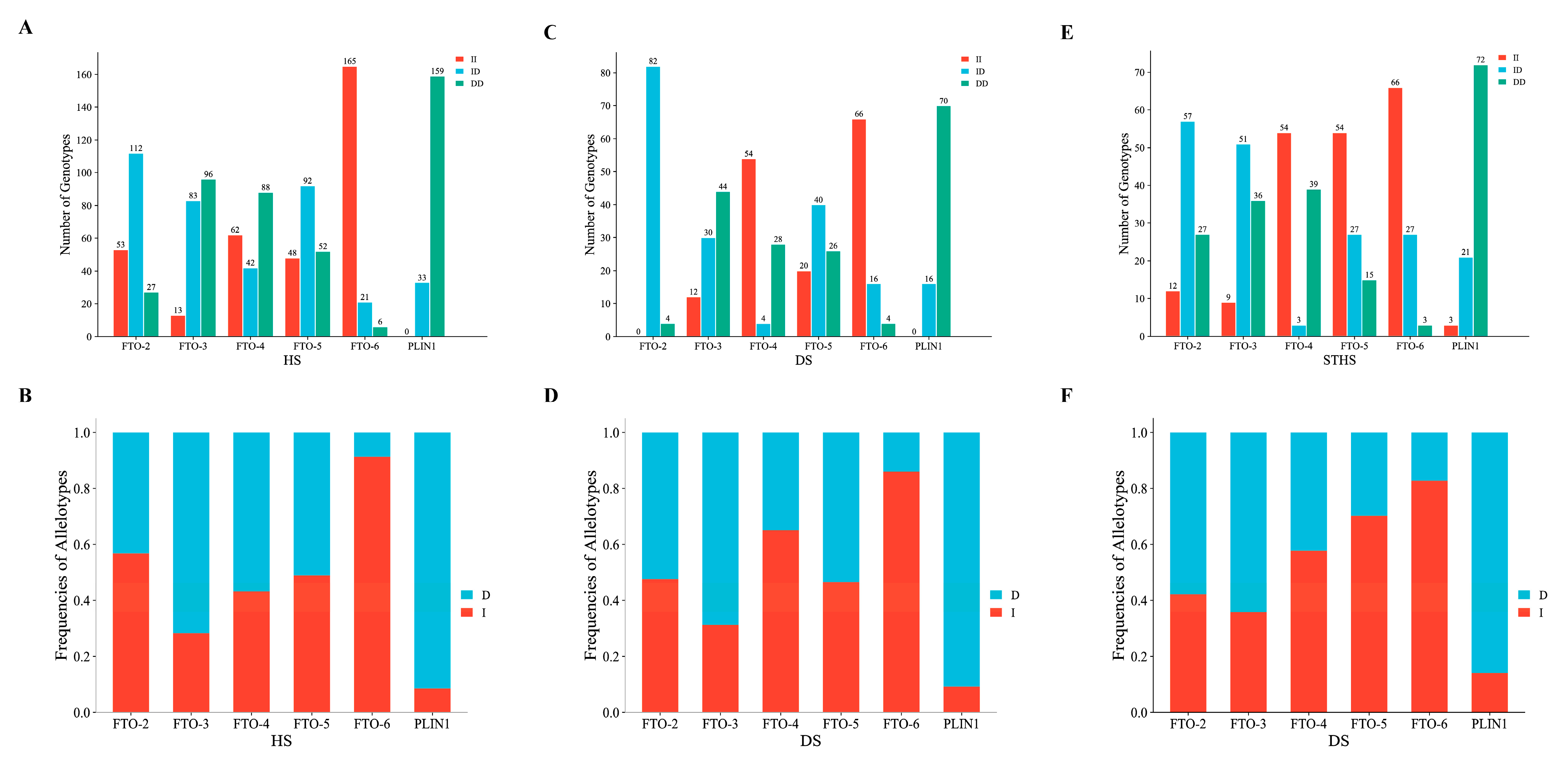
3.2. Estimation of Genetic Parameters for the InDels of the FTO and PLIN1 Genes
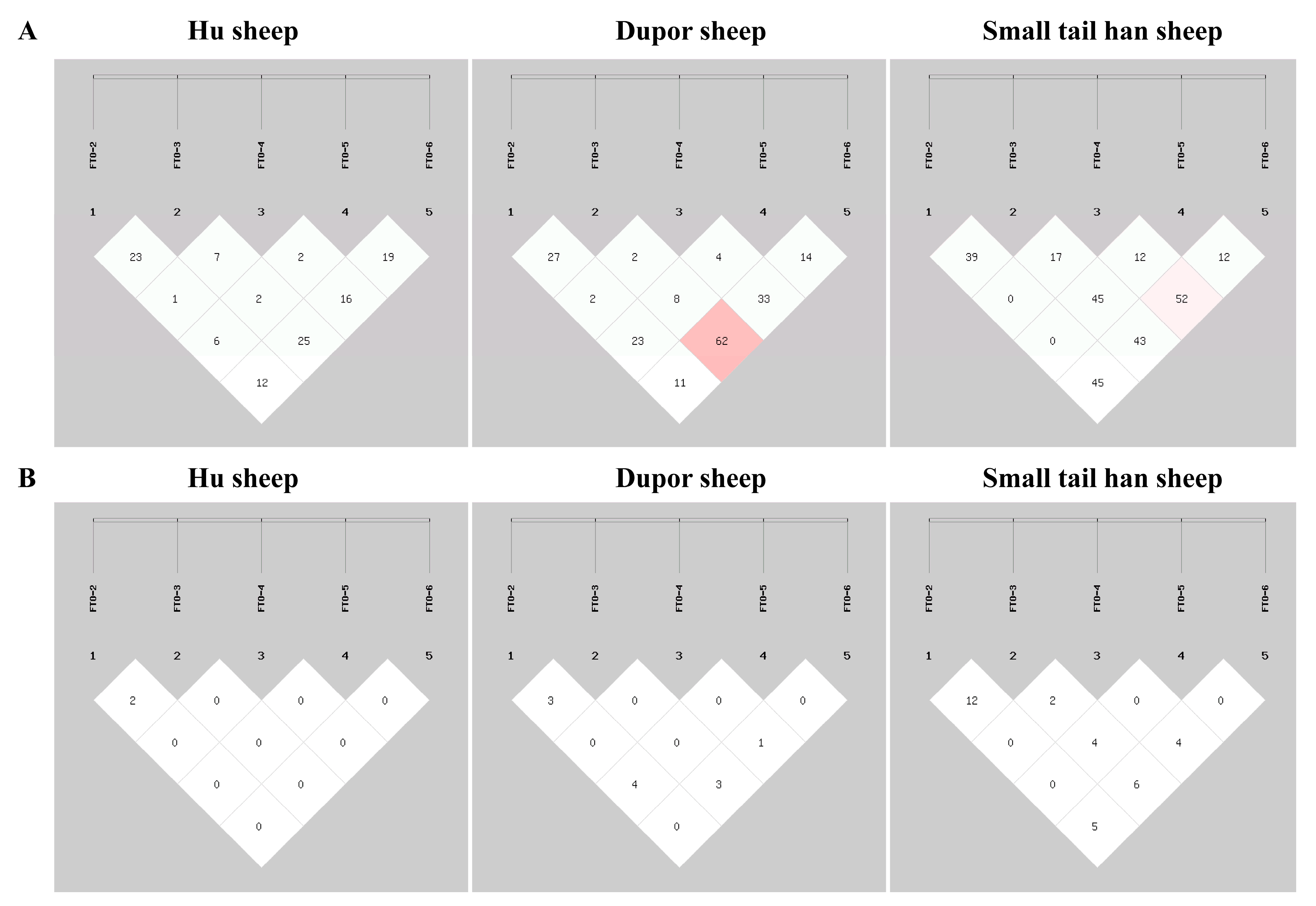
3.3. Linkage Disequilibrium Analysis for InDels in the FTO Gene
3.4. Association of InDels in the FTO and PLIN1 Genes with Morphometric Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gebreselassie, G.; Berihulay, H.; Jiang, L.; Ma, Y. Review on genomic regions and candidate genes associated with economically important production and reproduction traits in sheep (Ovies aries). Animals 2020, 10, 33. [Google Scholar] [CrossRef]
- Ren, T.; Zhang, Z.; Fu, R.; Yang, Y.; Li, W.; Liang, J.; Mo, G.; Luo, W.; Zhang, X. A 51 bp indel polymorphism within the PTH1R gene is significantly associated with chicken growth and carcass traits. Anim. Genet. 2020, 51, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhao, H.; Wu, M.; He, S.; Yuan, T.; Yi, X.; Liu, S.; Pan, Y.; Li, Q.; Wang, S.; et al. A novel 17 bp InDel polymorphism within the PPARGC1A gene is significantly associated with growth traits in sheep. Anim. Biotechnol. 2022, 33, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Y.; Liu, Y.; Gao, C.F.; Lan, X.Y.; Wu, X.F. A novel duplicated insertion/deletion (InDel) of the CPT1a gene and its effects on growth traits in goat. Anim. Biotechnol. 2021, 32, 343–351. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Shen, C.; Niu, Z.; Yang, H.; Zhang, K.; Liu, Z.; Wang, Y.; Lan, X. Indel mutations within the bovine HSD17B3 gene are significantly associated with ovary morphological traits and mature follicle number. J. Steroid Biochem. Mol. Biol. 2021, 209, 105833. [Google Scholar] [CrossRef]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Jevsinek Skok, D.; Kunej, T.; Kovac, M.; Malovrh, S.; Potocnik, K.; Petric, N.; Zgur, S.; Dovc, P.; Horvat, S. FTO gene variants are associated with growth and carcass traits in cattle. Anim. Genet. 2016, 47, 219–222. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, D.; Zhang, X.; Li, F.; Xu, D.; Zhao, L.; Li, X.; Zhang, Y.; Wang, J.; Yang, X.; et al. Expression features of the ovine FTO gene and association between FTO polymorphism and tail fat deposition related-traits in Hu sheep. Gene 2022, 826, 146451. [Google Scholar] [CrossRef]
- Wang, S.; Liu, S.; Yuan, T.; Sun, X. Genetic effects of FTO gene insertion/deletion (InDel) on fat-tail measurements and growth traits in Tong sheep. Anim. Biotechnol. 2021, 32, 229–239. [Google Scholar] [CrossRef]
- Bialesova, L.; Kulyté, A.; Petrus, P.; Sinha, I.; Laurencikiene, J.; Zhao, C.; Wright, K.D.; Arner, P.; Dahlman, I. Epigenetic regulation of PLIN 1 in obese women and its relation to lipolysis. Sci. Rep. 2017, 7, 10152. [Google Scholar] [CrossRef]
- Zhang, H.L.; Fan, H.J.; Liu, X.L.; Wu, Y.; Hou, S.S. Molecular cloning of the perilipin gene and its association with carcass and fat traits in Chinese ducks. Genet. Mol. Res. 2013, 12, 1582–1592. [Google Scholar] [CrossRef] [PubMed]
- Gol, S.; Ros-Freixedes, R.; Zambonelli, P.; Tor, M.; Pena, R.N.; Braglia, S.; Zappaterra, M.; Estany, J.; Davoli, R. Relationship between perilipin genes polymorphisms and growth, carcass and meat quality traits in pigs. J. Anim. Breed. Genet. 2016, 133, 24–30. [Google Scholar] [CrossRef]
- Zappaterra, M.; Mazzoni, M.; Zambonelli, P.; Davoli, R. Investigation of the Perilipin 5 gene expression and association study of its sequence polymorphism with meat and carcass quality traits in different pig breeds. Animal 2018, 12, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.H.A.; Shijun, L.; Khan, R.; Schreurs, N.M.; Manzari, Z.; Abd El-Aziz, A.H.; Ullah, I.; Kaster, N.; Shah, M.A.; Zan, L. Polymorphism of the PLIN1 gene and its association with body measures and ultrasound carcass traits in Qinchuan beef cattle. Genome 2020, 63, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, R.P.; Bailey, D.R.; Shannon, N.H. Body dimensions and carcass measurements of cattle selected for postweaning gain fed two different diets. J. Anim. Sci. 1993, 71, 1688–1698. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boyle, T. Population genetic analysis of codominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Shi, Y.Y.; He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005, 15, 97–98. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Gong, X.; Chen, P.; Ma, P.; Gao, J.; Yang, J.; Guo, H.; Yan, C.; Zhang, B.; He, Y. MICA polymorphisms associated with antithyroid drug-induced agranulocytosis in the Chinese Han population. Immun. Inflamm. Dis. 2020, 8, 695–703. [Google Scholar] [CrossRef]
- Loos, R.J.; Bouchard, C. FTO: The first gene contributing to common forms of human obesity. Obes. Rev. 2008, 9, 246–250. [Google Scholar] [CrossRef]
- Olszewski, P.K.; Fredriksson, R.; Olszewska, A.M.; Stephansson, O.; Alsiö, J.; Radomska, K.J.; Levine, A.S.; Schiöth, H.B. Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci. 2009, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.; Girard, C.A.; Tung, Y.C.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.; McDonough, M.A.; Cunliffe, S.; McNeill, L.A.; et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007, 318, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Brasaemle, D.L.; Subramanian, V.; Garcia, A.; Marcinkiewicz, A.; Rothenberg, A. Perilipin A and the control of triacylglycerol metabolism. Mol. Cell. Biochem. 2009, 326, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Zhao, H.; Tang, X.; Li, Q.; Yi, X.; Liu, S.; Sun, X. Novel InDels of GHR, GHRH, GHRHR and their association with growth traits in seven Chinese sheep breeds. Animals 2020, 10, 1883. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xian, Y.; Li, Z.; Wang, Z.; Nie, Q. G0S2 Gene polymorphism and its relationship with carcass traits in chicken. Animals 2022, 12, 916. [Google Scholar] [CrossRef]
- Sun, X.; Liang, Y.; Gao, Q.; Guo, J.; Tang, C.; Shi, K.; Yang, Z.; Mao, Y. AGPAT3 Gene polymorphisms are associated with milk production traits in Chinese Holstein cows. J. Dairy Res. 2021, 88, 247–252. [Google Scholar] [CrossRef]
- Huang, Y.; Su, P.; Akhatayeva, Z.; Pan, C.; Zhang, Q.; Lan, X. Novel InDel variations of the Cry2 gene are associated with litter size in Australian White sheep. Theriogenology 2022, 179, 155–161. [Google Scholar] [CrossRef]
- Liu, L.; Cui, Z.; Xiao, Q.; Zhang, H.; Zhao, X.; Wang, Y.; Yin, H.; Li, D.; Zhu, Q. Polymorphisms in the chicken growth differentiation factor 9 gene associated with reproductive traits. Biomed Res. Int. 2018, 2018, 9345473. [Google Scholar] [CrossRef]
- Khan, R.; Raza, S.H.A.; Guo, H.; Xiaoyu, W.; Sen, W.; Suhail, S.M.; Rahman, A.; Ullah, I.; Abd El-Aziz, A.H.; Manzari, Z.; et al. Genetic variants in the TORC2 gene promoter and their association with body measurement and carcass quality traits in Qinchuan cattle. PLoS ONE 2020, 15, e0227254. [Google Scholar] [CrossRef]
- Lin, X.; Li, B.; Chen, Y.; Chen, H.; Liu, M. KAT2B Gene Polymorphisms Are Associated with Body Measure Traits in Four Chinese Cattle Breeds. Animals 2022, 12, 1954. [Google Scholar] [CrossRef]
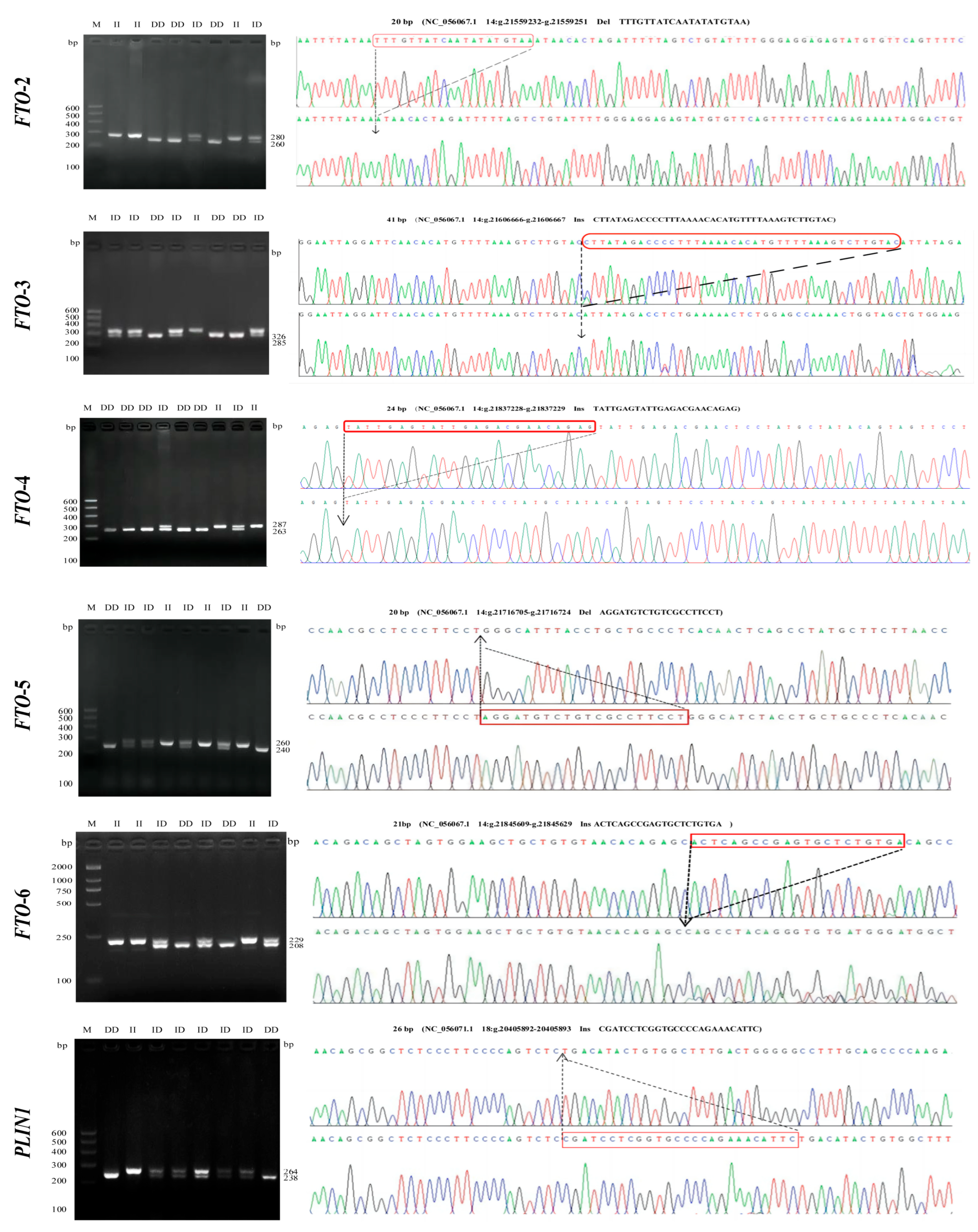
| Breed | Abbreviation | Sampling Source | Number | Sample Tissue |
|---|---|---|---|---|
| Hu | HS | Luoyang city | 192 | Venous blood |
| Dupor | DS | Luoyang city | 86 | Venous blood |
| Small Tail Han | STHS | Puyang city | 96 | Venous blood |
| Gene | Primer Sequence (5′-3′) | Product Size (bp) | InDel Sizes (bp) |
|---|---|---|---|
| FTO-1 | F: TAAGTTTACTAAGCCGAGGAGC R: TTTAGTCTGTATTTTGGGAGGA | 260 | 37 |
| FTO-2 | F: TTATTGGAGTTTATTGGAGTTAC R: GTCTGTGAGCCTATTTCTGTC | 280 | 20 |
| FTO-3 | F: CCACAACTCAGCAGCTACCAGT R: CTCAGCAATCATCAGCATCAGG | 326 | 41 |
| FTO-4 | F: AAGGAGGTAAAGGAGAAAACT R: TACAGCAAAATGAATCTGACA | 287 | 24 |
| FTO-5 | F: CCAAGGAGAAAGCTGCTGTAGTC R: GGATTTCAAAGAGATTTCACCGTTG | 260 | 20 |
| FTO-6 | F: CCATCCCATCCCACCCTGTA R: GCTGCCTTGCGGACCTTT | 208 | 21 |
| FTO-7 | F: CTGCATGTGCCTCGAGTCTT R: GGTCAGAGAGTTGGTGTCATGG | 225 | 24 |
| FTO-8 | F: TCTTCACGGTTCATGGTAGGC R: CACTAGTGGGAATGAAGGGGAC | 316 | 48 |
| FTO-9 | F: TGTGGAATTCATTCTTGTCGCT R: AGGGAGAGATGCTAAGGTCCA | 304 | 15 |
| FTO-10 | F: TGGGACAGTACAGTCTTGTGTT R: TCTGGATCAACCTCCACTGTT | 342 | 41 |
| PLIN1 | F: GCAGCAGGTAGGAAGGATGG R: TCACTGACAACGTGGTGGAC | 238 | 26 |
| Breed | Name | Loci | Allele Frequency | Genetic Parameter | HWE (P) | ||||
|---|---|---|---|---|---|---|---|---|---|
| I | D | Ho | He | PIC | Ne | ||||
| HS | FTO-2 (P2-Del-20 bp) | NC_056067.1 Chr:14 g.21559232-g.21559251 | 0.568 | 0.432 | 0.509 | 0.491 | 0.370 | 1.964 | 0.009 |
| FTO-3 (P3-Ins-41 bp) | NC_056067.1 Chr:14 g.21606666-g.21606667 | 0.284 | 0.716 | 0.593 | 0.407 | 0.324 | 1.685 | 0.381 | |
| FTO-4 (P4-Ins-24 bp) | NC_056067.1 Chr:14 g.21837228-g.21837229 | 0.432 | 0.568 | 0.509 | 0.491 | 0.370 | 1.964 | 0.000 | |
| FTO-5 (P5-Del-20 bp) | NC_056067.1 Chr:14 g.21716705-g.21716724 | 0.490 | 0.510 | 0.500 | 0.500 | 0.375 | 1.999 | 0.568 | |
| FTO-6 (P6-Ins-21 bp) | NC_056067.1 Chr:14 g.21845609-g.21845629 | 0.914 | 0.086 | 0.843 | 0.157 | 0.145 | 1.186 | 0.000 | |
| PLIN1 (P1-Ins-26 bp) | NC_056071.1 Chr:14 g.20405892-g.20405893 | 0.086 | 0.914 | 0.843 | 0.157 | 0.145 | 1.186 | 0.193 | |
| DS | FTO-2 (P2-Del-20 bp) | NC_056067.1 Chr:14 g.21559232-g.21559251 | 0.477 | 0.523 | 0.501 | 0.499 | 0.374 | 1.996 | 0.000 |
| FTO-3 (P3-Ins-41 bp) | NC_056067.1 Chr:14 g.21606666-g.21606667 | 0.314 | 0.686 | 0.569 | 0.431 | 0.338 | 1.757 | 0.078 | |
| FTO-4 (P4-Ins-24 bp) | NC_056067.1 Chr:14 g.21837228-g.21837229 | 0.651 | 0.349 | 0.546 | 0.454 | 0.351 | 1.833 | 1.000 | |
| FTO-5 (P5-Del-20 bp) | NC_056067.1 Chr:14 g.21716705-g.21716724 | 0.465 | 0.535 | 0.502 | 0.498 | 0.374 | 1.990 | 0.545 | |
| FTO-6 (P6-Ins-21 bp) | NC_056067.1 Chr:14 g.21845609-g.21845629 | 0.860 | 0.140 | 0.760 | 0.240 | 0.211 | 1.316 | 0.037 | |
| PLIN1 (P1-Ins-26 bp) | NC_056071.1 Chr:14 g.20405892-g.20405893 | 0.093 | 0.907 | 0.831 | 0.169 | 0.155 | 1.203 | 0.342 | |
| STHS | FTO-2 (P2-Del-20 bp) | NC_056067.1 Chr:14 g.21559232-g.21559251 | 0.422 | 0.578 | 0.512 | 0.488 | 0.369 | 1.952 | 0.033 |
| FTO-3 (P3-Ins-41 bp) | NC_056067.1 Chr:14 g.21606666-g.21606667 | 0.359 | 0.641 | 0.540 | 0.460 | 0.354 | 1.853 | 0.132 | |
| FTO-4 (P4-Ins-24 bp) | NC_056067.1 Chr:14 g.21837228-g.21837229 | 0.578 | 0.422 | 0.512 | 0.488 | 0.369 | 1.952 | 1.000 | |
| FTO-5 (P5-Del-20 bp) | NC_056067.1 Chr:14 g.21716705-g.21716724 | 0.703 | 0.297 | 0.583 | 0.417 | 0.330 | 1.717 | 0.001 | |
| FTO-6 (P6-Ins-21 bp) | NC_056067.1 Chr:14 g.21845609-g.21845629 | 0.828 | 0.172 | 0.715 | 0.285 | 0.244 | 1.398 | 0.906 | |
| PLIN1 (P1-Ins-26 bp) | NC_056071.1 Chr:14 g.20405892-g.20405893 | 0.141 | 0.859 | 0.758 | 0.242 | 0.212 | 1.319 | 0.352 | |
| Loci | Breeds | Growth Traits (cm) | Observed Genotypes (Mean ± SE) | ANOVA p Values | Unadjusted p Values | Adjusted p Values | ||
|---|---|---|---|---|---|---|---|---|
| II | ID | DD | ||||||
| FTO-2 | HS ram | Cannon circumference | 7.005 ± 0.168 ab | 7.331 ± 0.112 a | 6.480 ± 0.237 b | p = 0.007 | p = 0.008 | p = 0.003 |
| HS ewe | Body height | 67.359 ± 0.814 b | 68.369 ± 0.607 ab | 71.059 ± 1.362 a | p = 0.041 | p = 0.037 | p = 0.012 | |
| STHS | Chest width | 33.750 ± 1.548 a | 26.895 ± 1.095 b | 27.333 ± 1.481 ab | p = 0.033 | p = 0.031 | p = 0.010 | |
| STHS | Cannon circumference | 11.500 ± 0.646 a | 9.579 ± 0.289 b | 9.222 ± 0.465 b | p = 0.020 | p = 0.021 | p = 0.007 | |
| STHS | Head length | 23.750 ± 1.250 a | 19.368 ± 0.693 b | 20.778 ± 0.722 ab | p = 0.022 | p = 0.022 | p = 0.007 | |
| STHS | Coccyx length | 28.500 ± 2.102 a | 22.632 ± 0.714 b | 22.222 ± 1.064 b | p = 0.007 | p = 0.008 | p = 0.003 | |
| FTO-3 | HS ram | Body height | 54.714 ± 1.149 b | 58.647 ± 0.529 a | 58.049 ± 0.571 ab | p = 0.024 | p = 0.019 | p = 0.006 |
| HS ewe | Body height | 72.750 ± 2.105 a | 69.663 ± 0.613 a | 66.982 ± 0.673 b | p = 0.002 | p = 0.013 | p = 0.004 | |
| HS ewe | Back height | 72.167 ± 1.493 a | 69.643 ± 0.625 a | 67.364 ± 0.656 b | p = 0.008 | p = 0.039 | p = 0.013 | |
| HS ewe | Chest width | 26.333 ± 3.451 a | 22.000 ± 0.747 ab | 20.200 ± 0.583 b | p = 0.010 | p = 0.016 | p = 0.005 | |
| HS ewe | Chest depth | 38.333 ± 1.542 a | 35.490 ± 0.548 ab | 33.836 ± 0.510 b | p = 0.007 | p = 0.021 | p = 0.007 | |
| DS | Chest width | 20.700 ± 2.382 b | 26.433 ± 0.543 a | 24.659 ± 0.751 ab | p = 0.006 | p = 0.005 | p = 0.002 | |
| STHS | Chest width | 29.667 ± 2.963 ab | 25.765 ± 1.006 b | 30.417 ± 1.417 a | p = 0.030 | p = 0.032 | p = 0.011 | |
| FTO-5 | DS | Body weight | 57.780 ± 1.053 a | 52.935 ± 1.042 b | 53.873 ± 1.598 ab | p = 0.039 | p = 0.037 | p = 0.012 |
| STHS | Body height | 64.000 ± 2.457 b | 74.056 ± 2.337 a | 69.200 ± 4.042 ab | p = 0.043 | p = 0.042 | p = 0.014 | |
| PLIN1 | DS | Body weight | — | 57.631 ± 1.013 a | 53.594 ± 0.874 b | p = 0.040 | — | — |
| STHS | Coccyx length | — | 26.143 ± 1.682 a | 22.583 ± 0.645 b | p = 0.039 | — | — | |
| STHS | Forehead width | — | 15.000 ± 0.787 a | 12.521 ± 0.382 b | p = 0.008 | — | — | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Li, J.; Bai, J.; Chen, M.; Wang, L.; Fan, H.; Zeng, F.; Lu, X.; He, Y. Exploring the Impact of Insertion/Deletion in FTO and PLIN1 Genes on Morphometric Traits in Sheep. Animals 2023, 13, 3032. https://doi.org/10.3390/ani13193032
Wang X, Li J, Bai J, Chen M, Wang L, Fan H, Zeng F, Lu X, He Y. Exploring the Impact of Insertion/Deletion in FTO and PLIN1 Genes on Morphometric Traits in Sheep. Animals. 2023; 13(19):3032. https://doi.org/10.3390/ani13193032
Chicago/Turabian StyleWang, Xinle, Jingyun Li, Junyan Bai, Mengke Chen, Longwei Wang, Hongdeng Fan, Fanlin Zeng, Xiaoning Lu, and Yuhan He. 2023. "Exploring the Impact of Insertion/Deletion in FTO and PLIN1 Genes on Morphometric Traits in Sheep" Animals 13, no. 19: 3032. https://doi.org/10.3390/ani13193032
APA StyleWang, X., Li, J., Bai, J., Chen, M., Wang, L., Fan, H., Zeng, F., Lu, X., & He, Y. (2023). Exploring the Impact of Insertion/Deletion in FTO and PLIN1 Genes on Morphometric Traits in Sheep. Animals, 13(19), 3032. https://doi.org/10.3390/ani13193032





